Submitted:
04 January 2023
Posted:
20 January 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plant materials
2.2. Methods
2.2.1.Transcriptomic Sequencing and Library Construction
2.2.2. Identification and Functional Annotation of DEGs
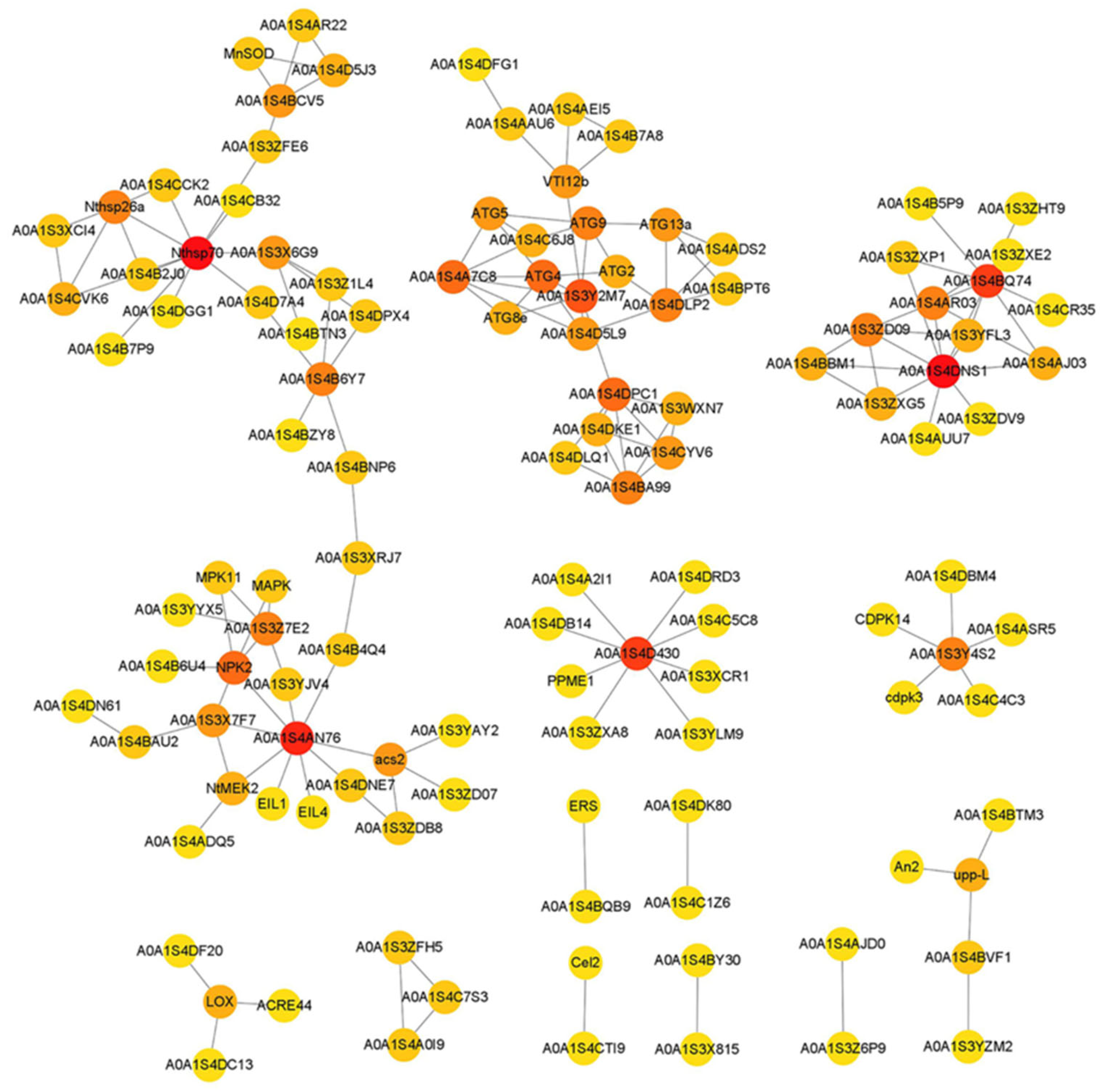
2.2.3. Protein-Protein Interaction Network Construction
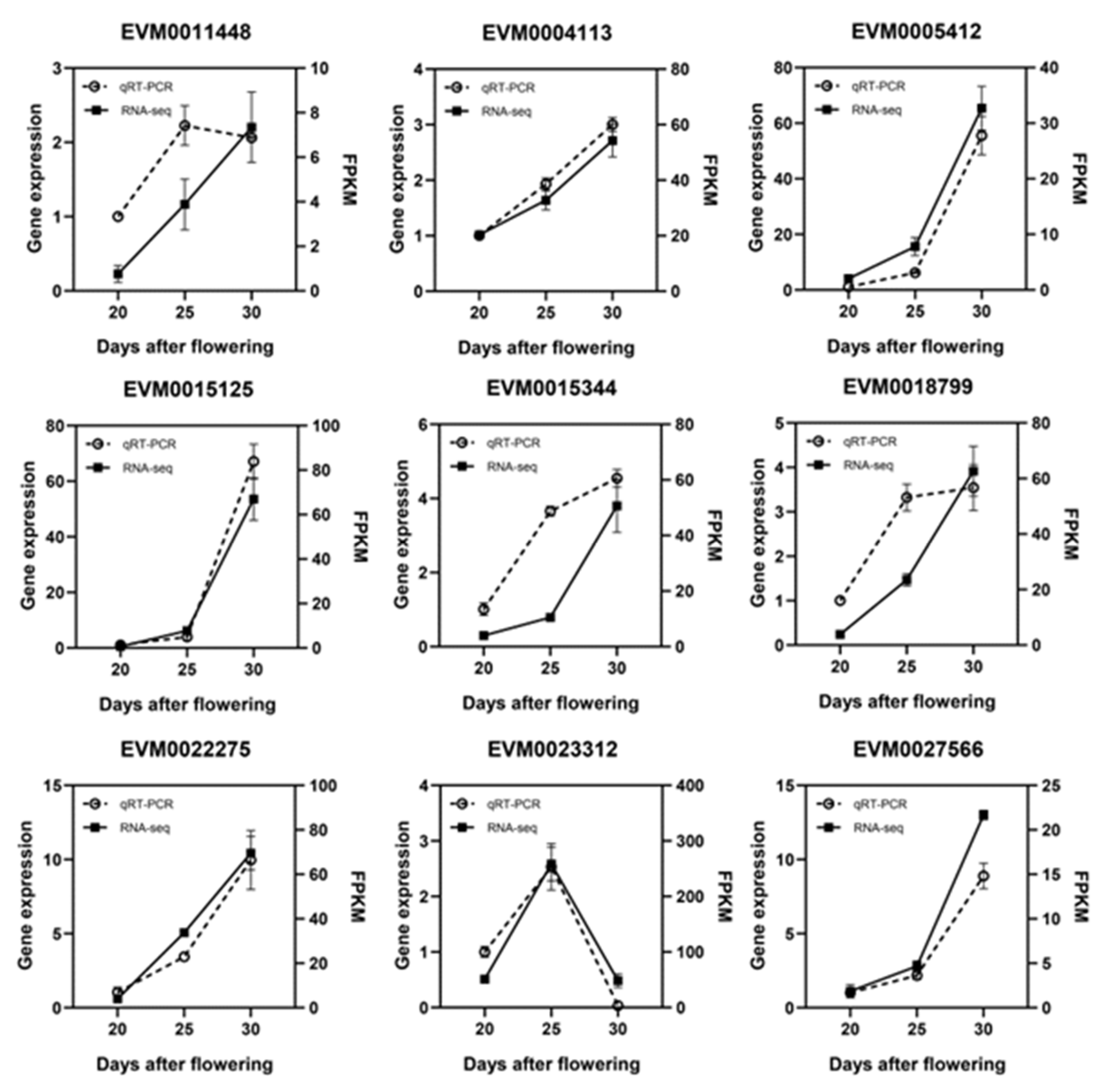
2.2.4. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Validation
3. Results
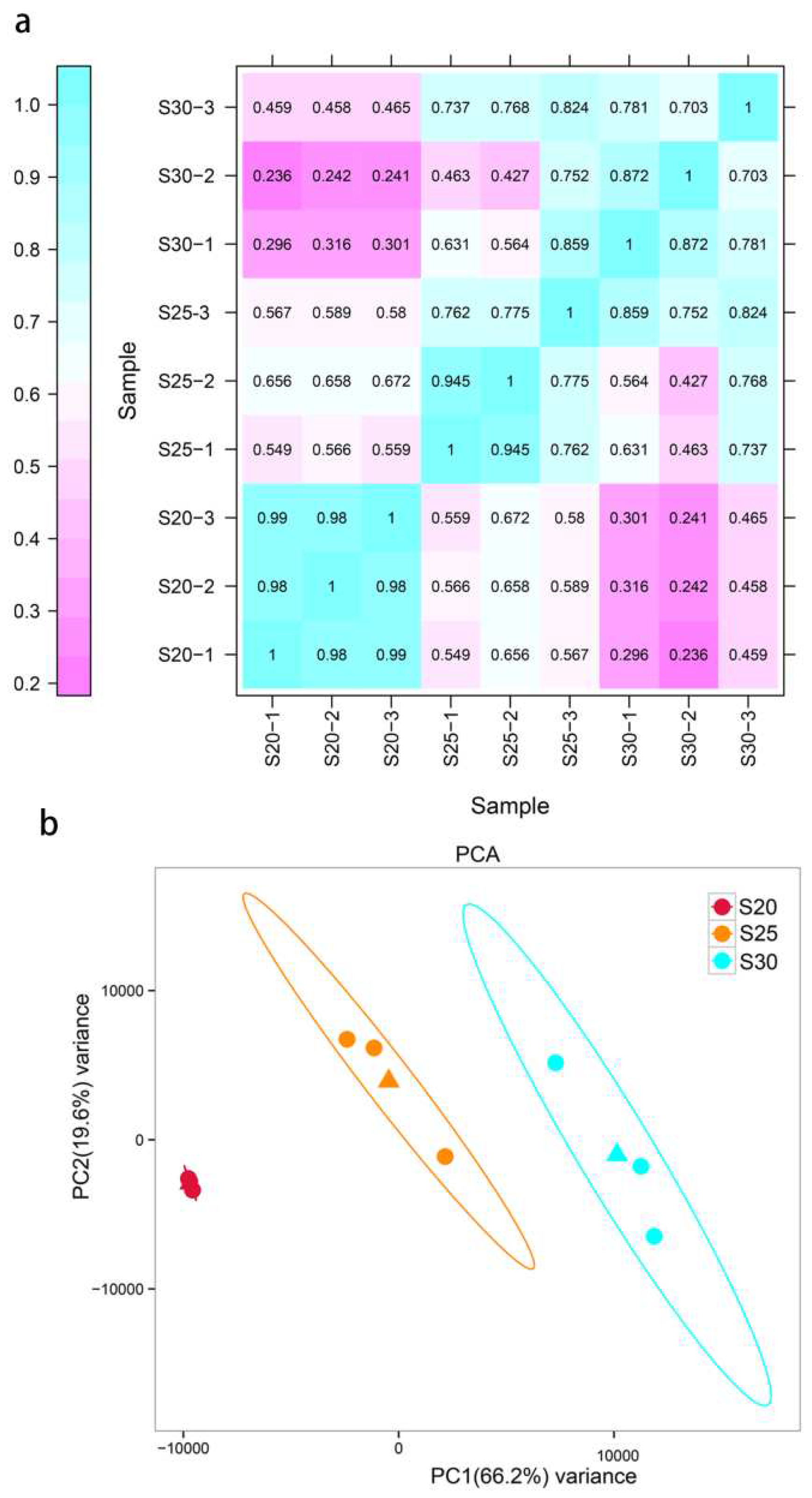
3.1. Transcriptomic Analysis
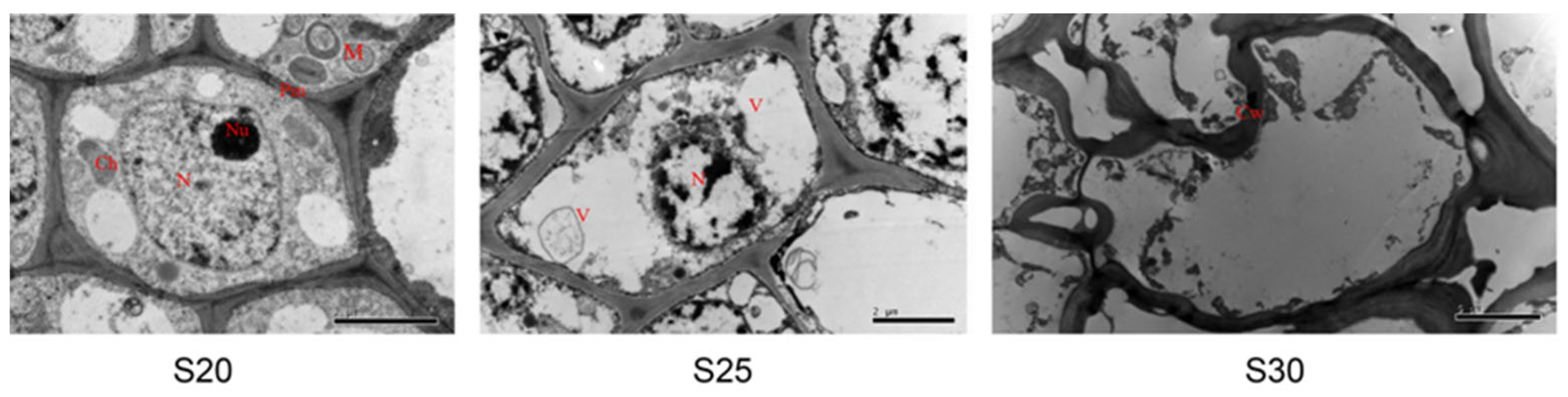
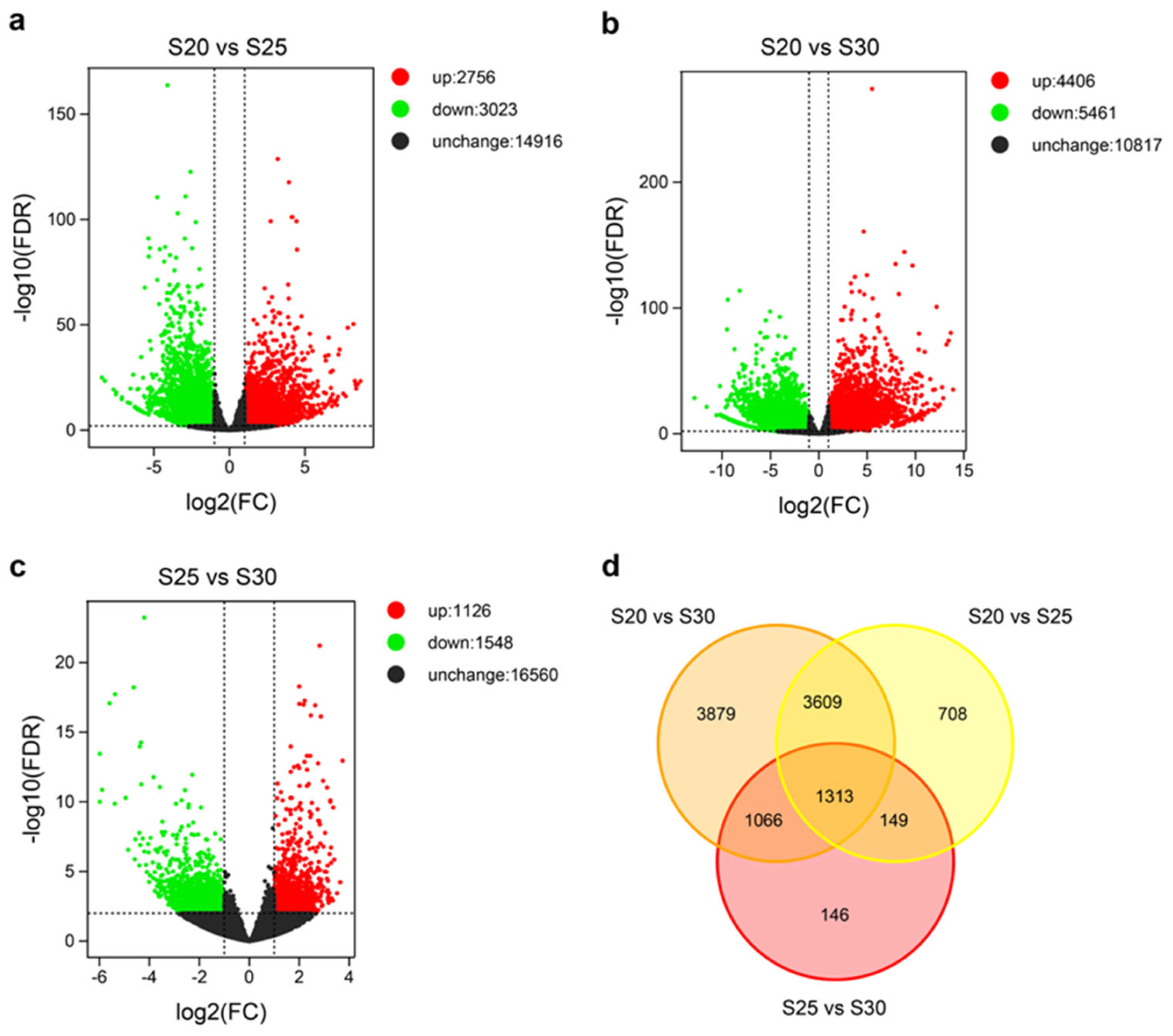
3.2. Global Transcriptional Changes before, during, and after PCD in Replaceable Buds of Chestnut cv. ‘Tima Zhenzhu’
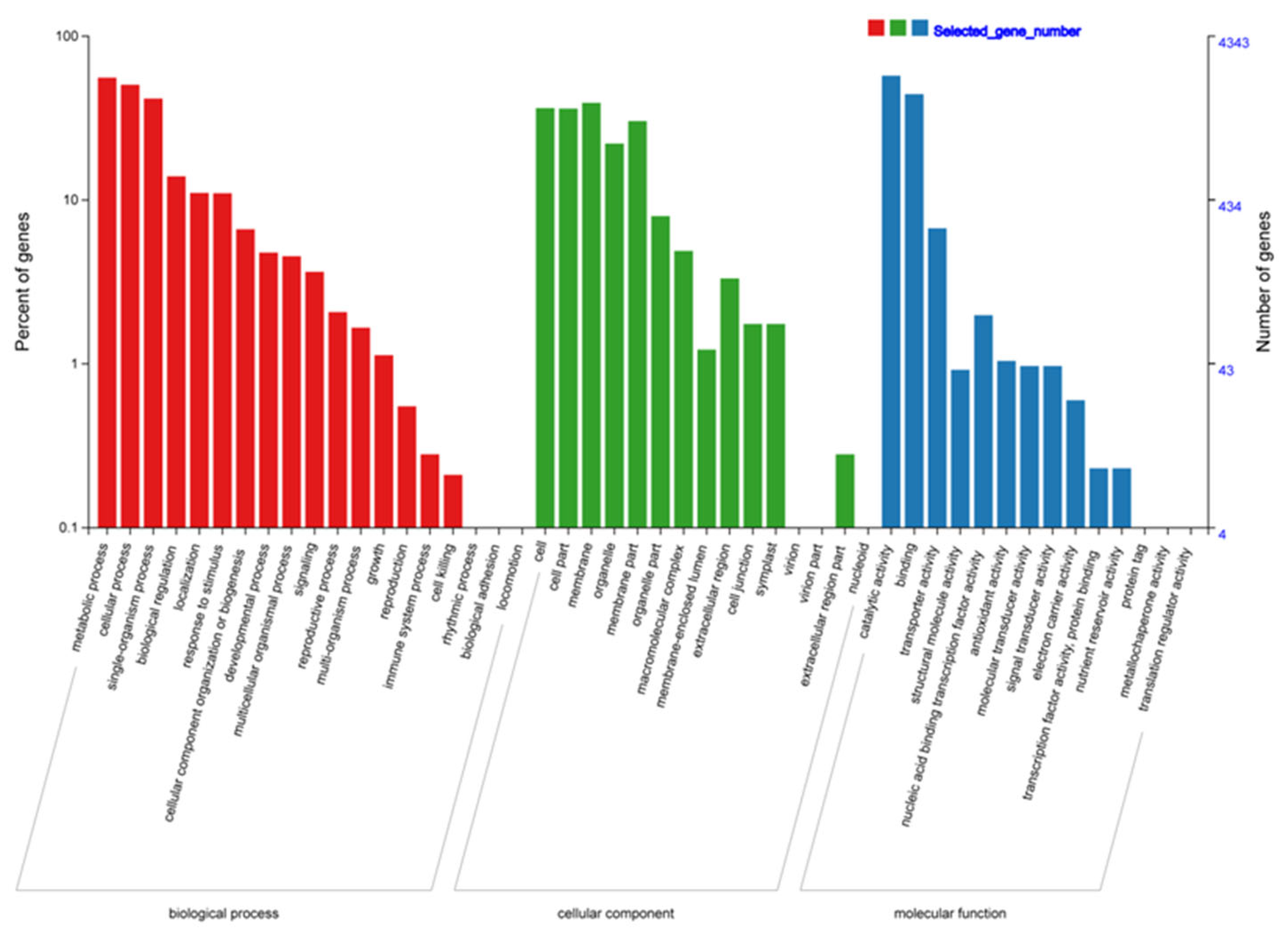
3.3. Functional Enrichment Analysis of the Common-DEGs
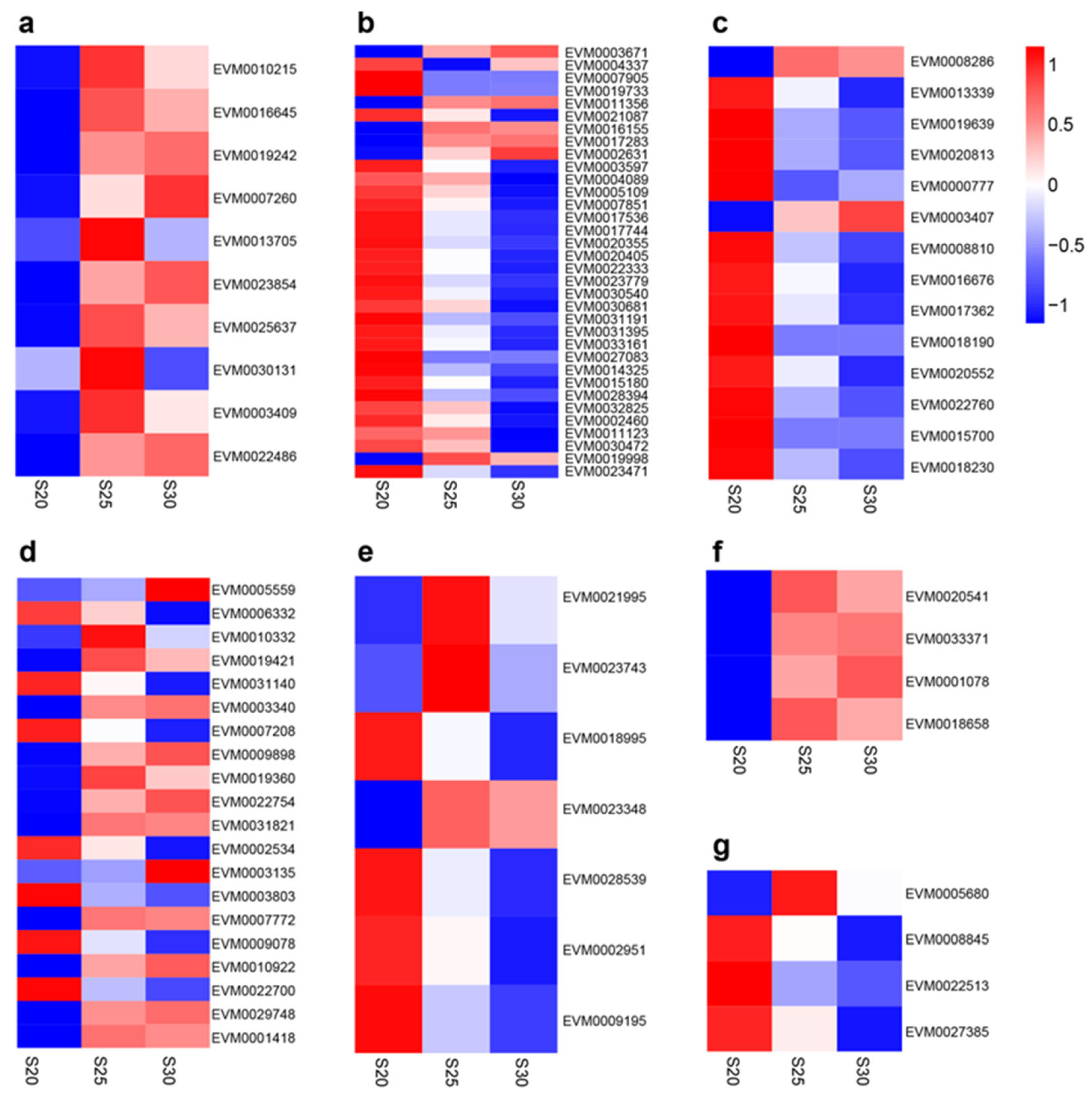
3.4. DEGs Related to Phytohormone Signal Transduction Pathways
3.5. DEGs Related to Replaceable Bud PCD Regulation in Chestnut cv. ‘Tima Zhenzhu’
4. Discussion
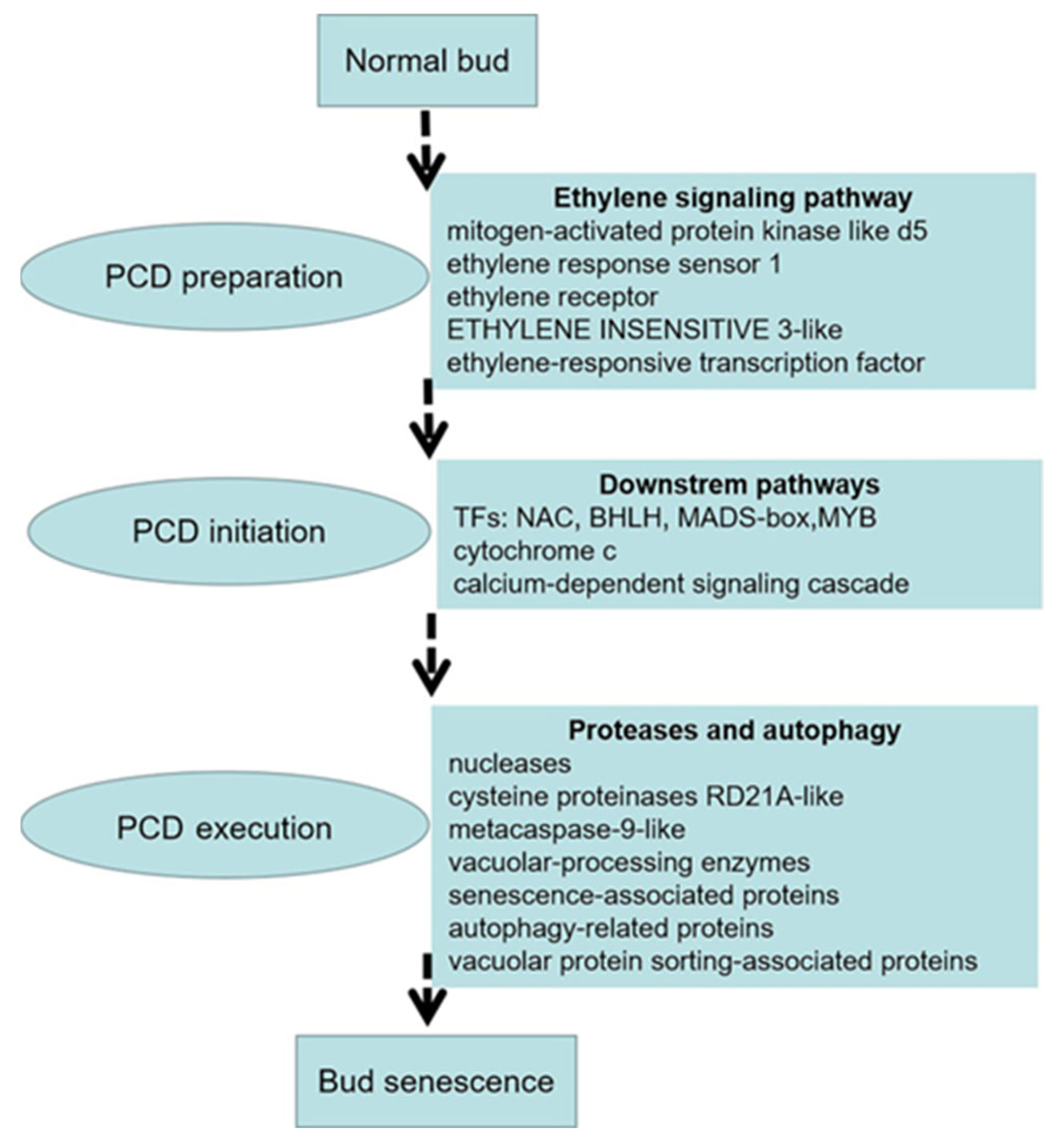
4.1. The role of the Ethylene Signaling in Replaceable Bud PCD of Chestnut cv. ‘Tima Zhenzhu’
4.2. PCD Initiation in Replaceable Buds of Chestnut cv. ‘Tima Zhenzhu’
4.3. PCD Execution in Replaceable Buds of Chestnut cv. ‘Tima Zhenzhu’
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, G.; Zhang, Z.; Kong, D.; Liu, Q.; Zhao, G. Programmed cell death is responsible for replaceable bud senescence in chestnut (Castanea mollissima BL.). Plant Cell Rep. 2012, 31, 1603–1610. [Google Scholar] [CrossRef]
- Liu, Q.; Kong, D.; Wang, G. A new chestnut variety ‘Tima Zhenzhu’. Acta Hort Sin 2004, 31, 698. [Google Scholar]
- Ye, C.; Zheng, S.; Jiang, D.; Lu, J.; Huang, Z.; Liu, Z.; Zhou, H.; Zhuang, C.; Li, J. Initiation and execution of programmed cell death and regulation of reactive oxygen species in plants. Int. J. Mol. Sci. 2021, 22. [Google Scholar] [CrossRef]
- Huang, D.; Huo, J.; Zhang, J.; Wang, CL.; Wang, B.; Fang, H.; Liao, W. Protein S-nitrosylation in programmed cell death in plants. Cell. Mol. Life Sci. 2019, 76, 1877–1887. [Google Scholar] [CrossRef]
- Hautegem, V.T.; Waters, A.J.; Goodrich, J.; Nowack, M.K. Only in dying, life: programmed cell death during plant development. Trends Plant Sci. 2015, 20, 102–113. [Google Scholar] [CrossRef]
- Daneva, A.; Gao, Z.; Van Durme, M.; Nowack, M.K. Functions and regulation of programmed cell death in plant development. Annu. Rev. Cell Dev. Biol. 2016, 32, 441–468. [Google Scholar] [CrossRef]
- Kabbage, M.; Kessens, R.; Bartholomay, L.C.; Williams, B. The life and death of a plant cell. Annu. Rev. Plant Biol. 2017, 68, 375–404. [Google Scholar] [CrossRef]
- Van Doorn, W.G. Classes of programmed cell death in plants, compared to those in animals. J. Exp. Bot. 2011, 62, 4749–4761. [Google Scholar] [CrossRef]
- Völz, R.; Heydlauff, J.; Ripper, D.; von Lyncker, L.; Groß-Hardt, R. Ethylene signaling is required for synergid degeneration and the establishment of a pollen tube block. Dev. Cell 2013, 25, 310–316. [Google Scholar] [CrossRef]
- Yamauchi, T.; Watanabe, K.; Fukazawa, A.; Mori, H.; Abe, F.; Kawaguchi, K.; Oyanagi, A.; Nakazono, M. Ethylene and reactive oxygen species are involved in root aerenchyma formation and adaptation of wheat seedlings to oxygendeficient conditions. J. Exp. Bot. 2014, 65, 261–273. [Google Scholar] [CrossRef]
- Grbic, V.; Beatrice, A.B. Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 1995, 8, 595–602. [Google Scholar] [CrossRef]
- Wang, H.; Wu, H.M.; Cheung, A.Y. Pollination induces mrna poly(A) tail-shortening and cell deterioration inflower transmitting tissue. Plant J. 1996, 9, 715–727. [Google Scholar] [CrossRef]
- Gunawardena, A.; Pearce, D.M.; Jackson, M.B.; Hawes, C.R.; Evans, D.E. Characterisation of programmed cell death during aerenchyma formation induced by ethylene or hypoxia in roots of maize (Zea mays L.). Planta 2001, 212, 205–214. [Google Scholar] [CrossRef]
- Plackett, A.R.; Thomas, S.G.; Wilson, Z.A.; Hedden, P. Gibberellin control of stamen development: a fertile field. Trends Plant Sci. 2011, 16, 568–578. [Google Scholar] [CrossRef]
- Fath, A.; Bethke, P. C.; Jones, R. L. Barley aleurone cell death is not apoptotic: characterization of nuclease activities and DNA degradation. Plant J. 1999, 20, 305–315. [Google Scholar] [CrossRef]
- Jiang, C.; Wang, J.W.; Leng, H.N.; Wang, X.Q.; Liu, Y.J.; Lu, H.W.; Lu, M.Z.; Zhang, J. Transcriptional regulation and signaling of developmental programmed cell death in plants. Front Plant Sci. 2021, 12, 702928. [Google Scholar] [CrossRef]
- Li, D.H.; Wu, D.; Li, S.Z.; Guo, N.; Gao, J.S.; Sun, X.; Cai, Y.P. Transcriptomic profiling identifies differentially expressed genes associated with programmed cell death of nucellar cells in ginkgo biloba L. BMC Plant Biol. 2019, 19, 1–17. [Google Scholar] [CrossRef]
- Anders, P.M.; Lee, P.Y.; Biesgen, C. The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 2000, 12, 1041–1061. [Google Scholar] [CrossRef]
- Sarwat, M.; Naqvi, A.R.; Ahmad, P. Phytohormones and microRNAs as sensors and regulators of leaf senescence: assigning macro roles to small molecules. Biotechnol. Adv. 2013, 31, 1153–71. [Google Scholar] [CrossRef]
- Huysmans, M.; Buono, R.A.; Skorzinski, N.; Radio, M.C.; Winter, F.D.; Parizot, B. NAC transcription factors ANAC087 and ANAC046 control distinct aspects of programmed cell death in the Arabidopsis columella and lateral root cap. Plant Cell 2018, 30, 2197–2213. [Google Scholar] [CrossRef]
- Petrov, V.; Hille, J.; Mueller-Roeber, B.; Gechev, T.S. ROS-mediated abiotic stressinduced programmed cell death in plants. Front Plant Sci 2015, 6, 69. [Google Scholar] [CrossRef]
- Fendrych, M.; Van Hautegem, T.; Van Durme, M.; Olvera-Carrillo, Y. Huysmans, M.; Karimi, M.; Lippens, S.; Guerin, C.J.; Krebs, M.; Schumacher, K.; Nowack, M.K. Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in Arabidopsis. Curr. Biol. 2014, 24, 931–940. [Google Scholar] [CrossRef]
- Ito, J.; Fukuda, H. ZEN1 is a key enzyme in the degradation of nuclear DNA during programmed cell death of tracheary elements. Plant Cell 2002, 34, 98–101. [Google Scholar] [CrossRef]
- Yin, L.; Xue, H. The MADS29 transcription factor regulates the degradation of the nucellus and the nucellar projection during rice seed development. Plant Cell 2012, 24, 1049–1065. [Google Scholar] [CrossRef]
- Niu, N.; Liang, W.; Yang, X.; Jin, W.; Wilson, ZA.; Hu, J.; Zhang, D. EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat. Commun. 2013, 4, 1445. [Google Scholar] [CrossRef] [PubMed]
- Buono, R.A.; Hudecek, R.; Nowack, M. K. Plant proteases during developmental programmed cell death(Review). J. Exp. Bot. 2019, 70, 2097–2112. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: a fast spliced aligner with low memory requirements [J]. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Wang, J.P.; Tian, S.L.; Sun, X.L.; Cheng, X.C.; Duan, N.B.; Tao, J.H.; Shen, G.N. Construction of pseudomolecules for the Chinese chestnut (Castanea mollissima)genome. G3: Genes, Genomes, Genet. 2020, 10, 3565–3574. [Google Scholar] [CrossRef] [PubMed]
- Florea, L.; Song, L.; Salzberg, S. L. Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Research 2013, 2, 188. [Google Scholar] [CrossRef]
- Love, M.I.; W Huber, S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lei, Q.S.; Zhang, S.J.; Kong, L.N.; Qing, B. Screening and identification of key biomarkers in hepatocellular carcinoma: Evidence from bioinformatic analysis. Oncol. Rep. 2017, 38, 2607–2618. [Google Scholar] [CrossRef]
- Denbigh, G.L.; Dauphinee, A.N.; Fraser, M.S.; Lacroix, C.R.; Gunawardena, A.H. The role of auxin in developmentally regulated programmed cell death in lace plant. Am. J. Bot. 2020, 107, 577–586. [Google Scholar] [CrossRef]
- Roberts, K.; Mc Cann, M.C. Xylogenesis: the birth of a corpse. Curr. Opin. Plant Biol. 2000, 3, 517–522. [Google Scholar] [CrossRef]
- Li, Z.; Peng, J.; Wen, X.; Guo, H. ETHYLENE-INSENSITIVE3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 2013, 25, 3311–3328. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Woo, H.R.; Kim, J.; Lim, P.O.; Lee, I.C.; Choi, S.H.; Hwang, D.; Nam, H.G. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 2009, 323, 1053–1057. [Google Scholar] [CrossRef] [PubMed]
- Wang, G. Studies on the characters and control signals of programmed cell death in mixed bud of apical-bud-senescence chestnut(Castanea mollissima BI.) Dissertation, Shenyang Agricultural University, Shenyang, China,2012.
- Kim, J.I.; Murphy, A.S.; Baek, D.; Lee, S.W.; Yun, D.J.; Bressan, R.A.; Narasimhan, M.L. YUCCA6 over-expressiondemonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 3981–3992. [Google Scholar] [CrossRef] [PubMed]
- Kacprzyk, J.; Burke, R.; Schwarze, J.; McCabe, P.F. Plant programmed cell death meets auxin signalling. The FEBS Journal 2022, 289, 1731–1745. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, S. Mitogen-activated protein kinase cascades in plant signaling. J. Integr. Plant Biol. 2022, 64, 301–341. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Han, X.F.; Yang, L.Y.; Deng, X.X.; Wu, H.J.; Zhang, M.M.; Liu, Y.D.; Zhang, S.Q.; Xu, J. Mitogen-activated protein kinases and calcium-dependent protein kinases are involved in wounding-induced ethylene biosynthesis in Arabidopsis thaliana. Plant, Cell Environ. 2018, 41, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Y.; Mei, Q.; Liang, W.F.; Sun, J.; Wang, X.M.; Zhou, J.; Wang, J.M.; Zhou, Y.H.; Zheng, B.S.; Yang, Y.; Chen, J.P. Gene mapping, genome-Wide transcriptome analysis, and WGCNA reveals the molecular mechanism for triggering programmed cell death in rice mutant pir1. Plants 2020, 9, 1607. [Google Scholar] [CrossRef]
- Li, S.T.; Samaj, J.; Franklin-Tong, V.E. A mitogen-activated protein kinase signals to programmed cell death induced by self-incompatibility in Papaver pollen. Plant physiol. 2007, 145, 236–245. [Google Scholar] [CrossRef]
- Zhou, C.J.; Cai, Z.H.; Guo, Y.F.; Gan, S.S. An Arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiol. 2009, 150, 167–177. [Google Scholar] [CrossRef]
- Huysmans, M.; Lema, A.S.; Coll, N.S.; Nowack, M.K. Dying two deaths-programmed cell death regulation in development and disease. Curr. Opin. Plant Biol. 2017, 35, 37–44. [Google Scholar] [CrossRef]
- Wang, F.; Mao, Y.L.; Guo, Y.J.; Gao, J.H.; Liu, X.Y.; Li, S.; Jimmy, Y.C.; Chen, H.; Wang, J.P.; Chiang, V.L.; Li, W. MYB transcription factor 161 mediates feedback regulation of Secondary wall-associated NAC-Domain 1 family genes for wood formation. Plant physiol. 2020, 184, 1389–1406. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Nam, H.G.; Lim, P.O. Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 2016, 33, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Plackett, A.R.; Ferguson, A.C.; Powers, S.J.; Wanchoo-Kohli, A.; Phillips, A.L.; Wilson, Z.A.; Hedden, P.; Thomas, S.G. DELLA activity is required for successful pollen development in the Columbia ecotype of Arabidopsis. New Phytol. 2014, 201, 825–836. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.S.; Li, M.J.; Ku, M.S.B.; Ho, Y.C.; Lin, Y.J.; Chuang, M.H.; Hsing, H.X.; Lien, Y.C.; Yang, H.T.; Chang, H.C.; Chan, M.T. The bHLH142 transcription factor coordinates with TDR1 to modulate the expression of EAT1 and regulate pollen development in rice. Plant Cell 2014, 26, 2486–2504. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Fiume, E.; Coen, O.; Pechoux, C.; Lepiniec, L.; Magnani, E. Endosperm and nucellus develop antagonistically in Arabidopsis seeds. Plant Cell 2016, 28, 1343–1360. [Google Scholar] [CrossRef] [PubMed]
- Durme, M.; Nowack, M.K. Mechanisms of developmentally controlled cell death in plants. Curr. Opin. Plant Biol. 2016, 29, 29–37. [Google Scholar] [CrossRef]
- Chen, H.Y.; Osuna, D.; Colville, L.; Lorenzo, O.; Graeber, K.; Kuster, H.; Leubner-Metzger, G.; Kranner, I. Transcriptome-wide mapping of pea seed ageing reveals a pivotal role for genes related to oxidative stress and programmed cell death. PLoS One 2013, 8, e78471. [Google Scholar] [CrossRef]
- Balk, J.; Leaver, C.J. The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 2001, 13, 1803–1818. [Google Scholar] [CrossRef] [PubMed]
- Lara, L.; Nello, C.; Piero, P.; Roberto, L. Caspase-like proteases involvement in programmed cell death of Phaseolus coccineus suspensor. Plant Sci. 2007, 172, 573–578. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhan, Q.D.; Shi, T.T.; Liu, J.; Zhao, K.J.; Gao, Y. The nuclear transporter SAD2 plays a role in calcium- and H2O2-mediated cell death in Arabidopsis. Plant J. 2020, 101, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.M.; Zhao, X.; Li, W.J.; Hussain, J.; Qi, G.; Liu, S.K. Calcium signaling in plant programmed cell death. Cells 2021, 10, 1089. [Google Scholar] [CrossRef]
- Kang, C.H.; Jung, W.Y.; Kang, Y.H.; Kim, J.Y.; Kim, D.G.; Jeong, J.C.; Baek, D.W.; Jin, J.B.; Lee, J.Y.; Kim, M.O.; Chung, W.S.; Mengiste, T.; Koiwa, H.; Kwak, S.S.; Bahk, J.D.; Lee, S.Y.; Nam, J.S.; Yun, D.J.; Cho, M.J. AtBAG6, a novel calmodulin-binding protein, induces programmed cell death in yeast and plants. Cell Death Differ. 2006, 13, 84–95. [Google Scholar] [CrossRef]
- Torre, F.D.L.; Gutiérrez-Beltrán, E.; Pareja-Jaime, Y.; Chakravarthy, S.; Martin, G.B.; Pozo, O.D. The tomato calcium sensor Cbl10 and its interacting protein kinase Cipk6 define a signaling Pathway in plant immunity. Plant Cell 2013, 25, 2748–2764. [Google Scholar]
- Lachaud, C.; Prigent, E.; Thuleau, P.; Grat, S.; Silva, D.D.; Brière, C.; Mazars, C.; Cotelle, V. 14-3-3-Regulated Ca2+-dependent protein kinase CPK3 is required for sphingolipid-induced cell death in Arabidopsis. Cell Death Differ. 2013, 20, 209–217. [Google Scholar] [CrossRef]
- Xin, X.; Lin, X.H.; Zhou, Y.C.; Chen, X.L.; Liu, X.; Lu, X.X. Proteome analysis of maize seeds: the effect of artificial ageing. Physiol. Plant. 2011, 143, 126–138. [Google Scholar] [CrossRef]
- Guo, Y.; Li, Y.; Zhang, S.H.; Zhang, X.F.; Wang, G.P. Dynamic changes of Ca2+ distribution during the process of programmed cell death in bud of apical-bud-senescence chestnut. J. Yunnan Agric. Univ. (Nat. Sci.) 2019, 34, 1–6. [Google Scholar]
- Nakaune, S.; Yamada, K.; Kondo, M.; Kato, T.; Tabata, S.; Nishimura, M.; Hara-Nishimura, I. A vacuolar processing enzyme, δVPE, is involved in seed coat formation at the early stage of seed development. Plant Cell 2005, 17, 876–887. [Google Scholar] [CrossRef]
- Andrade Buono, R.; Hudecek, R.; Nowack, M.K. The roles of proteases during developmental programmed cell death in plants. J. Exp. Bot. 2019, 70, 2097–2112. [Google Scholar] [CrossRef]
- Huang, X.; Li, F. Q. Roles of autophagy in plant programmed cell death. Chin. Bull. Bot. 2016, 51, 859–862. [Google Scholar]
- Bassham, D.C. Plant autophagy—more than a starvation response. Curr. Opin. Plant Biol. 2007, 10, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Paula, T.B.; Raz, D.; Hadas, P.Z.; Eduard, B.; Mohamad, A.A.; Tamar, A.W.; Einat, S.; Dani, E. Vacuolar processing enzyme translocates to the vacuole through the autophagy pathway to induce programmed cell death. Autophagy 2021, 17, 3109–3123. [Google Scholar] [CrossRef]
- Matilla, A.J. Role of metacaspases and autophagy in developmental programmed cell death in plants. Curr. Top. Phytochem. 2019, 15, 1–14. [Google Scholar]
- Guo, M.M.; Chen, M.; Liu, R.B.; Ma, Y.Z.; Li, L.C.; Xu, Z.S.; Zhang, X.H. Vacuolar protein sorting AtVPS25 regulates auxin responses in Arabidopsis thaliana. Sci.Agric.Sin. 2014, 47, 3501–3512. [Google Scholar]

| Sample | S20 | S25 | S30 |
| Total reads | 42,979,831 | 45,269,139 | 45,562,945 |
| Total mapped reads | 37,370,780 (86.94%) | 40,861,735 (90.25%) | 40,896,455 (89.77%) |
| Uniquely mapped reads | 35,620,696 (82.87%) | 38,678,320 (85.43%) | 37,872,455 (83.21%) |
| Multiple mapped Reads | 1,750,084 (4.07%) | 2,183,415 (4.81%) | 3,024,000 (6.56%) |
| GC content (%) | 44.92 | 44.82 | 45.11 |
| Percentage of Q30 base (%) | 93.24 | 94.29 | 93.31 |
| Pathway ID | Description | Number of annotated DEGs | p value | q value |
| ko00592 | alpha-Linolenic acid metabolism | 31 | 1.99E-06 | 9.82E-05 |
| ko00760 | Nicotinate and nicotinamide metabolism | 14 | 2.10E-06 | 9.82E-05 |
| ko04146 | Peroxisome | 36 | 5.04E-06 | 0.00013059 |
| ko00071 | Fatty acid degradation | 27 | 5.58E-06 | 0.00013059 |
| ko00591 | Linoleic acid metabolism | 15 | 5.43E-05 | 0.00101764 |
| ko00940 | Phenylpropanoid biosynthesis | 93 | 6.63E-05 | 0.001035256 |
| ko00480 | Glutathione metabolism | 46 | 8.73E-05 | 0.0011687 |
| ko04075 | Plant hormone signal transduction | 93 | 0.000115513 | 0.001352718 |
| ko00330 | Arginine and proline metabolism | 27 | 0.000175227 | 0.001823997 |
| ko00280 | Valine, leucine and isoleucine degradation | 26 | 0.000353738 | 0.003275552 |
| ko00564 | Glycerophospholipid metabolism | 36 | 0.000416394 | 0.003275552 |
| ko02010 | ABC transporters | 14 | 0.000419565 | 0.003275552 |
| ko00073 | Cutin, suberine and wax biosynthesis | 17 | 0.00079293 | 0.005714235 |
| ko00565 | Ether lipid metabolism | 16 | 0.000870345 | 0.005824112 |
| Gene ID | KEGG Orthology | Signaling Pathway | Annotation |
|---|---|---|---|
| EVM0010215 | K14512 | Ethylene | mitogen-activated protein kinase like d5 |
| EVM0016645 | K14509 | Ethylene | ERS1,ethylene response sensor 1 |
| EVM0019242 | K14509 | Ethylene | ethylene receptor 2-like |
| EVM0007260 | K14515 | Ethylene | EBF1EIN3-binding F-box protein 1-like |
| EVM0013705 | K14514 | Ethylene | EIN3,ETHYLENE INSENSITIVE 3-like 1 protein |
| EVM0023854 | K14514 | Ethylene | EIN3,ETHYLENE INSENSITIVE 3-like 3 protein |
| EVM0025637 | K14514 | Ethylene | EIN3,protein ETHYLENE INSENSITIVE 3-like |
| EVM0030131 | K14516 | Ethylene | ethylene-responsive transcription factor 1B-like |
| EVM0003409 | K14516 | Ethylene | ethylene-responsive transcription factor 1B |
| EVM0022486 | K14516 | Ethylene | ethylene-responsive transcription factor 1B |
| EVM0003671 | K14488 | Auxin | auxin-responsive protein SAUR36-like |
| EVM0004337 | K14488 | Auxin | auxin-responsive protein SAUR50-like |
| EVM0007905 | K14488 | Auxin | auxin-responsive protein SAUR50-like |
| EVM0019733 | K14488 | Auxin | auxin-responsive protein SAUR32 |
| EVM0011356 | K14488 | Auxin | auxin-responsive protein SAUR72-like |
| EVM0021087 | K14488 | Auxin | auxin-responsive protein SAUR50-like |
| EVM0016155 | K14488 | Auxin | auxin-responsive protein SAUR72-like |
| EVM0017283 | K14488 | Auxin | auxin-responsive protein SAUR32-like |
| EVM0002631 | K14488 | Auxin | auxin-responsive protein SAUR36 |
| EVM0003597 | K14484 | Auxin | auxin-responsive protein IAA27-like |
| EVM0004089 | K14484 | Auxin | auxin-responsive protein IAA29 isoform X1 |
| EVM0005109 | K14484 | Auxin | auxin-responsive protein IAA4-like |
| EVM0007851 | K14484 | Auxin | auxin-responsive protein IAA11 isoform X1 |
| EVM0017536 | K14484 | Auxin | auxin-responsive protein IAA4-like |
| EVM0017744 | K14484 | Auxin | auxin-responsive protein IAA13-like |
| EVM0020355 | K14484 | Auxin | auxin-responsive protein IAA27-like |
| EVM0020405 | K14484 | Auxin | auxin-responsive protein IAA29-like |
| EVM0022333 | K14484 | Auxin | auxin-responsive protein IAA26-like isoform X2 |
| EVM0023779 | K14484 | Auxin | auxin-responsive protein IAA32-like |
| EVM0030540 | K14484 | Auxin | auxin-responsive protein IAA4-like |
| EVM0030681 | K14484 | Auxin | auxin-responsive protein IAA16 |
| EVM0031191 | K14484 | Auxin | auxin-responsive protein IAA14-like |
| EVM0031395 | K14484 | Auxin | auxin-responsive protein IAA20-like |
| EVM0033161 | K14484 | Auxin | auxin-responsive protein IAA26-like isoform X2 |
| EVM0027083 | K14484 | Auxin | auxin-induced protein IAA6 |
| EVM0014325 | K14484 | Auxin | auxin-induced protein AUX28-like |
| EVM0015180 | K14486 | Auxin | auxin response factor 3 |
| EVM0028394 | K14486 | Auxin | auxin response factor 5 isoform X1 |
| EVM0032825 | K14486 | Auxin | auxin response factor 9-like |
| EVM0002460 | K14486 | Auxin | auxin response factor 18 |
| EVM0011123 | K13946 | Auxin | auxin transporter-like protein 1 |
| EVM0030472 | K13946 | Auxin | auxin transporter-like protein 5 |
| EVM0019998 | K13946 | Auxin | auxin influx transport protein |
| EVM0023471 | K14485 | Auxin | protein auxin signaling F-BOX 3-like |
| EVM0008286 | K14489 | Cytokinin | histidine kinase 3 |
| EVM0013339 | K14489 | Cytokinin | histidine kinase 2 |
| EVM0019639 | K14489 | Cytokinin | histidine kinase 4 |
| EVM0020813 | K14489 | Cytokinin | histidine kinase 4 |
| EVM0000777 | K14491 | Cytokinin | putative two-component response regulator ARR20 isoform X1 |
| EVM0003407 | K14492 | Cytokinin | two-component response regulator ORR9-like |
| EVM0008810 | K14492 | Cytokinin | two-component response regulator ARR8-like |
| EVM0016676 | K14492 | Cytokinin | two-component response regulator ORR9-like |
| EVM0017362 | K14492 | Cytokinin | two-component response regulator ARR5-like |
| EVM0018190 | K14492 | Cytokinin | two-component response regulator ORR9-like |
| EVM0020552 | K14491 | Cytokinin | two-component response regulator ARR12-like isoform X1 |
| EVM0022760 | K14492 | Cytokinin | two-component response regulator ARR5-like |
| EVM0015700 | K14490 | Cytokinin | AHP5,histidine-containing phosphotransfer protein 5 |
| EVM0018230 | K14490 | Cytokinin | histidine-containing phosphotransfer protein 1-like |
| EVM0005559 | K14496 | Abscisic acid | abscisic acid receptor PYR1 |
| EVM0006332 | K14432 | Abscisic acid | abscisic acid-insensitive 5-like protein 2 |
| EVM0010332 | K14432 | Abscisic acid | abscisic acid-insensitive 5-like protein 7 |
| EVM0019421 | K14432 | Abscisic acid | abscisic acid-insensitive 5-like protein 2 |
| EVM0031140 | K14432 | Abscisic acid | abscisic acid-insensitive 5-like protein 2 |
| EVM0003340 | K14497 | Abscisic acid | PP2C, probable protein phosphatase 2C 51 |
| EVM0007208 | K14497 | Abscisic acid | PP2C, probable protein phosphatase 2C 8 |
| EVM0009898 | K14497 | Abscisic acid | PP2C, protein phosphatase 2C 37-like |
| EVM0019360 | K14497 | Abscisic acid | PP2C, putative protein phosphatase 2C 24 |
| EVM0022754 | K14497 | Abscisic acid | PP2C, protein phosphatase 2C 77 |
| EVM0031821 | K14497 | Abscisic acid | PP2C, probable protein phosphatase 2C 75 isoform X1 |
| EVM0002534 | K14500 | Abscisic acid | putative serine/threonine-protein kinase |
| EVM0003135 | K14498 | Abscisic acid | serine/threonine-protein kinase SAPK2-like |
| EVM0003803 | K14498 | Abscisic acid | serine/threonine-protein kinase SAPK3-like isoform X2 |
| EVM0007772 | K14500 | Abscisic acid | probable serine/threonine-protein kinase At4g35230 |
| EVM0009078 | K14498 | Abscisic acid | serine/threonine-protein kinase SRK2A-like |
| EVM0010922 | K14500 | Abscisic acid | probable serine/threonine-protein kinase At4g35230 isoform X1 |
| EVM0022700 | K14500 | Abscisic acid | probable serine/threonine-protein kinase At4g35230 isoform X2 |
| EVM0029748 | K14498 | Abscisic acid | serine/threonine-protein kinase SRK2I |
| EVM0001418 | K14498 | Abscisic acid | serine/threonine-protein kinase SRK2A-like isoform X2 |
| EVM0021995 | K12126 | Gibberellin | transcription factor PIF3-like isoform X1 |
| EVM0023743 | K12126 | Gibberellin | transcription factor PIF3 |
| EVM0018995 | K14493 | Gibberellin | gibberellin receptor GID1C-like |
| EVM0023348 | K14493 | Gibberellin | gibberellin receptor GID1B-like |
| EVM0028539 | K14495 | Gibberellin | F-box protein GID2 |
| EVM0002951 | K14494 | Gibberellin | DELLA protein SLN1-like |
| EVM0009195 | K14494 | Gibberellin | DELLA protein GAIP-like |
| EVM0020541 | K13449 | Salicylic acid | pathogenesis-related protein 1-like |
| EVM0033371 | K13449 | Salicylic acid | pathogenesis-related protein 1-like |
| EVM0001078 | K13449 | Salicylic acid | basic form of pathogenesis-related protein 1-like |
| EVM0018658 | K14508 | Salicylic acid | regulatory protein NPR5 |
| EVM0005680 | K13416 | Brassinosteroid | brassinosteroid insensitive 1-associated receptor kinase 1 |
| EVM0008845 | K14505 | Brassinosteroid | cyclin-D3-2-like |
| EVM0022513 | K14505 | Brassinosteroid | cyclin-D3-1-like |
| EVM0027385 | K14505 | Brassinosteroid | cyclin-D3-3-like |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




