Submitted:
27 June 2023
Posted:
28 June 2023
Read the latest preprint version here
Abstract
Keywords:
1. Introduction
2. Methods


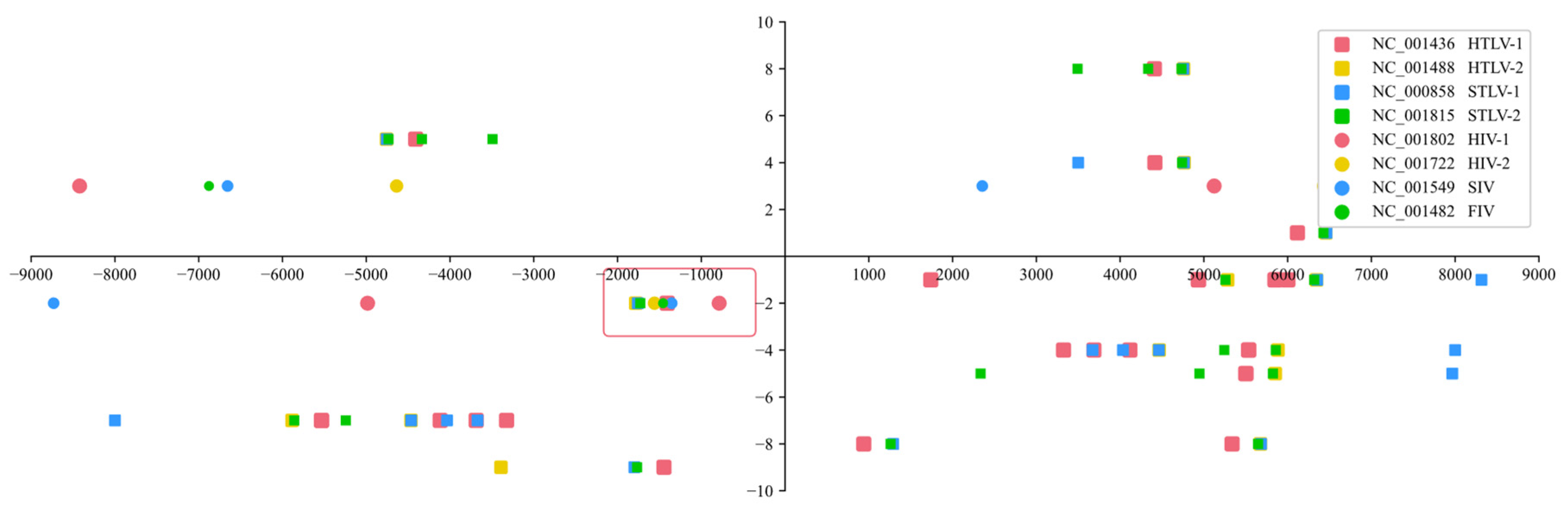
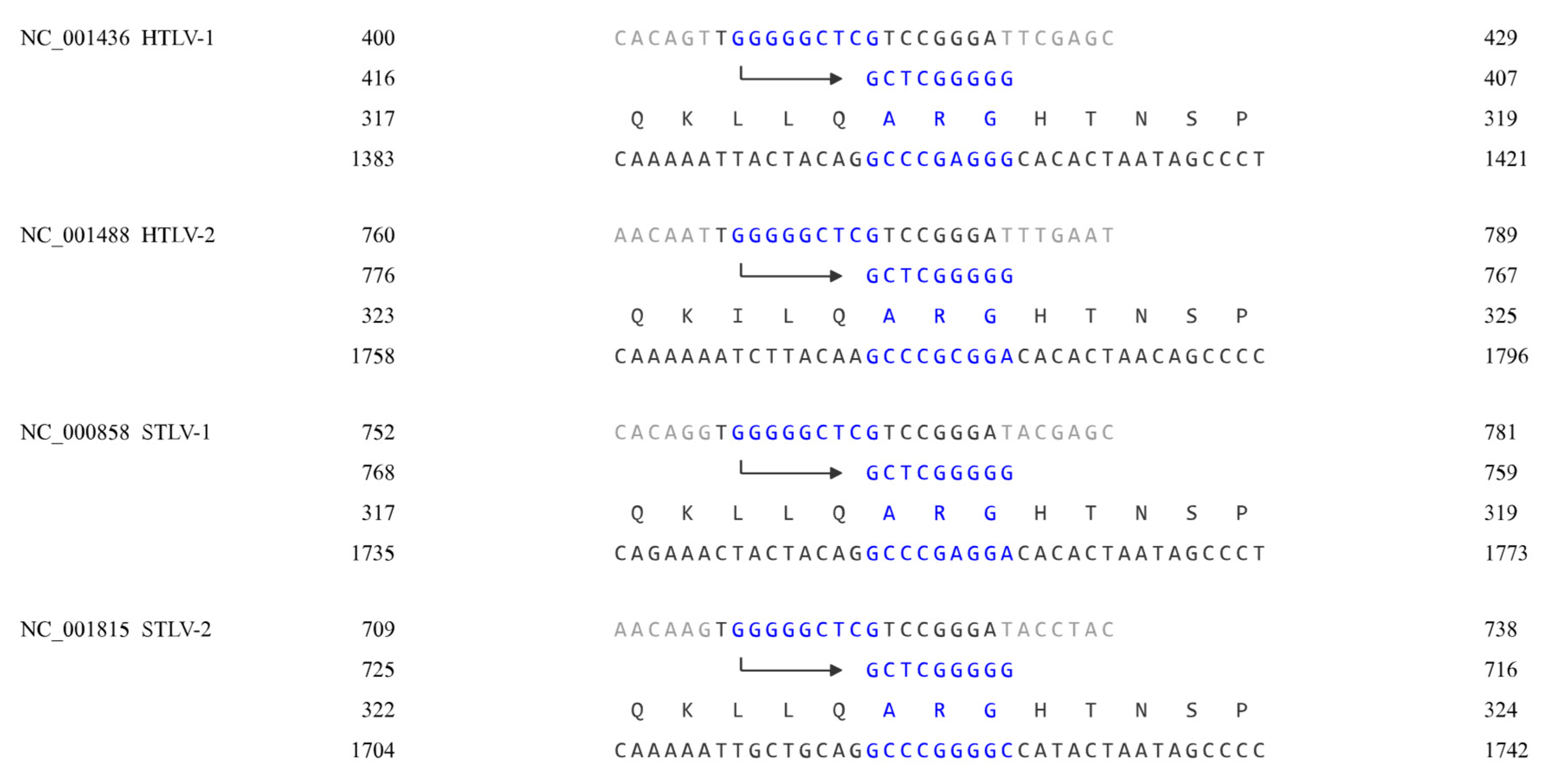
3. Results
4. Discussion
5. Conclusions
Abbreviations
| NF-κB | Nuclear factor kappa light chain enhancer of activated B cells |
| IGF1R | Insulin-like growth factor 1 receptor |
| CRISPRs | Clustered regularly interspaced short palindromic repeats |
| IAP | Inhibitor of apoptosis protein |
| cIAP1 | Cellular inhibitor of apoptosis protein 1 |
| SIK3 | Salt-inducible kinase 3 |
| HTLV | Human T-lymphotropic virus |
| STLV | Simian T-lymphotropic virus |
| HIV | Human immunodeficiency virus |
| SIV | Simian immunodeficiency virus |
| FIV | Feline immunodeficiency virus |
| LysRS | Lysyl-tRNA synthetase |
| PBS | primer binding site |
| CTD | C-terminal domain |
| NTD | N-terminal domain |
| Gag | Group-specific antigen |
| CA | Capsid |
References
- National Institute of Health. What is a Latent HIV Reservoir? NIH. 2021 August 4. https://hivinfo.nih.gov/understanding-hiv/fact-sheets/what-latent-hiv-reservoir.
- Siliciano RF. Drugs fail to reawaken dormant HIV infection. Johns Hopkins Medicine. 2014 March 24. https://hopkinsmedicine.org/news/media/releases/drugs_fail_to_reawaken_dormant_hiv_infection.
- Hiscott J, Kwon H, Génin P. Hostile takeovers: viral appropriation of the NF-kB pathway. The Journal of clinical investigation. 2001 Jan 15;107(2):143-51. [CrossRef]
- Seibert SA, Howell CY, Hughes MK, Hughes AL. Natural selection on the gag, pol, and env genes of human immunodeficiency virus 1 (HIV-1). Molecular biology and evolution. 1995 Sep 1;12(5):803-13. [CrossRef]
- Abbott AM, Bueno R, Pedrini MT, Murray JM, Smith RJ. Insulin-like growth factor I receptor gene structure. Journal of Biological Chemistry. 1992 ;267(15):10759-63. 25 May. [CrossRef]
- Pandini G, Mineo R, Frasca F, Roberts Jr CT, Marcelli M, Vigneri R, Belfiore A. Androgens up-regulate the insulin-like growth factor-I receptor in prostate cancer cells. Cancer research. 2005 Mar 1;65(5):1849-57. [CrossRef]
- Zhang F, Wen Y, Guo X. CRISPR/Cas9 for genome editing: progress, implications and challenges. Human molecular genetics. 2014 Sep 15;23(R1):R40-6. [CrossRef]
- Bak RO, Gomez-Ospina N, Porteus MH. Gene editing on center stage. Trends in Genetics. 2018 Aug 1;34(8):600-11. [CrossRef]
- Aleksic T, Gray N, Wu X, Rieunier G, Osher E, Mills J, Verrill C, Bryant RJ, Han C, Hutchinson K, Lambert AG. Nuclear IGF1R interacts with regulatory regions of chromatin to promote RNA polymerase II recruitment and gene expression associated with advanced tumor stage. Cancer research. 2018 Jul 1;78(13):3497-509. [CrossRef]
- Ott M, Geyer M, Zhou Q. The control of HIV transcription: keeping RNA polymerase II on track. Cell host & microbe. 2011 Nov 17;10(5):426-35. [CrossRef]
- Van Rossum G. An introduction to Python for UNIX/C programmers. Proceedings of the NLUUG najaarsconferentie (Dutch UNIX users group). 1993 Nov.
- Kwon H, Pelletier N, DeLuca C, Genin P, Cisternas S, Lin R, Wainberg MA, Hiscott J. Inducible expression of IκBα repressor mutants interferes with NF-κB activity and HIV-1 replication in Jurkat T cells. Journal of biological chemistry. 1998 Mar 27;273(13):7431-40. [CrossRef]
- Quinto I, Mallardo M, Baldassarre F, Scala G, Englund G, Jeang KT. Potent and stable attenuation of live-HIV-1 by gain of a proteolysis-resistant inhibitor of NF-κB (IκB-αS32/36A) and the implications for vaccine development. Journal of biological chemistry. 1999 Jun 18;274(25):17567-72. [CrossRef]
- National Institute of Health. NIH-supported scientists reverse HIV and SIV latency in two animal models. NIH. 2020 January 22. https://www.nih.gov/news-events/news-releases/nih-supported-scientists-reverse-hiv-siv-latency-two-animal-models.
- Nixon CC, Mavigner M, Sampey GC, Brooks AD, Spagnuolo RA, Irlbeck DM, Mattingly C, Ho PT, Schoof N, Cammon CG, Tharp GK. Systemic HIV and SIV latency reversal via non-canonical NF-κB signalling in vivo. Nature. 2020 Feb;578(7793):160-5. [CrossRef]
- McBrien JB, Mavigner M, Franchitti L, Smith SA, White E, Tharp GK, Walum H, Busman-Sahay K, Aguilera-Sandoval CR, Thayer WO, Spagnuolo RA. Robust and persistent reactivation of SIV and HIV by N-803 and depletion of CD8+ cells. Nature. 2020 Feb;578(7793):154-9. [CrossRef]
- Hennessy EJ, Adam A, Aquila BM, Castriotta LM, Cook D, Hattersley M, Hird AW, Huntington C, Kamhi VM, Laing NM, Li D. Discovery of a novel class of dimeric Smac mimetics as potent IAP antagonists resulting in a clinical candidate for the treatment of cancer (AZD5582). Journal of medicinal chemistry. 2013 Dec 27;56(24):9897-919. [CrossRef]
- Cuevas JM, Geller R, Garijo R, López-Aldeguer J, Sanjuán R. Extremely high mutation rate of HIV-1 in vivo. PLoS biology. 2015 Sep 16;13(9):e1002251. [CrossRef]
- Pache L, Marsden MD, Teriete P, Portillo AJ, Heimann D, Kim JT, Soliman MS, Dimapasoc M, Carmona C, Celeridad M, Spivak AM. Pharmacological activation of non-canonical NF-κB signaling activates latent HIV-1 reservoirs in vivo. Cell Reports Medicine. 2020 Jun 23;1(3):100037. [CrossRef]
- Bullen CK, Laird GM, Durand CM, Siliciano JD, Siliciano RF. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nature medicine. 2014 Apr;20(4):425-9. [CrossRef]
- Spivak AM, Andrade A, Eisele E, Hoh R, Bacchetti P, Bumpus NN, Emad F, Buckheit III R, McCance-Katz EF, Lai J, Kennedy M. A pilot study assessing the safety and latency-reversing activity of disulfiram in HIV-1–infected adults on antiretroviral therapy. Clinical infectious diseases. 2014 Mar 15;58(6):883-90. [CrossRef]
- Darcis G, Kula A, Bouchat S, Fujinaga K, Corazza F, Ait-Ammar A, Delacourt N, Melard A, Kabeya K, Vanhulle C, Van Driessche B. An in-depth comparison of latency-reversing agent combinations in various in vitro and ex vivo HIV-1 latency models identified bryostatin-1+ JQ1 and ingenol-B+ JQ1 to potently reactivate viral gene expression. PLoS pathogens. 2015 Jul 30;11(7):e1005063. [CrossRef]
- Laird GM, Bullen CK, Rosenbloom DI, Martin AR, Hill AL, Durand CM, Siliciano JD, Siliciano RF. Ex vivo analysis identifies effective HIV-1 latency–reversing drug combinations. The Journal of clinical investigation. 2015 ;125(5):1901-12. 1 May. [CrossRef]
- Lin J, Haffner MC, Zhang Y, Lee BH, Brennen WN, Britton J, Kachhap SK, Shim JS, Liu JO, Nelson WG, Yegnasubramanian S. Disulfiram is a DNA demethylating agent and inhibits prostate cancer cell growth. The Prostate. 2011 Mar 1;71(4):333-43. [CrossRef]
- Philip PA, Rea D, Thavasu P, Carmichael J, Stuart NS, Rockett H, Talbot DC, Ganesan T, Pettit GR, Balkwill F, Harris AL. Phase I study of bryostatin 1: assessment of interleukin 6 and tumor necrosis factor α induction in vivo. JNCI: Journal of the National Cancer Institute. 1993 Nov 17;85(22):1812-8. [CrossRef]
- Silva VA, Rosa MN, Tansini A, Lima JP, Jones C, Pianowski LF, Reis RM. Cytotoxic activity of semi-synthetic ingenol derived from Euphorbia tirucalli on a large panel of human cancer cell lines. 2013; e13559-e13559. [CrossRef]
- Alotaibi D, Amara S, Johnson TL, Tiriveedhi V. Potential anticancer effect of prostratin through SIK3 inhibition. Oncology letters. 2018 Mar 1;15(3):3252-8. [CrossRef]
- Duchon AA, St. Gelais C, Titkemeier N, Hatterschide J, Wu L, Musier-Forsyth K. HIV-1 exploits a dynamic multi-aminoacyl-tRNA synthetase complex to enhance viral replication. Journal of virology. 2017 Oct 13;91(21):e01240-17. [CrossRef]
- Weiner AM, Maizels N. tRNA-like structures tag the 3'ends of genomic RNA molecules for replication: implications for the origin of protein synthesis. Proceedings of the National Academy of Sciences. 1987 Nov;84(21):7383-7. [CrossRef]
- Hu WS, Temin HM. Retroviral recombination and reverse transcription. Science. 1990 Nov 30;250(4985):1227-33. [CrossRef]
- Negroni M, Buc H. Copy-choice recombination by reverse transcriptases: reshuffling of genetic markers mediated by RNA chaperones. Proceedings of the National Academy of Sciences. 2000 Jun 6;97(12):6385-90. [CrossRef]
- Petropoulos, C. Retroviral taxonomy, protein structures, sequences, and genetic maps. Retroviruses; Cold Spring Harbor Laboratory Press, Cold Spring Harbor: NY, 1997; pp. 757–805. [Google Scholar]
- Shimotohno K, Takahashi Y, Shimizu N, Gojobori T, Golde DW, Chen IS, Miwa M, Sugimura T. Complete nucleotide sequence of an infectious clone of human T-cell leukemia virus type II: an open reading frame for the protease gene. Proceedings of the National Academy of Sciences. 1985 May;82(10):3101-5. [CrossRef]
- Saksena NK, Hervé V, Sherman MP, Durand JP, Mathiot C, Müller M, Love JL, Leguenno B, Sinoussi FB, Dube DK, Poiesz BJ. Sequence and phylogenetic analyses of a new STLV-I from a naturally infected tantalus monkey from Central Africa. Virology. 1993 Jan 1;192(1):312-20. [CrossRef]
- Van Brussel M, Salemi M, Liu HF, Gabriëls J, Goubau P, Desmyter J, Vandamme AM. The Simian T-Lymphotropic Virus STLV-PP1664 fromPan paniscusIs Distinctly Related to HTLV-2 but Differs in Genomic Organization. Virology. 1998 Apr 10;243(2):366-79. [CrossRef]
- Martoglio B, Graf R, Dobberstein B. Signal peptide fragments of preprolactin and HIV-1 p-gp160 interact with calmodulin. The EMBO journal. 1997 Nov 15;16(22):6636-45. [CrossRef]
- Kirchhoff F, Jentsch KD, Bachmann B, Stuke A, Laloux C, Luke W, Stahl-Hennig C, Schneider J, Nieselt K, Eigen M, Hunsmann G. A novel proviral clone of HIV-2: biological and phylogenetic relationship to other primate immunodeficiency viruses. Virology. 1990 Jul 1;177(1):305-11. [CrossRef]
- Fomsgaard A, Hirsch VM, Allan JS, Johnson PR. A highly divergent proviral DNA clone of SIV from a distinct species of African green monkey. Virology. 1991 ;182(1):397-402. 1 May. [CrossRef]
- Olmsted RA, Hirsch VM, Purcell RH, Johnson PR. Nucleotide sequence analysis of feline immunodeficiency virus: genome organization and relationship to other lentiviruses. Proceedings of the National Academy of Sciences. 1989 Oct;86(20):8088-92. [CrossRef]
- Jin D, Musier-Forsyth K. Role of host tRNAs and aminoacyl-tRNA synthetases in retroviral replication. Journal of Biological Chemistry. 2019 Apr 5;294(14):5352-64. [CrossRef]
- Mak J, Jiang M, Wainberg MA, Hammarskjöld ML, Rekosh D, Kleiman L. Role of Pr160gag-pol in mediating the selective incorporation of tRNA (Lys) into human immunodeficiency virus type 1 particles. Journal of virology, 1994, 68(4): 2065-2072. [CrossRef]
- Kovaleski BJ, Kennedy R, Hong MK, Datta SA, Kleiman L, Rein A, Musier-Forsyth K. In vitro characterization of the interaction between HIV-1 Gag and human lysyl-tRNA synthetase. Journal of Biological Chemistry, 2006, 281(28): 19449-19456. [CrossRef]
- Rhim H, Park J, Morrow CD. Deletions in the tRNA(Lys) primer-binding site of human immunodeficiency virus type 1 identify essential regions for reverse transcription. Journal of virology 65.9 (1991): 4555-4564. [CrossRef]
- Bell NM, Lever AM. HIV Gag polyprotein: processing and early viral particle assembly. Trends in microbiology 21.3 (2013): 136-144. [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




