Submitted:
20 April 2023
Posted:
20 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
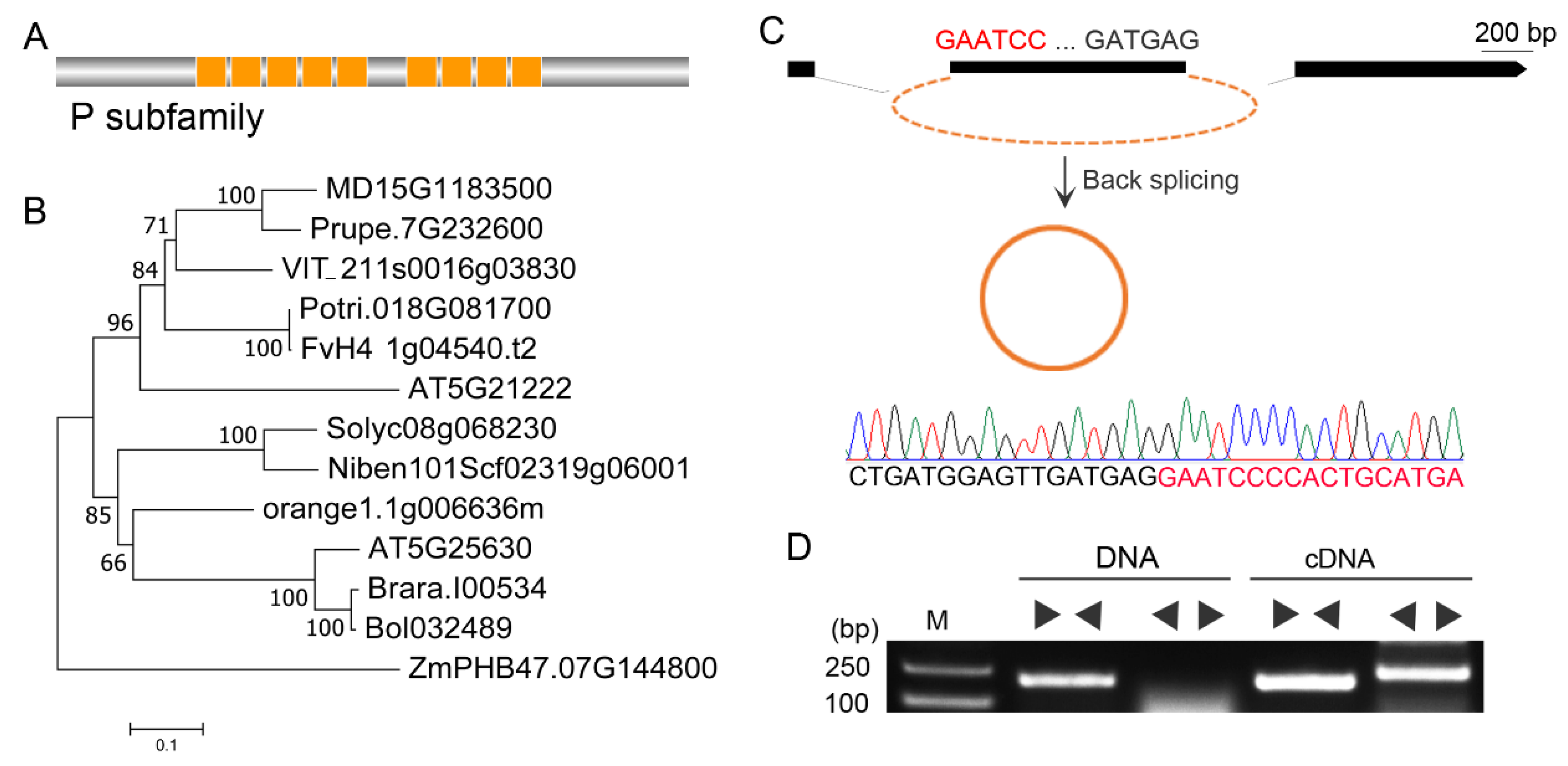
2.1. Identification and verification of Vv-circPTCD1
2.2. Nonconservative back-splicing of Vv-circPTCD1 in plants
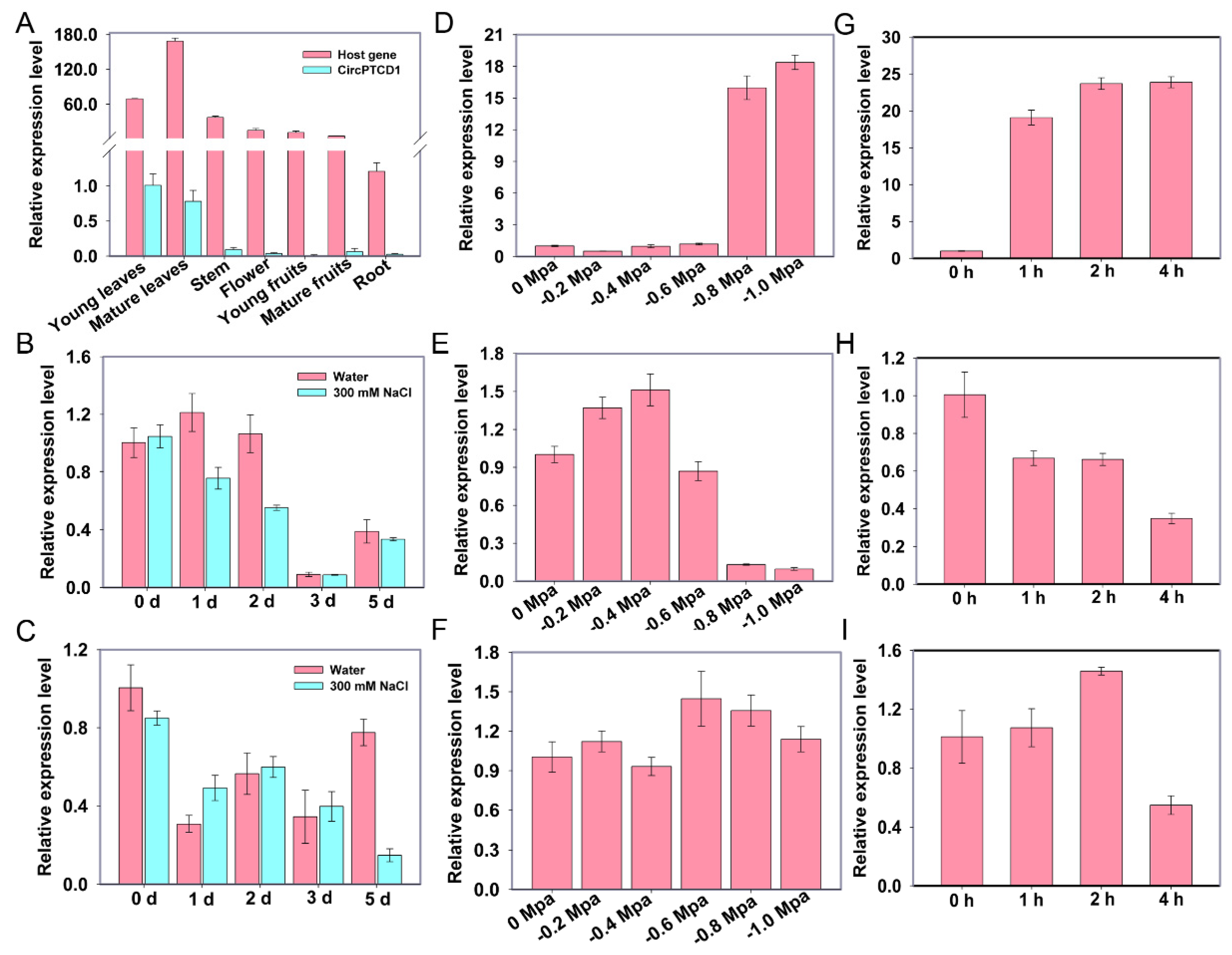
2.3. The expression analyses of PTCD1 and Vv-circPTCD1 under abiotic stress
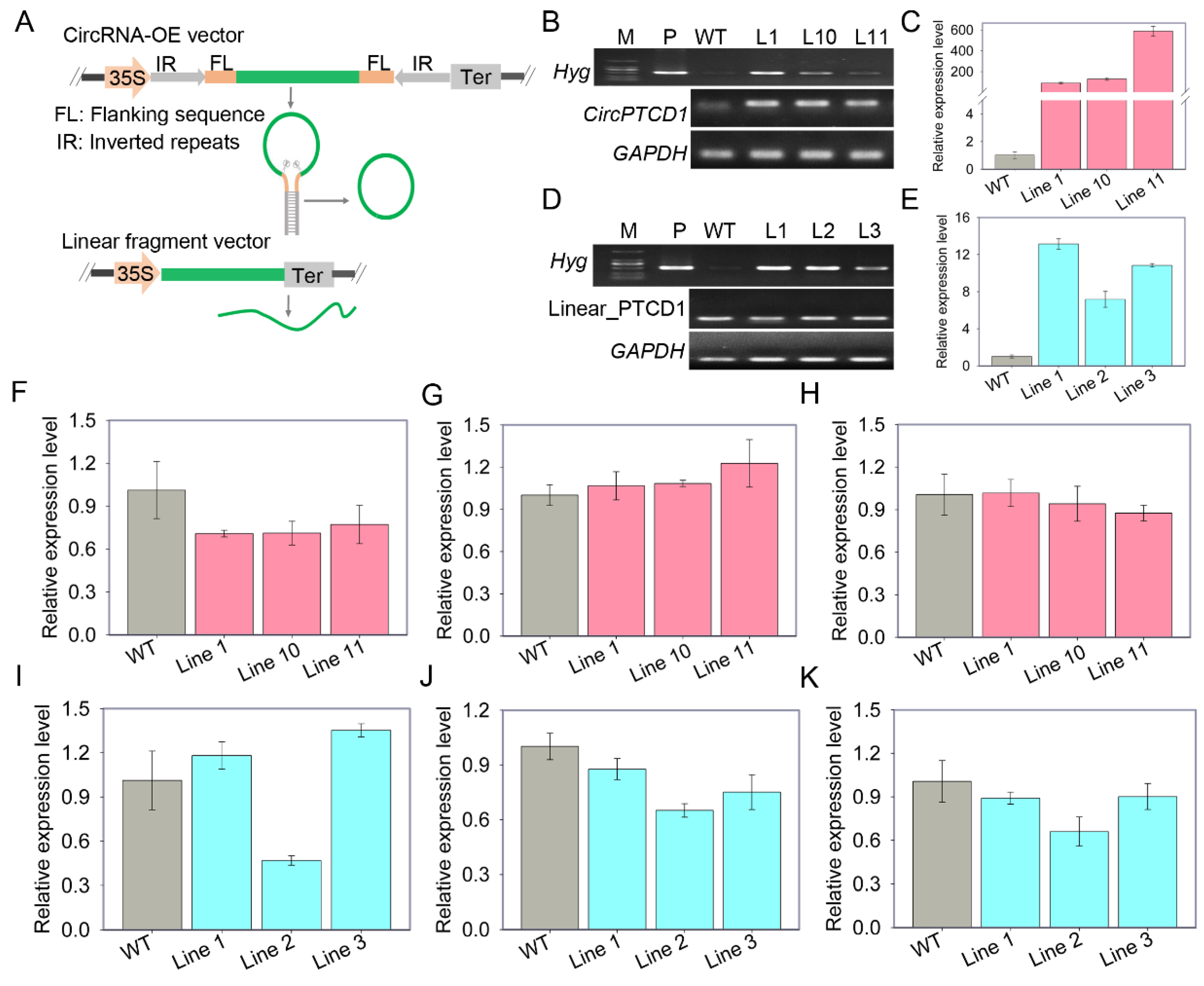
2.4. Overexpression of Vv-CircPTCD1 in grapevine callus and Arabidopsis
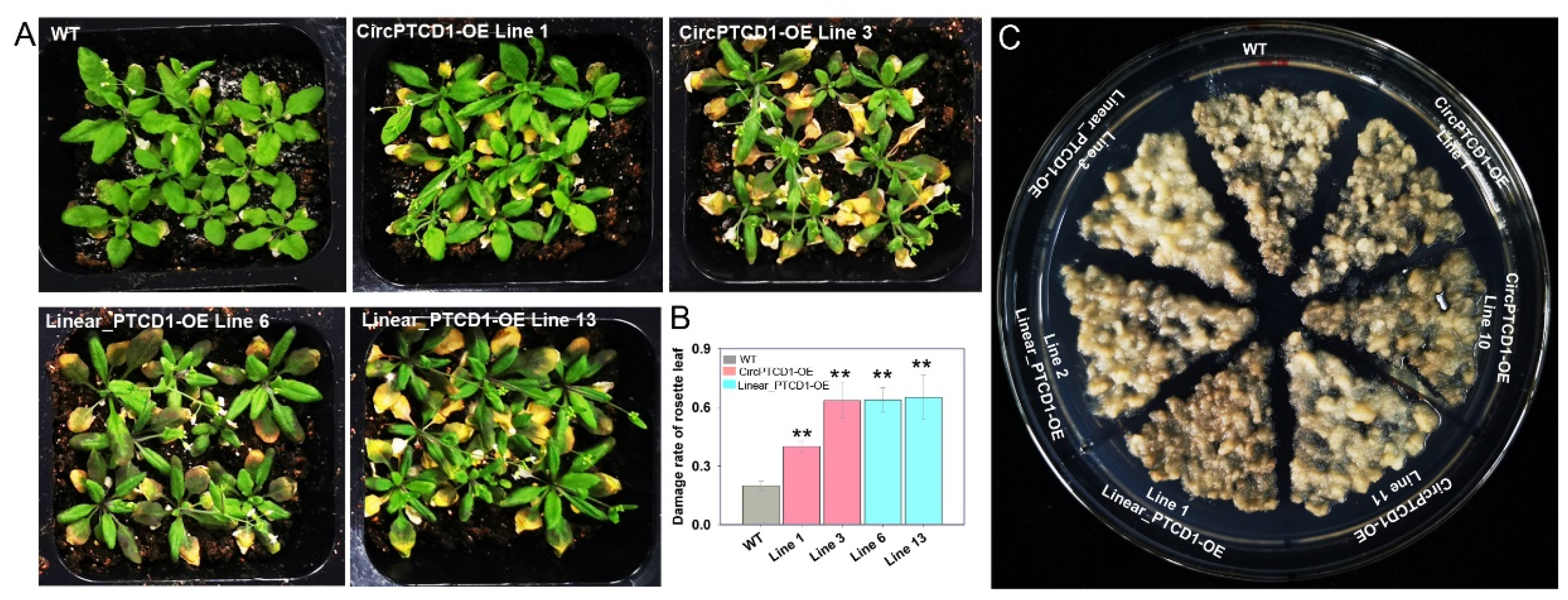
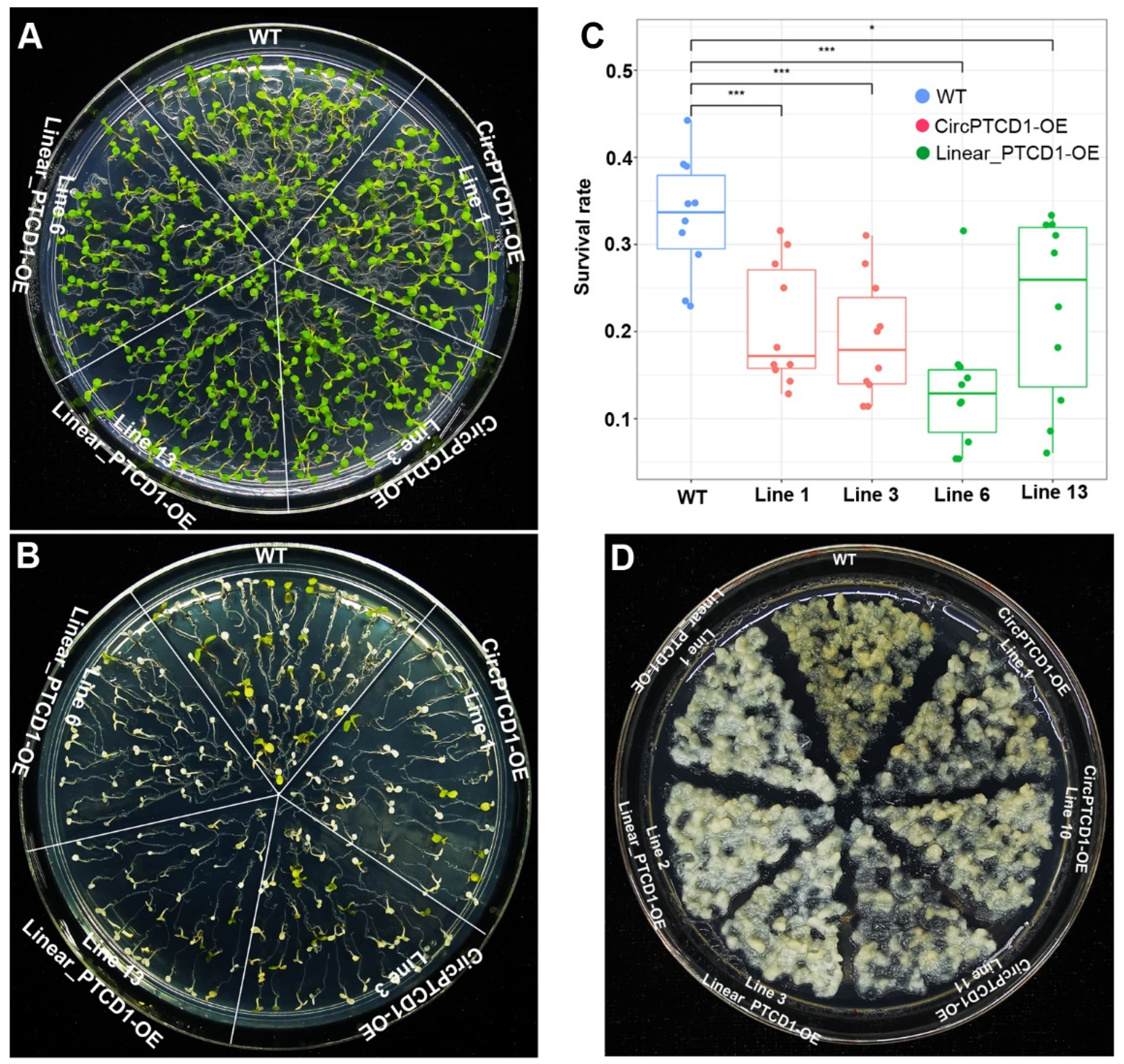
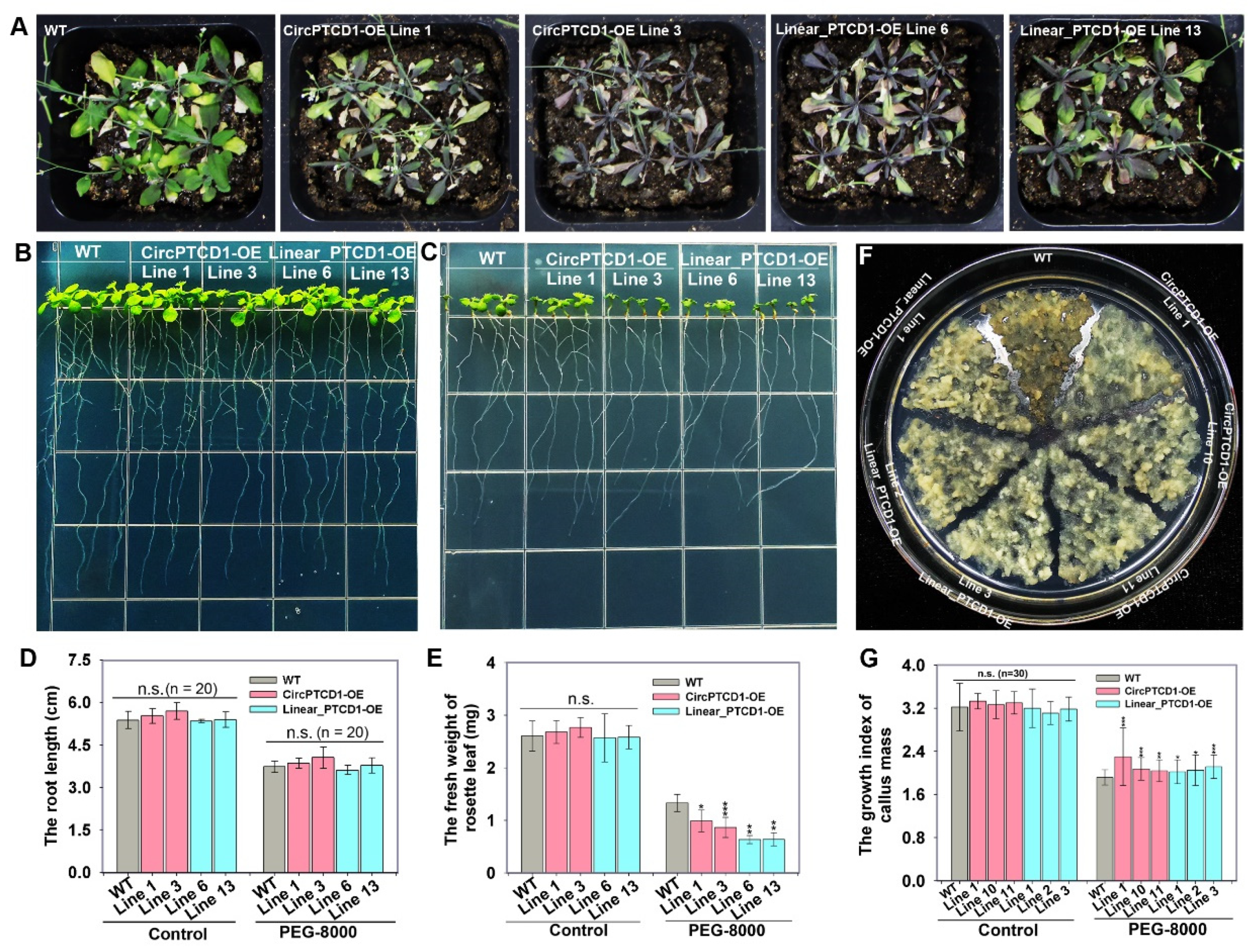
2.5. Phenotype of OE lines under heat stress
2.6. Phenotype of OE lines under salt stress
2.7. Phenotype of OE lines under drought stress
3. Discussion
4. Materials and methods
4.1. Plant materials and treatments
4.2. Validation of circRNA and RT-qPCR
4.3. Vector construction
4.4. Arabidopsis transformation and verification
4.5. Callus transformation of ‘Thompson Seedless’ and treatment
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Y.; Yang, M.; Wei, S.; Qin, F.; Zhao, H.; Suo, B. Identification of circular RNAs and their targets in leaves of Triticum aestivum L. under dehydration stress. Front Plant Sci 2016, 7, 2024. [CrossRef]
- Zhou, Y.; Wang, X.; Qi, K.; Bao, J.; Zhang, S.; Gu, C. Involvement of long non-coding RNAs in pear fruit senescence under high- and low-temperature conditions. Hortic Plant J 2022, 198, 112251. [CrossRef]
- Chen, L. L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol 2020, 21, 475-490. [CrossRef]
- Chu, Q.; Zhang, X.; Zhu, X.; Liu, C.; Mao, L.; Ye, C.; Zhu, Q. H.; Fan, L. PlantcircBase: a database for plant circular RNAs. Mol Plant 2017, 10, 1126-1128. [CrossRef]
- Zhang, P.; Meng, X.; Chen, H.; Liu, Y.; Xue, J.; Zhou, Y.; Chen, M. PlantCircNet: a database for plant circRNA-miRNA-mRNA regulatory networks. Database (Oxford) 2017, 2017, bax089. [CrossRef]
- Chu, Q.; Bai, P.; Zhu, X.; Zhang, X.; Mao, L.; Zhu, Q. H.; Fan, L.; Ye, C. Y. Characteristics of plant circular RNAs. Brief Bioinform 2020, 21, 135-143. [CrossRef]
- Hansen, T. B.; Jensen, T. I.; Clausen, B. H.; Bramsen, J. B.; Finsen, B.; Damgaard, C. K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384-8. [CrossRef]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; Laneve, P.; Rajewsky, N.; Bozzoni, I. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell 2017, 66, 22-37. [CrossRef]
- Yang, Y.; Gao, X.; Zhang, M.; Yan, S.; Sun, C.; Xiao, F.; Huang, N.; Yang, X.; Zhao, K.; Zhou, H.; Huang, S.; Xie, B.; Zhang, N. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst 2018, 110, 304-315. [CrossRef]
- Li, Q.; Wang, Y.; Wu, S.; Zhou, Z.; Ding, X.; Shi, R.; Thorne, R. F.; Zhang, X. D.; Hu, W.; Wu, M. CircACC1 regulates assembly and activation of AMPK complex under metabolic stress. Cell Metab 2019, 30, 157-173. [CrossRef]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; Zhu, P.; Chang, Z.; Wu, Q.; Zhao, Y.; Jia, Y.; Xu, P.; Liu, H.; Shan, G. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015, 22, 256-264. [CrossRef]
- Tan, J.; Zhou, Z.; Niu, Y.; Sun, X.; Deng, Z. Identification and functional characterization of tomato circRNAs derived from genes involved in fruit pigment accumulation. Sci Rep 2017, 7, 8594. [CrossRef]
- Conn, V. M.; Hugouvieux, V.; Nayak, A.; Conos, S. A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C.; Conn, S. J. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants 2017, 3, 17053. [CrossRef]
- Zhou, J.; Yuan, M.; Zhao, Y.; Quan, Q.; Yu, D.; Yang, H.; Tang, X.; Xin, X.; Cai, G.; Qian, Q.; Qi, Y.; Zhang, Y. Efficient deletion of multiple circle RNA loci by CRISPR-Cas9 reveals Os06circ02797 as a putative sponge for OsMIR408 in rice. Plant Biotechnol J 2021, 19, 1240-1252. [CrossRef]
- Song, Y.; Bu, C.; Chen, P.; Liu, P.; Zhang, D. Miniature inverted repeat transposable elements cis-regulate circular RNA expression and promote ethylene biosynthesis, reducing heat tolerance in Populus tomentosa. J Exp Bot 2021, 72, , 1978-1994. [CrossRef]
- Baroi, A. M.; Popitiu, M.; Fierascu, I.; Sardarescu, I. D.; Fierascu, R. C. Grapevine wastes: a rich source of antioxidants and other biologically active compounds. Antioxidants (Basel) 2022, 11, 393. [CrossRef]
- Venios, X.; Korkas, E.; Nisiotou, A.; Banilas, G. Grapevine responses to heat stress and global warming. Plants (Basel) 2020, 9,1754. [CrossRef]
- Zhu, Z.; Quan, R.; Chen, G.; Yu, G.; Li, X.; Han, Z.; Xu, W.; Li, G.; Shi, J.; Li, B. An R2R3-MYB transcription factor VyMYB24, isolated from wild grape Vitis yanshanesis J. X. Chen., regulates the plant development and confers the tolerance to drought. Front Plant Sci 2022, 13, 966641. [CrossRef]
- Ju, Y. L.; Yue, X. F.; Min, Z.; Wang, X. H.; Fang, Y. L.; Zhang, J. X. VvNAC17, a novel stress-responsive grapevine (Vitis vinifera L.) NAC transcription factor, increases sensitivity to abscisic acid and enhances salinity, freezing, and drought tolerance in transgenic Arabidopsis. Plant Physiol Biochem 2020, 146, 98-111. [CrossRef]
- Zha, Q.; Xi, X. J.; He, Y. N.; Jiang, A. L. Transcriptomic analysis of the leaves of two grapevine cultivars under high-temperature stress. Sci Hortic 2020, 265, 109265. [CrossRef]
- Merrill, N. K.; García de Cortázar-Atauri, I.; Parker, A. K.; Walker, M. A.; Wolkovich, E. M. Exploring grapevine phenology and high temperatures response under controlled conditions. Front Env Sci 2020, 8, 516527. [CrossRef]
- Ryu, S.; Han, J. H.; Cho, J. G.; Jeong, J. H.; Lee, S. K.; Lee, H. J. High temperature at veraison inhibits anthocyanin biosynthesis in berry skins during ripening in 'Kyoho' grapevines. Plant Physiol Biochem 2020, 157, 219-228. [CrossRef]
- Jiang, J.; Liu, X.; Liu, C.; Liu, G.; Li, S.; Wang, L. Integrating omics and alternative splicing reveals insights into grape response to high temperature. Plant Physiol 2017, 173, 1502-1518. [CrossRef]
- Liu, G. T.; Jiang, J. F.; Liu, X. N.; Jiang, J. Z.; Sun, L.; Duan, W.; Li, R. M.; Wang, Y.; Lecourieux, D.; Liu, C. H.; Li, S. H.; Wang, L. J. New insights into the heat responses of grape leaves via combined phosphoproteomic and acetylproteomic analyses. Hortic Res 2019, 6, 100. [CrossRef]
- Haider, S.; Iqbal, J.; Naseer, S.; Yaseen, T.; Shaukat, M.; Bibi, H.; Ahmad, Y.; Daud, H.; Abbasi, N. L.; Mahmood, T. Molecular mechanisms of plant tolerance to heat stress: current landscape and future perspectives. Plant Cell Rep 2021, 40, 2247-2271. [CrossRef]
- Pillet, J.; Egert, A.; Pieri, P.; Lecourieux, F.; Kappel, C.; Charon, J.; Gomes, E.; Keller, F.; Delrot, S.; Lecourieux, D. VvGOLS1 and VvHsfA2 are involved in the heat stress responses in grapevine berries. Plant Cell Physiol 2012, 53, 1776-92. [CrossRef]
- Liu, G.; Chai, F.; Wang, Y.; Jiang, J.; Duan, W.; Wang, Y.; Wang, F.; Li, S.; Wang, L. Genome-wide identification and classification of HSF family in grape, and their transcriptional analysis under heat acclimation and heat stress. Hortic Plant J 2018, 4,133-143. [CrossRef]
- Gambetta, G. A.; Herrera, J. C.; Dayer, S.; Feng, Q.; Hochberg, U.; Castellarin, S. D. The physiology of drought stress in grapevine: towards an integrative definition of drought tolerance. J Exp Bot 2020, 71, 4658-4676. [CrossRef]
- Mao, H.; Jian, C.; Cheng, X.; Chen, B.; Mei, F.; Li, F.; Zhang, Y.; Li, S.; Du, L.; Li, T.; Hao, C.; Wang, X.; Zhang, X.; Kang, Z. The wheat ABA receptor gene TaPYL1-1B contributes to drought tolerance and grain yield by increasing water-use efficiency. Plant Biotechnol J 2022, 20, 846-861. [CrossRef]
- Qin, T.; Zhao, H.; Cui, P.; Albesher, N.; Xiong, L. A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance. Plant Physiol 2017, 175, 1321-1336. [CrossRef]
- Li, J.; Duan, Y.; Sun, N.; Wang, L.; Feng, S.; Fang, Y.; Wang, Y. The miR169n-NF-YA8 regulation module involved in drought resistance in Brassica napus L. Plant Sci 2021, 313, 111062. [CrossRef]
- Pagliarani, C.; Vitali, M.; Ferrero, M.; Vitulo, N.; Incarbone, M.; Lovisolo, C.; Valle, G.; Schubert, A. The accumulation of miRNAs differentially modulated by drought stress is affected by grafting in grapevine. Plant Physiol 2017, 173, 2180-2195. [CrossRef]
- Yang, Y.; Guo, Y. Unraveling salt stress signaling in plants. J Integr Plant Biol 2018, 60, 796-804. [CrossRef]
- Yu, Y.; Ni, Y.; Qiao, T.; Ji, X.; Xu, J.; Li, B.; Sun, Q. Overexpression of VvASMT1 from grapevine enhanced salt and osmotic stress tolerance in Nicotiana benthamiana. PLoS One 2022, 17, e0269028. [CrossRef]
- Mzid, R.; Zorrig, W.; Ben Ayed, R.; Ben Hamed, K.; Ayadi, M.; Damak, Y.; Lauvergeat, V.; Hanana, M. The grapevine VvWRKY2 gene enhances salt and osmotic stress tolerance in transgenic Nicotiana tabacum. 3 Biotech 2018, 8, 277. [CrossRef]
- Gao, Z.; Li, J.; Luo, M.; Li, H.; Chen, Q.; Wang, L.; Song, S.; Zhao, L.; Xu, W.; Zhang, C.; Wang, S.; Ma, C. Characterization and cloning of Grape Circular RNAs Identified the Cold Resistance-Related Vv-circATS1. Plant Physiol 2019, 180,966-985. [CrossRef]
- Xing, H.; Fu, X.; Yang, C.; Tang, X.; Guo, L.; Li, C.; Xu, C.; Luo, K. Genome-wide investigation of pentatricopeptide repeat gene family in poplar and their expression analysis in response to biotic and abiotic stresses. Sci Rep 2018, 8, 2817. [CrossRef]
- Verslues, P. E.; Agarwal, M.; Katiyar-Agarwal, S.; Zhu, J.; Zhu, J. K. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 2006, 45, 523-39. [CrossRef]
- Livak, K. J.; Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-∆∆CT) Method. Methods 2001, 25, 402-408. [CrossRef]
- Clough, S. J.; Bent, A. F. Floral dip a simplified method for Agrobacterium -mediated transformation of Arabidopsis thaliana. Plant J 1998, 16, 9. [CrossRef]
- Gambino, G.; Ruffa, P.; Vallania, R.; Gribaudo, I. Somatic embryogenesis from whole flowers, anthers and ovaries of grapevine (Vitis spp.). Plant Cell Tiss Org 2007, 90, 79-83. [CrossRef]
- Fujita, Y.; Nakashima, K.; Yoshida, T.; Katagiri, T.; Kidokoro, S.; Kanamori, N.; Umezawa, T.; Fujita, M.; Maruyama, K.; Ishiyama, K.; Kobayashi, M.; Nakasone, S.; Yamada, K.; Ito, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol 2009, 50, 2123-2132. [CrossRef]
- Chu, Q.; Ding, Y.; Xu, X.; Ye, C. Y.; Zhu, Q. H.; Guo, L.; Fan, L. Recent origination of circular RNAs in plants. New Phytol 2022, 233, 515-525. [CrossRef]
- Liao, X.; Li, X. J.; Zheng, G. T.; Chang, F. R.; Fang, L.; Yu, H.; Huang, J.; Zhang, Y. F. Mitochondrion-encoded circular RNAs are widespread and translatable in plants. Plant Physiol 2022, 51, 1-6. [CrossRef]
- Lurin, C.; Andres, C.; Aubourg, S.; Bellaoui, M.; Bitton, F.; Bruyere, C.; Caboche, M.; Debast, C.; Gualberto, J.; Hoffmann, B.; Lecharny, A.; Le Ret, M.; Martin-Magniette, M. L.; Mireau, H.; Peeters, N.; Renou, J. P.; Szurek, B.; Taconnat, L.; Small, I. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 2004, 16, 2089-103. [CrossRef]
- Liu, Z.; Dong, F.; Wang, X.; Wang, T.; Su, R.; Hong, D.; Yang, G. A pentatricopeptide repeat protein restores nap cytoplasmic male sterility in Brassica napus. J Exp Bot 2017, 68, 4115-4123. [CrossRef]
- Sosso, D.; Canut, M.; Gendrot, G.; Dedieu, A.; Chambrier, P.; Barkan, A.; Consonni, G.; Rogowsky, P. M. PPR8522 encodes a chloroplast-targeted pentatricopeptide repeat protein necessary for maize embryogenesis and vegetative development. J Exp Bot 2012, 63, 5843-57. [CrossRef]
- Li, X.; Gu, W.; Sun, S.; Chen, Z.; Chen, J.; Song, W.; Zhao, H.; Lai, J. Defective Kernel 39 encodes a PPR protein required for seed development in maize. J Integr Plant Biol 2018, 60, 45-64. [CrossRef]
- Laluk, K.; Abuqamar, S.; Mengiste, T. The Arabidopsis mitochondria-localized pentatricopeptide repeat protein PGN functions in defense against necrotrophic fungi and abiotic stress tolerance. Plant Physiol 2011, 156, 2053-2068. [CrossRef]
- Xu, C.; Zhang, J. Mammalian circular RNAs result largely from splicing errors. Cell Rep 2021, 36, 09439. [CrossRef]
- Cocquerelle, C.; Mascrez, B.; Hétuin, D.; Bailleul, B. Mis-splicing yields circular RNA molecules. FASEB J 1993, 7, 6. [CrossRef]
- Zhang, X. O.; Wang, H. B.; Zhang, Y.; Lu, X.; Chen, L. L.; Yang, L., Complementary sequence-mediated exon circularization. Cell 2014, 159, 134-147. [CrossRef]
- Liu, C. X.; Chen, L. L. Circular RNAs: Characterization, cellular roles, and applications. Cell 2022, 185, 2016-2034. [CrossRef]
- Zhang, P.; Fan, Y.; Sun, X.; Chen, L.; Terzaghi, W.; Bucher, E.; Li, L.; Dai, M. A large-scale circular RNA profiling reveals universal molecular mechanisms responsive to drought stress in maize and Arabidopsis. Plant J 2019, 98, 697-713. [CrossRef]
- Liang, D.; Wilusz, J. E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev 2014, 28, 2233-2247. [CrossRef]
- Liu, Y.; Su, H.; Zhang, J.; Liu, Y.; Feng, C.; Han, F. Back-spliced RNA from retrotransposon binds to centromere and regulates centromeric chromatin loops in maize. PLoS Biol 2020, 18, e3000582. [CrossRef]
- Fan, J.; Quan, W.; Li, G. B.; Hu, X. H.; Wang, Q.; Wang, H.; Li, X. P.; Luo, X.; Feng, Q.; Hu, Z. J.; Feng, H.; Pu, M.; Zhao, J. Q.; Huang, Y. Y.; Li, Y.; Zhang, Y.; Wang, W. M. circRNAs Are Involved in the Rice-Magnaporthe oryzae Interaction. Plant Physiology 2020, 182, 272-286. [CrossRef]
- Liu, R.; Ma, Y.; Guo, T.; Li, G. Identification, biogenesis, function, and mechanism of action of circular RNAs in plants. Plant Commun 2022, 4, 100430. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




