Submitted:
09 January 2026
Posted:
12 January 2026
You are already at the latest version
Abstract
Keywords:
1. Introduction
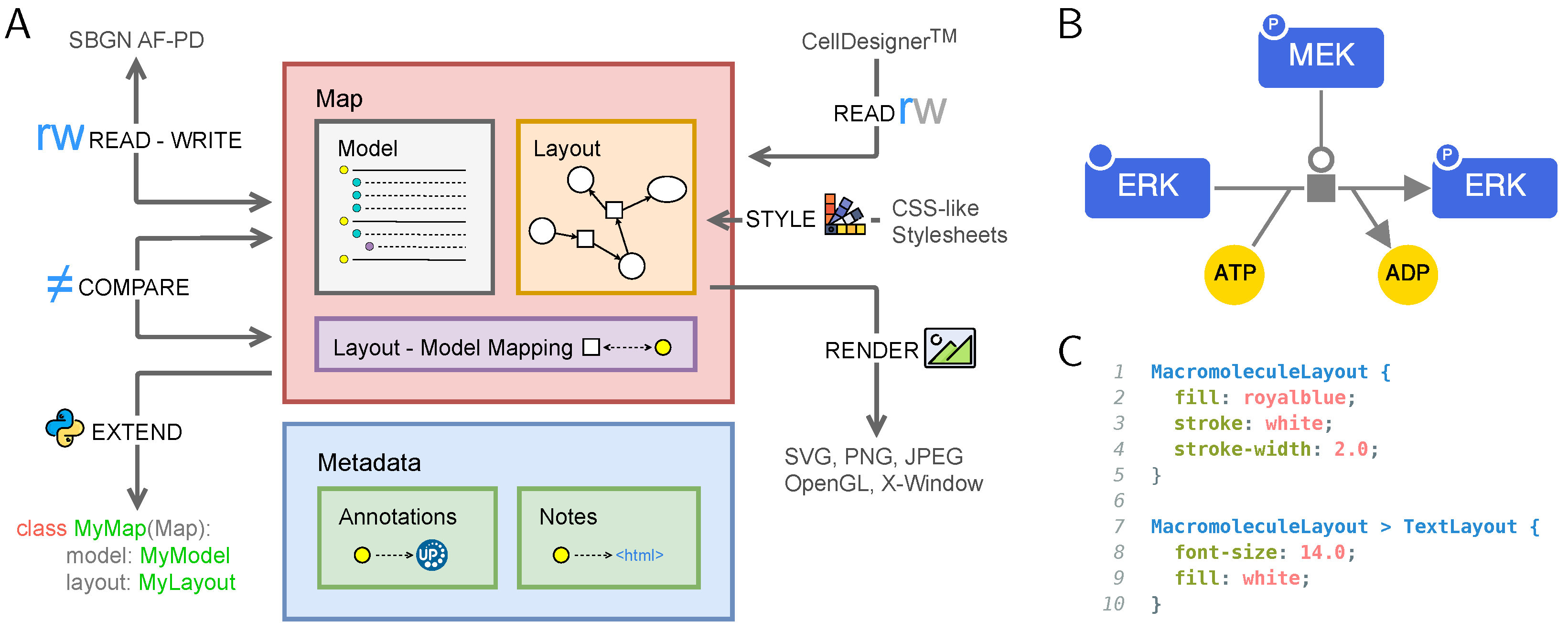
2. A momapy Map: A Model, a Layout, and a Layout-Model Mapping
3. momapy’s Features
3.1. Supported Map Formats
3.2. Exploring and Manipulating Maps
3.3. Comparing Maps
3.4. Styling and Style Sheets
3.5. Rendering of Maps
3.6. Extending momapy with New Formats and Functionalities
4. Software Implementation and Availability
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| SBGN | Systems Biology Graphical Notation |
| SBML | Systems Biology Markup Language |
| BEL | Biological Expression Language |
References
- Ostaszewski, M.; Gebel, S.; Kuperstein, I.; Mazein, A.; Zinovyev, A.; Dogrusoz, U.; Hasenauer, J.; Fleming, R.M.T.; Le Novère, N.; Gawron, P.; et al. Community-driven roadmap for integrated disease maps. Briefings in Bioinformatics 2019, 20, 659–670. [CrossRef]
- Oda, K.; Matsuoka, Y.; Funahashi, A.; Kitano, H. A comprehensive pathway map of epidermal growth factor receptor signaling. Molecular Systems Biology 2005, 1, 2005.0010. Publisher: John Wiley & Sons, Ltd. [CrossRef]
- Calzone, L.; Gelay, A.; Zinovyev, A.; Radvanyi, F.; Barillot, E. A comprehensive modular map of molecular interactions in RB/E2F pathway. Molecular Systems Biology 2008, 4. Publisher: EMBO Press.
- Niarakis, A.; Ostaszewski, M.; Mazein, A.; Kuperstein, I.; Kutmon, M.; Gillespie, M.E.; Funahashi, A.; Acencio, M.L.; Hemedan, A.; Aichem, M.; et al. Drug-target identification in COVID-19 disease mechanisms using computational systems biology approaches. Frontiers in Immunology 2024, 14. Publisher: Frontiers. [CrossRef]
- Funahashi, A.; Matsuoka, Y.; Jouraku, A.; Morohashi, M.; Kikuchi, N.; Kitano, H. CellDesigner 3.5: a versatile modeling tool for biochemical networks. Proceedings of the IEEE 2008, 96, 1254–1265. Publisher: IEEE.
- Czauderna, T.; Klukas, C.; Schreiber, F. Editing, validating and translating of SBGN maps. Bioinformatics 2010, 26, 2340–2341. [CrossRef]
- Balci, H.; Siper, M.C.; Saleh, N.; Safarli, I.; Roy, L.; Kilicarslan, M.; Ozaydin, R.; Mazein, A.; Auffray, C.; Babur, Ã.; et al. Newt: a comprehensive web-based tool for viewing, constructing and analyzing biological maps. Bioinformatics 2021, 37, 1475–1477. [CrossRef]
- Gawron, P.; Ostaszewski, M.; Satagopam, V.; Gebel, S.; Mazein, A.; Kuzma, M.; Zorzan, S.; McGee, F.; Otjacques, B.; Balling, R.; et al. MINERVA: a platform for visualization and curation of molecular interaction networks. npj Systems Biology and Applications 2016, 2, 16020. Publisher: Nature Publishing Group. [CrossRef]
- Xu, J.; Jiang, J.; Sauro, H.M. SBMLDiagrams: a python package to process and visualize SBML layout and render. Bioinformatics 2023, 39, btac730. [CrossRef]
- van Iersel, M.P.; Villéger, A.C.; Czauderna, T.; Boyd, S.E.; Bergmann, F.T.; Luna, A.; Demir, E.; Sorokin, A.; Dogrusoz, U.; Matsuoka, Y.; et al. Software support for SBGN maps: SBGN-ML and LibSBGN. Bioinformatics 2012, 28, 2016–2021. [CrossRef]
- Balaur, I.; Roy, L.; Mazein, A.; Karaca, S.G.; Dogrusoz, U.; Barillot, E.; Zinovyev, A. cd2sbgnml: bidirectional conversion between CellDesigner and SBGN formats. Bioinformatics 2020, 36, 2620–2622. Publisher: Oxford University Press.
- Rougny, A.; Balaur, I.; Luna, A.; Mazein, A. StonPy: a tool to parse and query collections of SBGN maps in a graph database. Bioinformatics 2023, 39, btad100. [CrossRef]
- Rougny, A.; Froidevaux, C.; Calzone, L.; Paulevé, L. Qualitative dynamics semantics for SBGN process description. BMC Systems Biology 2016, 10, 42. [CrossRef]
- Aghamiri, S.S.; Singh, V.; Naldi, A.; Helikar, T.; Soliman, S.; Niarakis, A. Automated inference of Boolean models from molecular interaction maps using CaSQ. Bioinformatics 2020, 36, 4473–4482. Publisher: Oxford University Press (OUP). [CrossRef]
- Hucka, M.; Finney, A.; Sauro, H.M.; Bolouri, H.; Doyle, J.C.; Kitano, H.; Arkin, A.P.; Bornstein, B.J.; Bray, D.; Cornish-Bowden, A.; et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 2003, 19, 524–531. Publisher: Oxford Univ Press.
- Gauges, R.; Rost, U.; Sahle, S.; Wengler, K.; Bergmann, F.T. The Systems Biology Markup Language (SBML) Level 3 Package: Layout, Version 1 Core. Journal of Integrative Bioinformatics 2015, 12, 550–602. Publisher: De Gruyter. [CrossRef]
- Bergmann, F.T.; Keating, S.M.; Gauges, R.; Sahle, S.; Wengler, K. SBML Level 3 package: Render, Version 1, Release 1. Journal of Integrative Bioinformatics 2018, 15. Publisher: De Gruyter. [CrossRef]
- Le Novère, N.; Hucka, M.; Mi, H.; Moodie, S.; Schreiber, F.; Sorokin, A.; Demir, E.; Wegner, K.; Aladjem, M.I.; Wimalaratne, S.M.; et al. The systems biology graphical notation. Nature Biotechnology 2009, 27, 735–741. Publisher: Nature Publishing Group.
- Rougny, A.; Touré, V.; Moodie, S.; Balaur, I.; Czauderna, T.; Borlinghaus, H.; Dogrusoz, U.; Mazein, A.; Dräger, A.; Blinov, M.L.; et al. Systems Biology Graphical Notation: Process Description language Level 1 Version 2.0. Journal of Integrative Bioinformatics 2019, 16. Publisher: De Gruyter. [CrossRef]
- Mi, H.; Schreiber, F.; Moodie, S.; Czauderna, T.; Demir, E.; Haw, R.; Luna, A.; Le Novère, N.; Sorokin, A.; Villéger, A. Systems biology graphical notation: activity flow language level 1 version 1.2. Journal of integrative bioinformatics 2015, 12, 340–381. Publisher: De Gruyter.
- Juty, N.; le Novère, N. Systems biology ontology. Encyclopedia of Systems Biology 2013, pp. 2063–2063. Publisher: Springer.
- Balaur, I.; Welter, D.; Rougny, A.; Inau, E.T.; Mazein, A.; Ghosh, S.; Schneider, R.; Waltemath, D.; Ostaszewski, M.; Satagopam, V. FAIR assessment of Disease Maps fosters open science and scientific crowdsourcing in systems biomedicine. Scientific Data 2025, 12, 851. Publisher: Nature Publishing Group. [CrossRef]
- Rougny, A.; Touré, V.; Albanese, J.; Waltemath, D.; Shirshov, D.; Sorokin, A.; Bader, G.D.; Blinov, M.L.; Mazein, A. SBGN Bricks Ontology as a tool to describe recurring concepts in molecular networks. Briefings in Bioinformatics 2021, 22, bbab049. [CrossRef]
- Hoyt, C.T.; Domingo-Fernández, D.; Hofmann-Apitius, M. BEL Commons: an environment for exploration and analysis of networks encoded in Biological Expression Language. Database 2018, 2018, bay126. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




