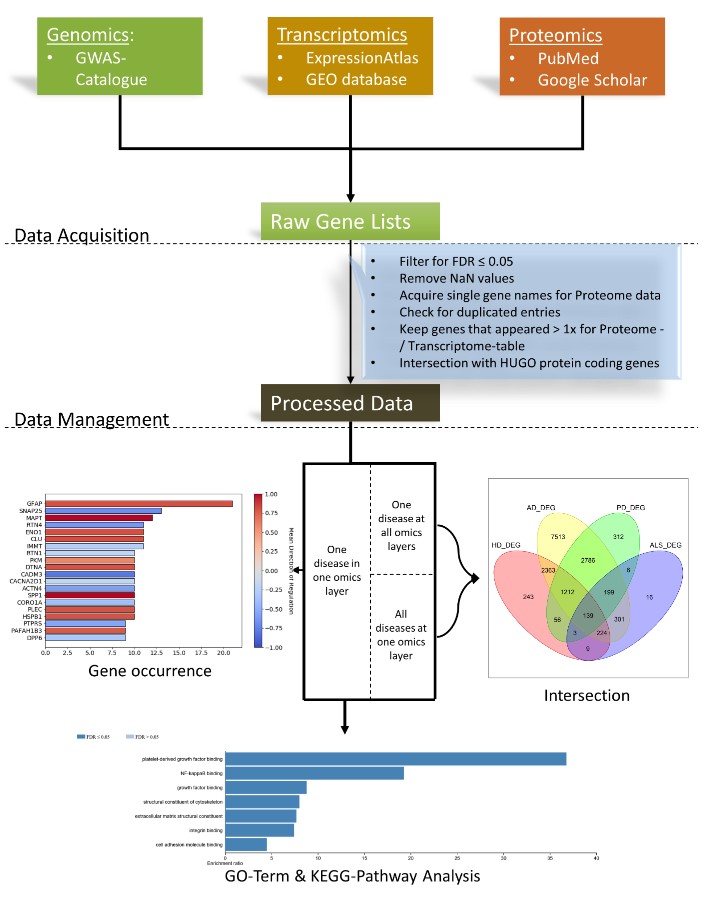
Neurodegenerative diseases such as Alzheimer's disease, Parkinson's disease, Huntington's disease, and Amyotrophic Lateral Sclerosis are heterogeneous, progressive diseases with frequently overlapping symptoms characterized by a loss of neurons. Studies suggested relations between neurodegenerative diseases for many years, e.g., regarding the aggregation of toxic proteins or triggering endogenous cell death pathways. Within this study, publicly available genomic, transcriptomic and proteomic data were gathered from 188 studies and more than one million patients to detect shared genetic patterns between the neurodegenerative diseases and the analyzed omics-layers within conditions. The results show a remarkably high number of shared genes between the transcriptomic and proteomic levels for all diseases while showing a significant relation between genomic and proteomic data only in some cases. A set of 139 genes was found to be differentially expressed in several transcriptomic experiments of all four diseases. These 139 genes showed overrepresented GO-Terms and pathways mainly involved in stress response, cell development, cell adhesion, and the cytoskeleton. Furthermore, the overlap of two and three omics-layers per disease were used to search for overrepresented pathways and GO-Terms. Taken together, we could confirm the existence of many relations between Alzheimer's disease, Parkinson's disease, Huntington's disease, and Amyotrophic Lateral Sclerosis on the transcriptomic and proteomic level by analyzing the pathways and GO-Terms arising in these intersections. The significance of the connection between the transcriptomic and proteomic data for all four analyzed neurodegenerative diseases showed that exploring these omics-layers simultaneously holds new insights that do not emerge from analyzing these omics-layers separately. Our data therefore suggests addressing human patients with neurodegenerative diseases as complex biological systems by integrating multiple underlying data sources.