Submitted:
08 January 2026
Posted:
09 January 2026
You are already at the latest version
Abstract
Keywords:
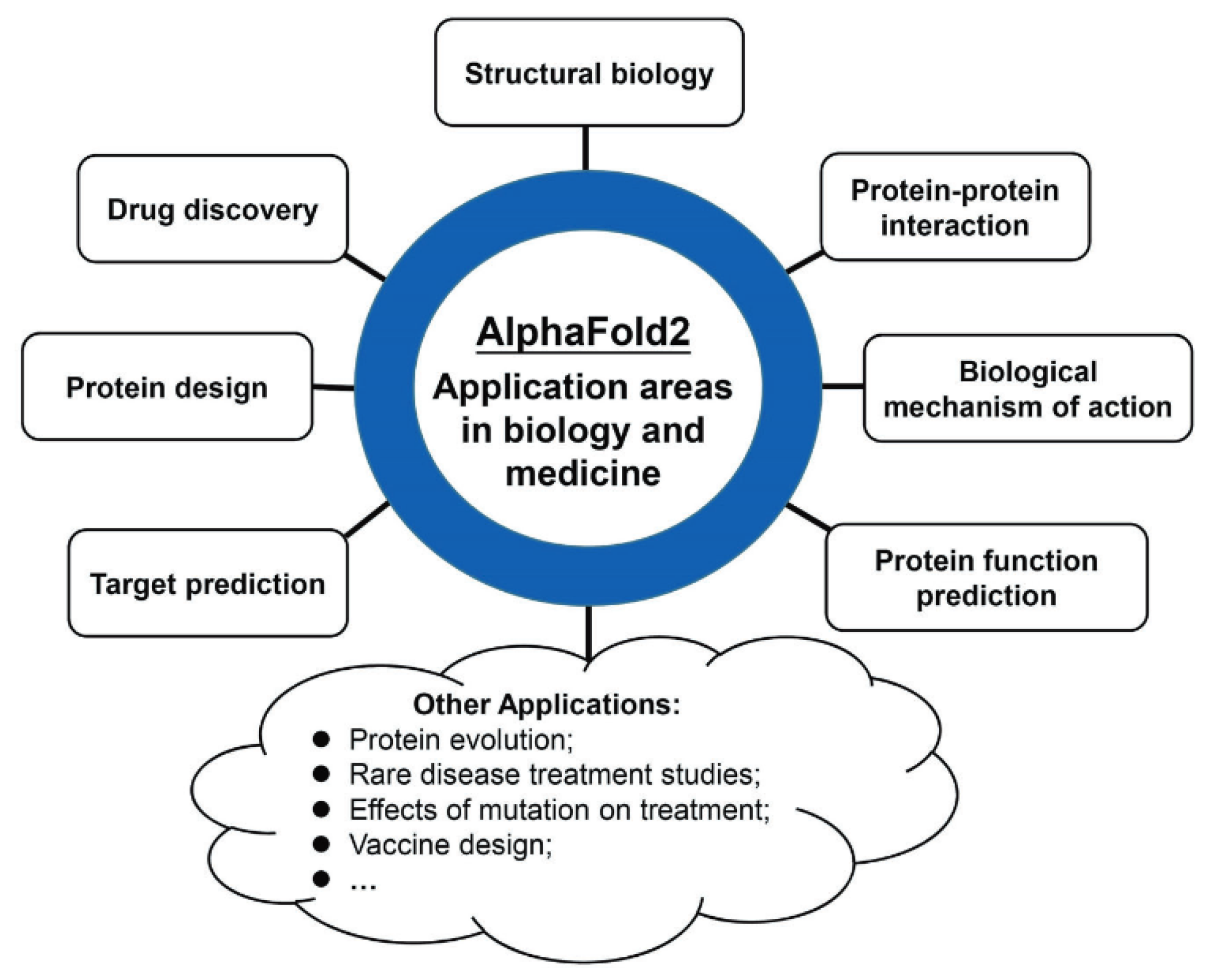
1. Introduction
“The rest of the paper is organized as follows: Section 2reviews the evolution of AlphaFold Architecture. Section 3describes the core limitations and challenges of the AlphaFold2 Framework. Section 4details the emerging solutions and the future. Section 5presents the conclusion.”
2. The Evolution of AlphaFold Architecture
2.1. The Predecessor: AlphaFold
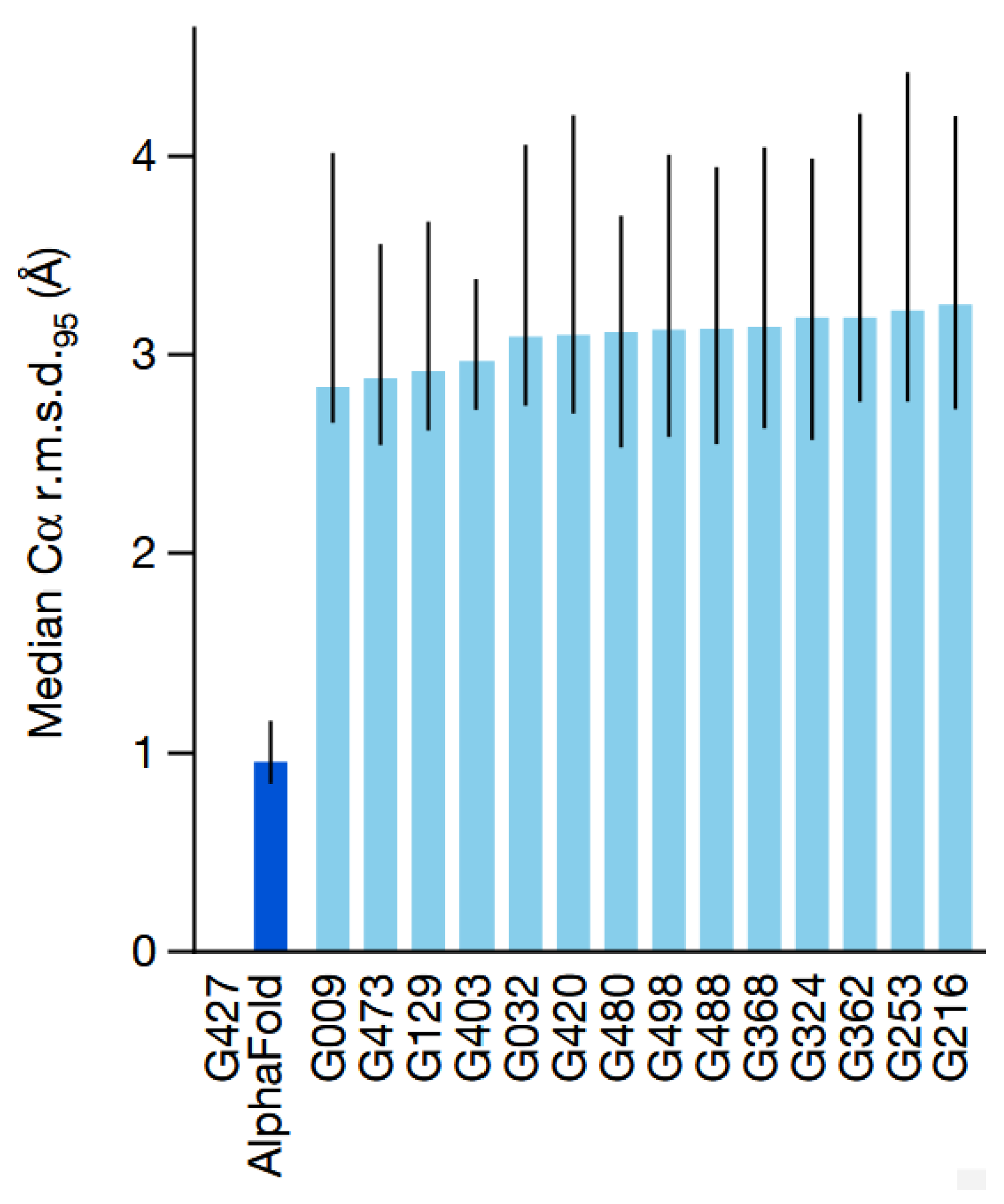
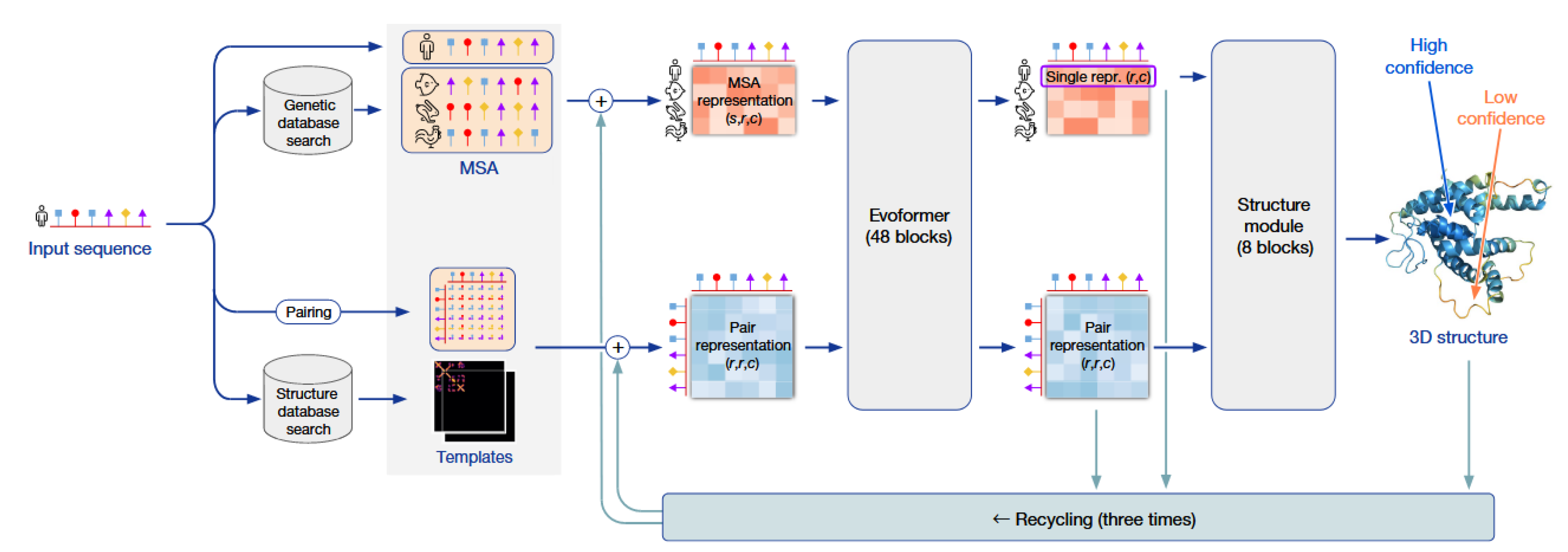
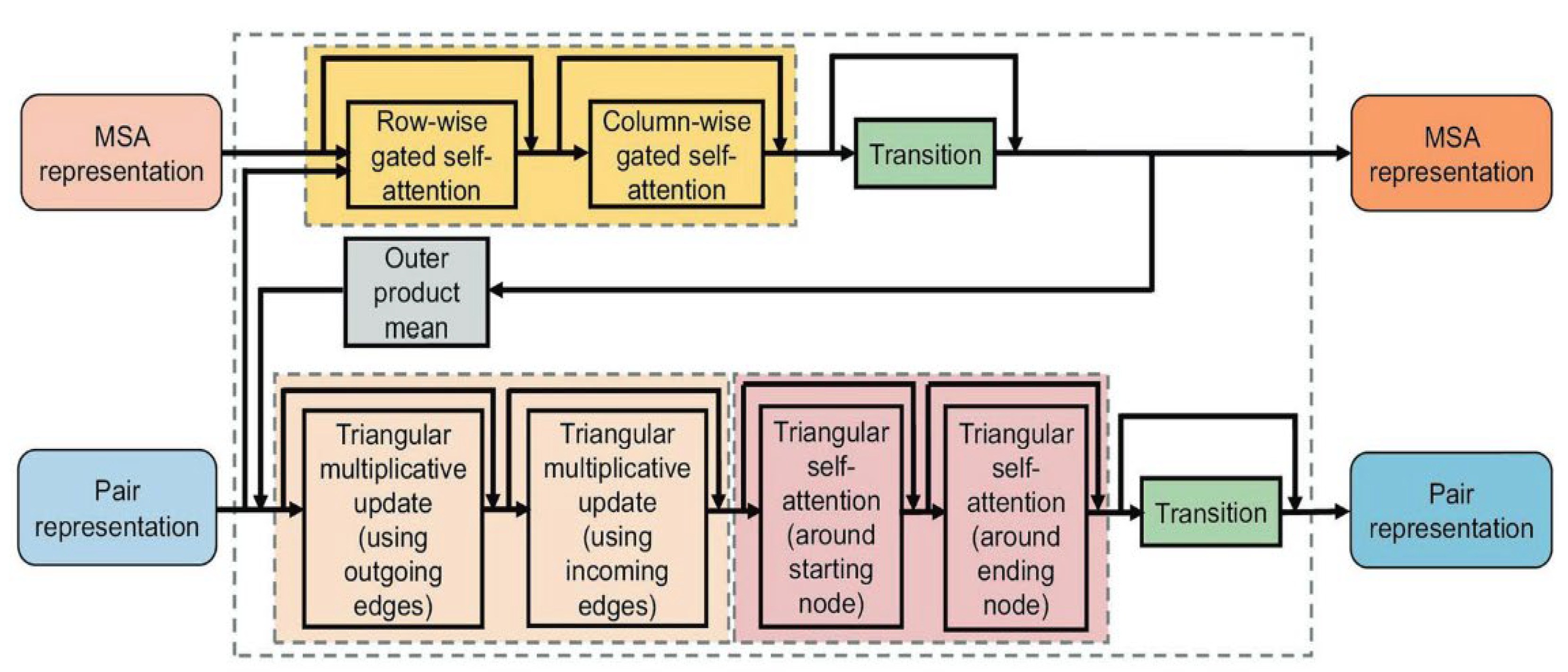
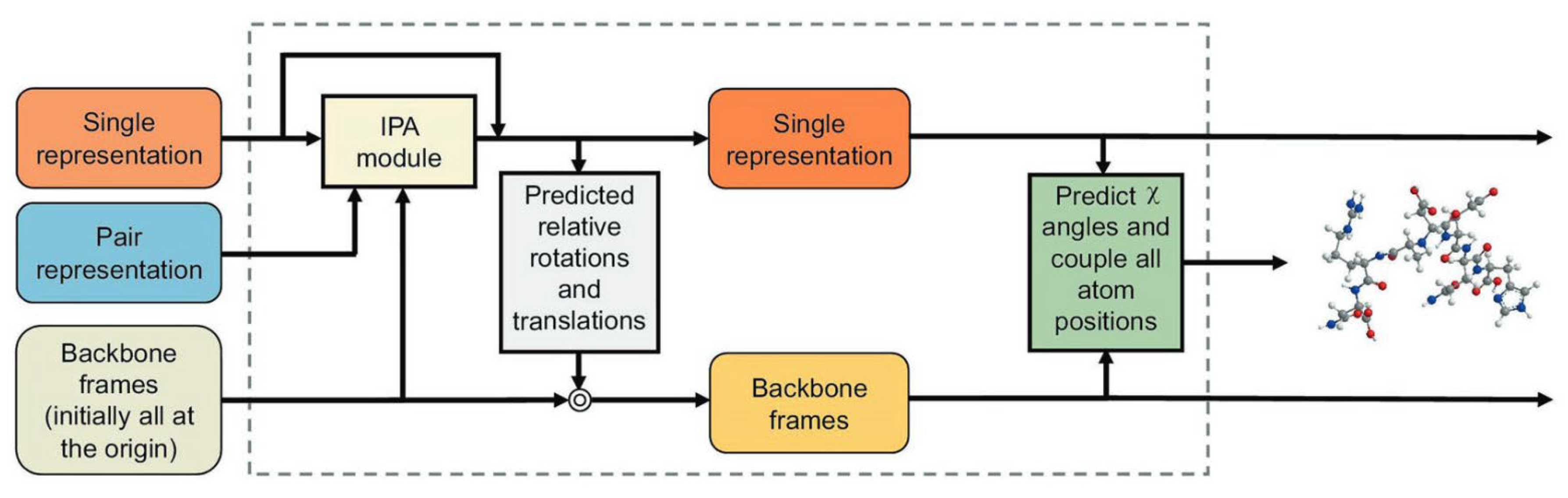
2.2. The Breakthrough: The End-to-End Architecture of AlphaFold2
3. Core Limitations And Unsolved Challenges Of The Static Model
3.1. The Static Structure Problem: Modeling Conformational Dynamics
3.2. Challenges in Predicting Biomolecular Interactions
3.3. Insensitivity to Point Mutations
4. Future Directions and Outlook
4.1. From Static Snapshots to Dynamic Ensembles
4.2. The Challenge of "Structural Hallucination"
4.3. Unified Biomolecular Modeling for Drug Discovery
4.4. The Synergy of AI and Experimentation
5. Conclusions
References
- Y. LeCun, Y. Bengio, and G. Hinton, “Deep learning,”. Nature 2015, vol. 521(no. 7553), 436–444. [CrossRef]
- A. Krizhevsky, I. Sutskever, and G. E. Hinton, “ImageNet classification with deep convolutional neural networks,”. Commun. ACM 2017, vol. 60(no. 6), 84–90. [CrossRef]
- Vaswani et al., “Attention Is All You Need,” Aug. 02, 2023, arXiv:1706.03762. 2023. [CrossRef]
- R. Nussinov, M. Zhang, Y. Liu, and H. Jang, “AlphaFold, Artificial Intelligence (AI), and Allostery,”. J. Phys. Chem. B 2022, vol. 126(no. 34), 6372–6383. [CrossRef]
- L. M. F. Bertoline, A. N. Lima, J. E. Krieger, and S. K. Teixeira, “Before and after AlphaFold2: An overview of protein structure prediction,”. Front. Bioinforma. 2023, vol. 3, 1120370. [CrossRef]
- J. Jumper et al., “Highly accurate protein structure prediction with AlphaFold,”. Nature 2021, vol. 596(no. 7873), 583–589. [CrossRef] [PubMed]
- Z. Yang, X. Zeng, Y. Zhao, and R. Chen, “AlphaFold2 and its applications in the fields of biology and medicine,”. Signal Transduct. Target. Ther. 2023, vol. 8(no. 1), 115. [CrossRef] [PubMed]
- A. W. Senior et al., “Improved protein structure prediction using potentials from deep learning,”. Nature 2020, vol. 577(no. 7792), 706–710. [CrossRef]
- W. M. Fitch, “Distinguishing Homologous from Analogous Proteins,”. Syst. Zool. 1970, vol. 19(no. 2), 99. [CrossRef]
- Chothia and A. M. Lesk, “The relation between the divergence of sequence and structure in proteins.,”. EMBO J. 1986, vol. 5(no. 4), 823–826. [CrossRef]
- Q. Xie, M.-T. Luong, E. Hovy, and Q. V. Le, “Self-training with Noisy Student improves ImageNet classification,”. arXiv 2019. [CrossRef]
- V. Agarwal and A. C. McShan, “The power and pitfalls of AlphaFold2 for structure prediction beyond rigid globular proteins,”. Nat. Chem. Biol. 2024, vol. 20(no. 8), 950–959. [CrossRef]
- N. Bordin et al., “AlphaFold2 reveals commonalities and novelties in protein structure space for 21 model organisms,”. Commun. Biol. 2023, vol. 6(no. 1), 160. [CrossRef]
- Barrio-Hernandez et al., “Clustering predicted structures at the scale of the known protein universe,”. Nature 2023, vol. 622(no. 7983), 637–645. [CrossRef] [PubMed]
- Durairaj et al., “Uncovering new families and folds in the natural protein universe,”. Nature 2023, vol. 622(no. 7983), 646–653. [CrossRef] [PubMed]
- M. Dobson, “Protein folding and misfolding,”. Nature 2003, vol. 426(no. 6968), 884–890. [CrossRef] [PubMed]
- V. Daggett and A. R. Fersht, “Is there a unifying mechanism for protein folding?,”. Trends Biochem. Sci. 2003, vol. 28(no. 1), 18–25. [CrossRef]
- S. Glazer, R. J. Radmer, and R. B. Altman, “Improving Structure-Based Function Prediction Using Molecular Dynamics,”. Structure 2009, vol. 17(no. 7), 919–929. [CrossRef]
- M. C. Childers and V. Daggett, “Validating Molecular Dynamics Simulations against Experimental Observables in Light of Underlying Conformational Ensembles,”. J. Phys. Chem. B 2018, vol. 122(no. 26), 6673–6689. [CrossRef]
- Y. I. Yang, Q. Shao, J. Zhang, L. Yang, and Y. Q. Gao, “Enhanced sampling in molecular dynamics,”. J. Chem. Phys. 2019, vol. 151(no. 7), 070902. [CrossRef]
- H. K. Wayment-Steele et al., “Predicting multiple conformations via sequence clustering and AlphaFold2,”. Nature 2024, vol. 625(no. 7996), 832–839. [CrossRef]
- L. Watson et al., “De novo design of protein structure and function with RFdiffusion,”. Nature 2023, vol. 620(no. 7976), 1089–1100. [CrossRef]
- J. Abramson et al., “Accurate structure prediction of biomolecular interactions with AlphaFold 3,”. Nature 2024, vol. 630(no. 8016), 493–500. [CrossRef]
- T. C. Terwilliger et al., “AlphaFold predictions are valuable hypotheses and accelerate but do not replace experimental structure determination,”. Nat. Methods 2024, vol. 21(no. 1), 110–116. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.



