Submitted:
26 August 2023
Posted:
29 August 2023
You are already at the latest version
Abstract
Keywords:
Introduction
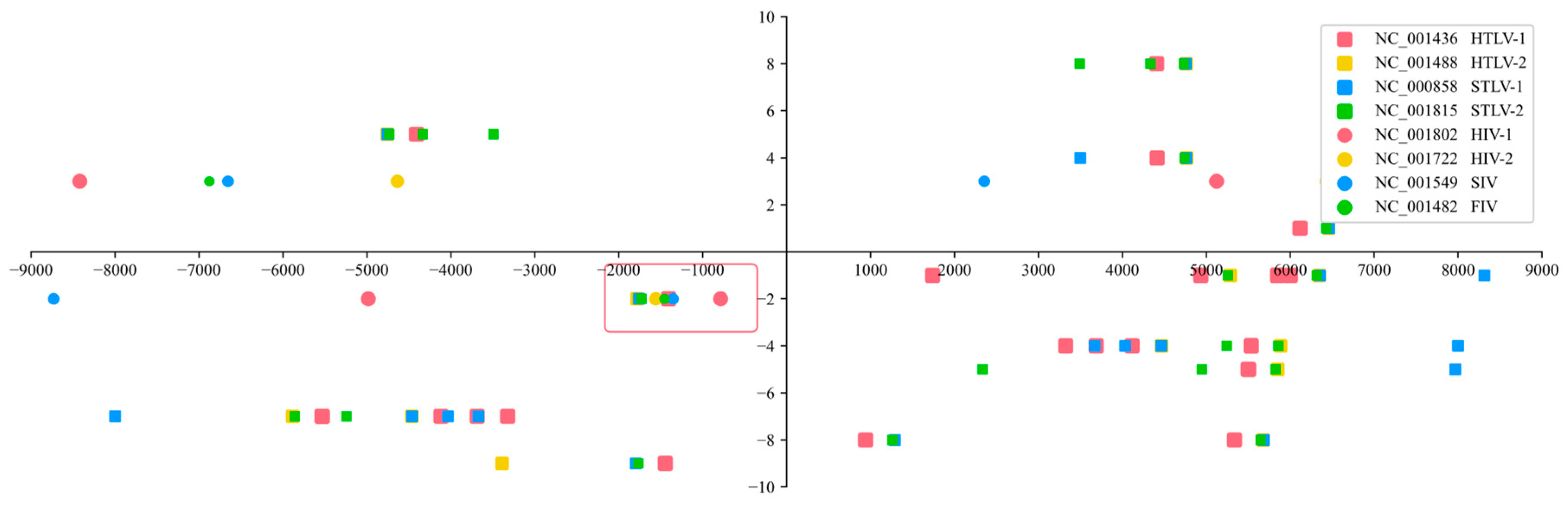
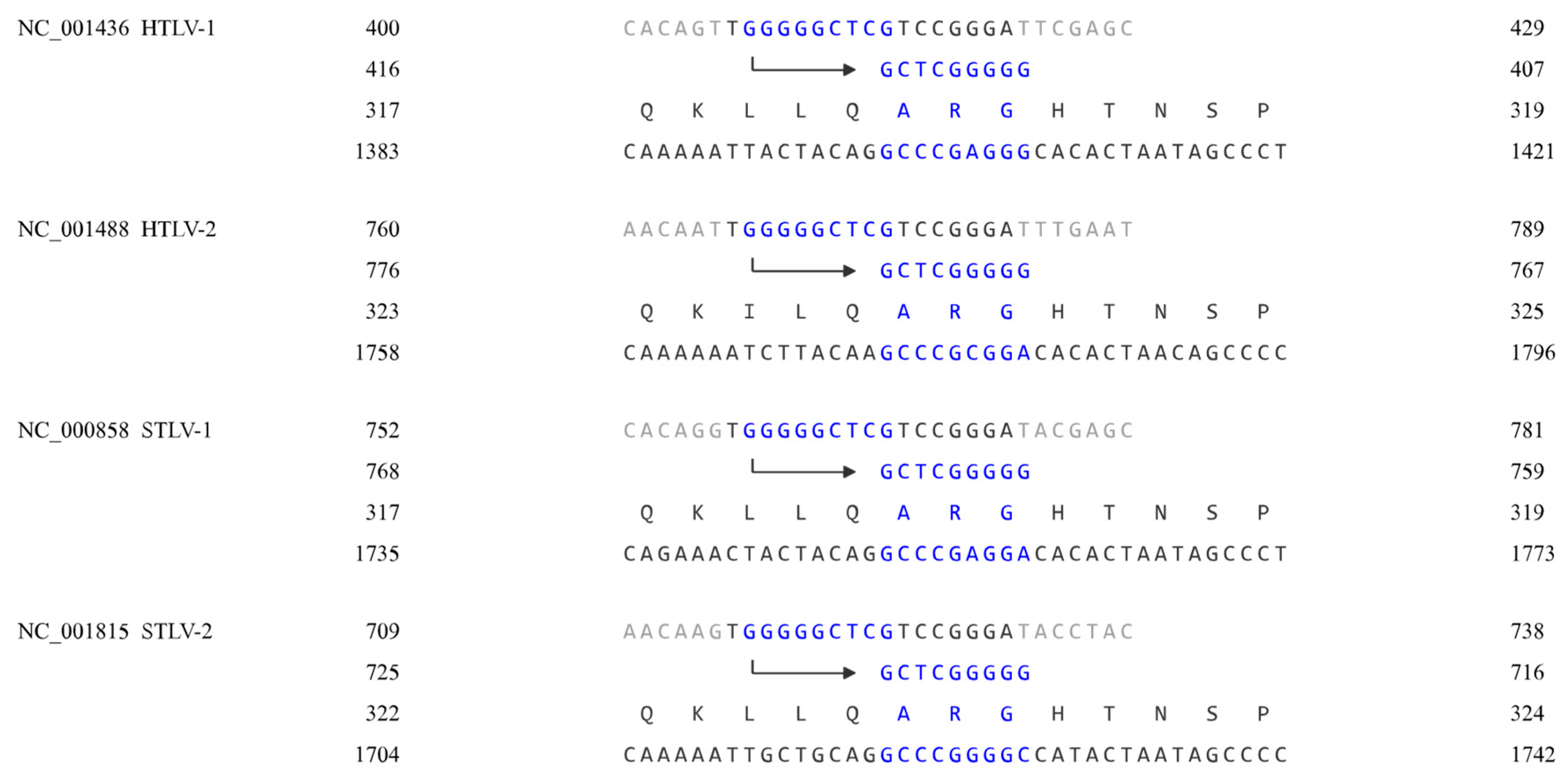
Methods
Results
Discussion
Conclusion
List of abbreviations
| LRAs | Latency-reversing agents |
| CD4 | Cluster of differentiation 4 |
| NF-κB | Nuclear factor kappa light chain enhancer of activated B cells |
| CRISPR | Clustered regularly interspaced short palindromic repeat |
| IAP | Inhibitor of apoptosis protein |
| cIAP1 | Cellular inhibitor of apoptosis protein 1 |
| SIK3 | Salt-inducible kinase 3 |
| HTLV | Human T-lymphotropic virus |
| STLV | Simian T-lymphotropic virus |
| HIV | Human immunodeficiency virus |
| SIV | Simian immunodeficiency virus |
| FIV | Feline immunodeficiency virus |
| LysRS | Lysyl-tRNA synthetase |
| PBS | Primer binding site |
| LTR | Long terminal repeat |
| CTD | C-terminal domain |
| NTD | N-terminal domain |
| Gag | Group-specific antigen |
| CA | Capsid |
Funding
Ethics approval and consent to participate
Consent to publish
Availability of data and materials
Contributions
Acknowledgments
Competing interests
References
- National Institute of Health. What is a Latent HIV Reservoir? NIH. 2021. Available online: https://hivinfo.nih.gov/understanding-hiv/fact-sheets/what-latent-hiv-reservoir.
- National Institute of Health. Latency-Reversing Agents. NIH. 2021. Available online: https://clinicalinfo.hiv.gov/en/glossary/latency-reversing-agents.
- Siliciano, R.F. Drugs fail to reawaken dormant HIV infection. Johns Hopkins Medicine. 2014. Available online: https://hopkinsmedicine.org/news/media/releases/drugs_fail_to_reawaken_dormant_hiv_infection.
- Hiscott, J.; Kwon, H.; Génin, P. Hostile takeovers: viral appropriation of the NF-κB pathway. The Journal of clinical investigation 2001, 107, 143–151. [Google Scholar] [CrossRef]
- Kwon, H.; Pelletier, N.; DeLuca, C.; et al. Inducible expression of IκBα repressor mutants interferes with NF-κB activity and HIV-1 replication in Jurkat T cells. Journal of biological chemistry 1998, 273, 7431–7440. [Google Scholar] [CrossRef] [PubMed]
- Quinto, I.; Mallardo, M.; Baldassarre, F.; et al. Potent and stable attenuation of live-HIV-1 by gain of a proteolysis-resistant inhibitor of NF-κB (IκB-αS32/36A) and the implications for vaccine development. Journal of biological chemistry 1999, 274, 17567–17572. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Verma, I. NF-κB regulation in the immune system. Nature reviews immunology 2002, 2, 725–734. [Google Scholar] [CrossRef]
- Makropoulos, V.; Brüning, T.; Schulze-Osthoff, K. Selenium-mediated inhibition of transcription factor NF-κ B and HIV-1 LTR promoter activity. Archives of toxicology 1996, 70, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Ait-Ammar, A.; Kula, A.; Darcis, G.; et al. Current status of latency reversing agents facing the heterogeneity of HIV-1 cellular and tissue reservoirs. Frontiers in microbiology 2020, 10, 3060. [Google Scholar] [CrossRef] [PubMed]
- Archin, N.M.; Liberty, A.L.; Kashuba, A.D.;Choudhary; et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 2012, 487, 482–485. [Google Scholar] [CrossRef]
- Spivak, A.M.; Andrade, A.; Eisele, E.; et al. A pilot study assessing the safety and latency-reversing activity of disulfiram in HIV-1–infected adults on antiretroviral therapy. Clinical infectious diseases 2014, 58, 883–890. [Google Scholar] [CrossRef]
- Elliott, J.H.; McMahon, J.H.; Chang, C.C.; et al. Short-term administration of disulfiram for reversal of latent HIV infection: a phase 2 dose-escalation study. Lancet HIV 2015, 2, e520–e529. [Google Scholar] [CrossRef]
- Norton, N.J.; Mok, H.P.; Sharif, F.; et al. HIV silencing and inducibility are heterogeneous and are affected by factors intrinsic to the virus. Mbio 2019, 10, 10–1128. [Google Scholar] [CrossRef]
- Spina, C.A.; Anderson, J.; Archin, N.M.; et al. An in-depth comparison of latent HIV-1 reactivation in multiple cell model systems and resting CD4+ T cells from aviremic patients. PLoS pathogens 2013, 9, e1003834. [Google Scholar] [CrossRef] [PubMed]
- Baxter, A.E.; Niessl, J.; Fromentin, R.; et al. Single-cell characterization of viral translation-competent reservoirs in HIV-infected individuals. Cell host & microbe 2016, 20, 368–380. [Google Scholar] [CrossRef]
- Grau-Expósito, J.; Luque-Ballesteros, L.; Navarro, J.; et al. Latency reversal agents affect differently the latent reservoir present in distinct CD4+ T subpopulations. PLoS pathogens 2019, 15, e1007991. [Google Scholar] [CrossRef]
- Kula, A.; Delacourt, N.; Bouchat, S.; et al. Heterogeneous HIV-1 reactivation patterns of disulfiram and combined disulfiram+ romidepsin treatments. JAIDS Journal of Acquired Immune Deficiency Syndromes 2019, 80, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Elliott, J.H.; Wightman, F.; Solomon, A.; et al. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS pathogens 2014, 10, e1004473. [Google Scholar] [CrossRef] [PubMed]
- Yukl, S.A.; Kaiser, P.; Kim, P.; et al. HIV latency in isolated patient CD4+ T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Science translational medicine 2018, 10, eaap9927. [Google Scholar] [CrossRef]
- Telwatte, S.; Lee, S.; Somsouk, M.A.; et al. Gut and blood differ in constitutive blocks to HIV transcription, suggesting tissue-specific differences in the mechanisms that govern HIV latency. PLoS pathogens 2018, 14, e1007357. [Google Scholar] [CrossRef]
- Chen, H.C.; Martinez, J.P.; Zorita, E.; et al. Position effects influence HIV latency reversal. Nature structural & molecular biology 2017, 24, 47–54. [Google Scholar] [CrossRef]
- Abner, E.; Stoszko, M.; Zeng, L.; et al. A new quinoline BRD4 inhibitor targets a distinct latent HIV-1 reservoir for reactivation from other “shock” drugs. Journal of Virology 2018, 92, 10–1128. [Google Scholar] [CrossRef]
- Battivelli, E.; Dahabieh, M.S.; Abdel-Mohsen, M.; et al. Distinct chromatin functional states correlate with HIV latency reactivation in infected primary CD4+ T cells. Elife 2018, 7, e34655. [Google Scholar] [CrossRef]
- Darcis, G.; Bouchat, S.; Kula, A.; et al. Reactivation capacity by latency-reversing agents ex vivo correlates with the size of the HIV-1 reservoir. Aids 2017, 31, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Das, B.; Dobrowolski, C.; Luttge, B.; et al. Estrogen receptor-1 is a key regulator of HIV-1 latency that imparts gender-specific restrictions on the latent reservoir. Proceedings of the National Academy of Sciences 2018, 115, E7795–E7804. [Google Scholar] [CrossRef] [PubMed]
- Garrido, C.; Spivak, A.M.; Soriano-Sarabia, N.; et al. HIV latency-reversing agents have diverse effects on natural killer cell function. Frontiers in immunology 2016, 7, 356. [Google Scholar] [CrossRef] [PubMed]
- Walker-Sperling, V.E.; Pohlmeyer, C.W.; Tarwater, P.M.; et al. The effect of latency reversal agents on primary CD8+ T cells: implications for shock and kill strategies for human immunodeficiency virus eradication. EBioMedicine 2016, 8, 217–229. [Google Scholar] [CrossRef]
- Jones, R.B.; O'Connor, R.; Mueller, S.; et al. Histone deacetylase inhibitors impair the elimination of HIV-infected cells by cytotoxic T-lymphocytes. PLoS pathogens 2014, 10, e1004287. [Google Scholar] [CrossRef]
- Clutton, G.; Xu, Y.; Baldoni, P.L.; et al. The differential short-and long-term effects of HIV-1 latency-reversing agents on T cell function. Scientific reports 2016, 6, 30749. [Google Scholar] [CrossRef]
- Desimio, M.G.; Giuliani, E.; Ferraro, A.S.; et al. In vitro exposure to prostratin but not bryostatin-1 improves natural killer cell functions including killing of CD4+ T cells harboring reactivated human immunodeficiency virus. Frontiers in immunology 2018, 9, 1514. [Google Scholar] [CrossRef]
- National Institute of Health. NIH-supported scientists reverse HIV and SIV latency in two animal models. NIH. 2020. Available online: https://www.nih.gov/news-events/news-releases/nih-supported-scientists-reverse-hiv-siv-latency-two-animal-models.
- Nixon, C.C.; Mavigner, M.; Sampey, G.C.; et al. Systemic HIV and SIV latency reversal via non-canonical NF-κB signalling in vivo. Nature 2020, 578, 160–165. [Google Scholar] [CrossRef]
- McBrien, J.B.; Mavigner, M.; Franchitti, L.; et al. Robust and persistent reactivation of SIV and HIV by N-803 and depletion of CD8+ cells. Nature 2020, 578, 154–159. [Google Scholar] [CrossRef]
- Hennessy, E.J.; Adam, A.; Aquila, B.M.; et al. Discovery of a novel class of dimeric Smac mimetics as potent IAP antagonists resulting in a clinical candidate for the treatment of cancer (AZD5582). Journal of medicinal chemistry 2013, 56, 9897–9919. [Google Scholar] [CrossRef]
- Pache, L.; Marsden, M.D.; Teriete, P.; et al. Pharmacological activation of non-canonical NF-κB signaling activates latent HIV-1 reservoirs in vivo. Cell Reports Medicine 2020, 1. [Google Scholar] [CrossRef] [PubMed]
- Bullen, C.K.; Laird, G.M.; Durand, C.M.; et al. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nature medicine 2014, 20, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Spivak, A.M.; Andrade, A.; Eisele, E.; et al. A pilot study assessing the safety and latency-reversing activity of disulfiram in HIV-1–infected adults on antiretroviral therapy. Clinical infectious diseases 2014, 58, 883–890. [Google Scholar] [CrossRef]
- Darcis, G.; Kula, A.; Bouchat, S.; et al. An in-depth comparison of latency-reversing agent combinations in various in vitro and ex vivo HIV-1 latency models identified bryostatin-1+ JQ1 and ingenol-B+ JQ1 to potently reactivate viral gene expression. PLoS pathogens 2015, 11, e1005063. [Google Scholar] [CrossRef]
- Laird, G.M.; Bullen, C.K.; Rosenbloom, D.I.S.; et al. Ex vivo analysis identifies effective HIV-1 latency–reversing drug combinations. The Journal of clinical investigation 2015, 125, 1901–1912. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Haffner, M.C.; Zhang, Y.; et al. Disulfiram is a DNA demethylating agent and inhibits prostate cancer cell growth. The Prostate 2011, 71, 333–343. [Google Scholar] [CrossRef]
- Philip, P.A.; Rea, D.; Thavasu, P.; et al. Phase I study of bryostatin 1: assessment of interleukin 6 and tumor necrosis factor α induction in vivo. JNCI: Journal of the National Cancer Institute 1993, 85, 1812–1818. [Google Scholar] [CrossRef]
- Silva, V.A.O.; Rosa, M.N.; Tansini, A.; et al. Cytotoxic activity of semi-synthetic ingenol derived from Euphorbia tirucalli on a large panel of human cancer cell lines. 2013. [Google Scholar] [CrossRef]
- Alotaibi, D.; Amara, S.; Johnson, T.L.; et al. Potential anticancer effect of prostratin through SIK3 inhibition. Oncology letters 2018, 15, 3252–3258. [Google Scholar] [CrossRef]
- Duchon, A.A.; St. Gelais, C.; Titkemeier, N.; et al. HIV-1 exploits a dynamic multi-aminoacyl-tRNA synthetase complex to enhance viral replication. Journal of virology 2017, 91, 10–1128. [Google Scholar] [CrossRef]
- Weiner, A.M.; Maizels, N. tRNA-like structures tag the 3'ends of genomic RNA molecules for replication: implications for the origin of protein synthesis. Proceedings of the National Academy of Sciences 1987, 84, 7383–7387. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.S.; Temin, H.M. Retroviral recombination and reverse transcription. Science 1990, 250, 1227–1233. [Google Scholar] [CrossRef] [PubMed]
- Negroni, M.; Buc, H. Copy-choice recombination by reverse transcriptases: reshuffling of genetic markers mediated by RNA chaperones. Proceedings of the National Academy of Sciences 2000, 97, 6385–6390. [Google Scholar] [CrossRef] [PubMed]
- Petropoulos, C. Retroviral taxonomy, protein structures, sequences, and genetic maps. In Retroviruses; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, 1997; pp. 757–805. [Google Scholar]
- Shimotohno, K.; Takahashi, Y.; Shimizu, N.; et al. Complete nucleotide sequence of an infectious clone of human T-cell leukemia virus type II: an open reading frame for the protease gene. Proceedings of the National Academy of Sciences 1985, 82, 3101–3105. [Google Scholar] [CrossRef]
- Saksena, N.K.; Hervé, V.; Sherman, M.P.; et al. Sequence and phylogenetic analyses of a new STLV-I from a naturally infected tantalus monkey from Central Africa. Virology 1993, 192, 312–320. [Google Scholar] [CrossRef]
- Van Brussel, M.; Salemi, M.; Liu, H.F.; et al. The Simian T-Lymphotropic Virus STLV-PP1664 fromPan paniscusIs Distinctly Related to HTLV-2 but Differs in Genomic Organization. Virology 1998, 243, 366–379. [Google Scholar] [CrossRef] [PubMed]
- Martoglio, B.; Graf, R.; Dobberstein, B. Signal peptide fragments of preprolactin and HIV-1 p-gp160 interact with calmodulin. The EMBO journal 1997, 16, 6636–6645. [Google Scholar] [CrossRef]
- Kirchhoff, F.; Jentsch, K.D.; Bachmann, B.; et al. A novel proviral clone of HIV-2: biological and phylogenetic relationship to other primate immunodeficiency viruses. Virology 1990, 177, 305–311. [Google Scholar] [CrossRef]
- Fomsgaard, A.; Hirsch, V.M.; Allan, J.S.; et al. A highly divergent proviral DNA clone of SIV from a distinct species of African green monkey. Virology 1991, 182, 397–402. [Google Scholar] [CrossRef]
- Olmsted, R.A.; Hirsch, V.M.; Purcell, R.H.; et al. Nucleotide sequence analysis of feline immunodeficiency virus: genome organization and relationship to other lentiviruses. Proceedings of the National Academy of Sciences 1989, 86, 8088–8092. [Google Scholar] [CrossRef]
- Jin, D.; Musier-Forsyth, K. Role of host tRNAs and aminoacyl-tRNA synthetases in retroviral replication. Journal of Biological Chemistry 2019, 294, 5352–5364. [Google Scholar] [CrossRef]
- Mak, J.; Jiang, M.; Wainberg, M.A.; et al. Role of Pr160gag-pol in mediating the selective incorporation of tRNA (Lys) into human immunodeficiency virus type 1 particles. Journal of virology 1994, 68, 2065–2072. [Google Scholar] [CrossRef] [PubMed]
- Kovaleski, B.J.; Kennedy, R.; Hong, M.K.; et al. In vitro characterization of the interaction between HIV-1 Gag and human lysyl-tRNA synthetase. Journal of Biological Chemistry 2006, 281, 19449–19456. [Google Scholar] [CrossRef] [PubMed]
- Bell, N.M.; Lever, A.M.L. HIV Gag polyprotein: processing and early viral particle assembly. Trends in microbiology 2013, 21, 136–144. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




