1. Introduction
Mango is a widely consumed tropical fruit worldwide. With its rising global consumption, there has been a corresponding increase in reports of mango allergy [
1,
2,
3,
4]. FAO had reported eight major categories of allergenic foods, which primarily include high-protein foods such as milk and eggs. Although mango, as a fruit, is not listed among them, numerous studies reporting its high allergenic prevalence necessitate increased attention [
5]. Six major allergenic proteins in mango have been identified based on literature and the Allergome database (allergome.org), including glyceraldehyde-3-phosphate dehydrogenase (Man i 1), Man i 2 protein, profilin (Man i 3), class I chitinase, β-1,3-glucanase, lipoxygenase, and the Bet v 1-like homologous protein (Man i 14kD). The current research of mango epitopes is still limited. While bioinformatic predictions have been conducted for mango allergens, these allergens have yet to be substantiated by robust experimental validation, such as serological testing or animal models.
Food cross-reactivity refers to the phenomenon where sensitization to one food allergen triggers allergic reactions upon consumption of other homologous or non-homologous foods [
6]. Currently, studies on mango cross-reactive allergens rely mainly on bioinformatic predictions. Nevertheless, the precise mechanisms governing cross-reactivity among different foods are not yet fully elucidated. Consequently, with growing recognition of the importance of allergenic cross-reactivity, research into its mechanisms has become imperative. Studies on mango cross-reactivity will establish a theoretical foundation for guiding dietary choices among mango-allergic individuals, preventing cross-reactive allergies, and ultimately contributing to public health assurance.
This study first utilized BLAST to predict potential cross-reactive allergens between mango allergens and other food allergens. Subsequently, cross-reactive allergens were identified serologically using IgE from mango-allergic patients as a probe, with Western blotting employed to detect food proteins that interacted with the patients’ serum antibodies. Furthermore, similar sequences of the identified cross-reactive allergens were predicted using BLAST. These sequences were then synthesized via solid-phase peptide synthesis method. Finally, the cross-reactive linear epitopes were identified by assessing their IgE-binding capacity with the patients’ serum.
2. Materials and Methods
2.1. Materials and Reagents
Bovine serum albumin (BSA), 4-chloro-1-naphthol, acrylamide, N,N’-methylenebisacrylamide, β-mercaptoethanol, Tris base, glycine, sodium dodecyl sulfate (SDS), dimethyl sulfoxide (DMSO), Tween-20, and 3,3’,5,5’-tetramethylbenzidine (TMB) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Coomassie Brilliant Blue R-250 was obtained from Beijing Banxia Science & Technology Development Co., Ltd. (Beijing, China). The low-molecular-weight protein standard was procured from TIANGEN Biotech (Beijing, China). Polyvinylidene fluoride (PVDF) membranes were sourced from Millipore (Billerica, MA, USA. Sera from patients with mango allergy were provided by the hospital of Zhejiang Gongshang University).
2.2. Instruments and Equipment
The following instruments were used in this study: an electrophoresis gel documentation and analysis system (Bio-Rad, Hercules, CA, USA); a SHZ-C water bath constant temperature shaker (Shanghai Longyue Instrument Equipment Co., Ltd., Shanghai, China); a PHS-3C pH meter (Shanghai Yidian Scientific Instrument Co., Ltd., Shanghai, China); a DYY-7C electrophoresis apparatus (LiuYi Instrument Factory, Beijing, China); and a KHB ST-360 microplate reader (Shanghai Kehua Bio-engineering Co., Ltd., Shanghai, China).
2.3. Methods
2.3.1. BLAST Analysis of the Similarity Between Mango and Its Cross-Reactive Allergens
Potential cross-reactive proteins were predicted using the protein-protein BLAST (BLASTP) tool. The amino acid sequences of known mango allergens – including glyceraldehyde-3-phosphate dehydrogenase, class I chitinase, profilin, lipoxygenase, and β-1,3-glucanase – were individually aligned against the amino acid sequences of allergenic proteins from other common foods (cow’s milk, egg, fish, crustaceans, mollusks, tree nuts, peanut, wheat, soybean, peach, banana, lychee, apple, pear, and carrot).
2.3.2. Extraction for Mango and Other Food Proteins and Determination of Protein Concentration
2.3.2.1. Extraction of Mango Proteins
Mango pulp was homogenized using a blender to create a homogenate for subsequent use. The method for extracting mango proteins was slightly modified from the protocols described by Dube et al. [
7] and Hou et al. [
8]. Briefly, the homogenate was mixed with acetone at a 1:3 ratio (w/v) and stirred for 2 hours. The mixture was then centrifuged at 8000 rpm for 20 minutes. After the acetone was evaporated from the precipitate, 2 grams of the resulting powder were stirred with 30 mL of 0.02 M phosphate buffer (pH 7.4) containing 0.13 M sodium chloride for 6 hours for extraction. The suspension was centrifuged again at 8000 rpm for 10 minutes. The supernatant was collected and dialyzed overnight against 10 mM sodium phosphate buffer (pH 7.4). The final protein extract was aliquoted and stored. The protein concentration was determined at 595 nm using the Coomassie Brilliant Blue assay.
2.3.2.3. Extraction of Peanut Proteins
According to the extraction method of Zhang [
10], after removing the seed coat, peanut kernels were ground into powder using a milling machine. The powder was added to acetone at a ratio of 1:10 (m/v) and shaken for defatting at room temperature for 6 h, then centrifuged for 10 min (12,000 × g). After removing acetone from the precipitate, 20 mM phosphate buffer (containing 1 M NaCl, pH 7.8) was added at a ratio of 1:20 (m/v), followed by shaking extraction for 4 h. The mixture was then centrifuged (14,000 × g, 10 min). The supernatant was dialyzed against ultrapure water at 4 °C for 4 h and then stored frozen for later use. The protein concentration was determined using the Coomassie Brilliant Blue method at 595 nm.
2.3.2.4. Extraction of Shrimp Proteins
According to the method of Yin et al. [
11], 4 g of Antarctic krill meat was weighed, and deionized water was added at a ratio of 3.15:1 (m/v). Subsequently, the pH was adjusted to 11.38 using 2 mol/L sodium hydroxide, and the mixture was left to stand for 1.5 h. It was then centrifuged at 10,000 × g for 10 min at 4 °C to obtain the supernatant. The entire extraction process was repeated three times. Finally, the collected supernatant was adjusted to pH 4.5 using 2 mol/L phosphoric acid, left to stand for 1.5 h, and centrifuged at 10,000 × g for 10 min at 4 °C to collect the precipitate. The protein concentration was determined using the Coomassie Brilliant Blue method at 595 nm.
2.3.2.5. Extraction of Almond Proteins
According to the method in reference [
12], raw almonds were ground into fine powder, then defatted by shaking with n-hexane at a ratio of 1:5 (w/v) for 3 h. After removing n-hexane from the defatted powder, it was shaken with carbonate buffer (0.05 mol/L, pH 9.6) at a ratio of 1:250 (w/v) for extraction for 1 h at 50 °C. The crude extract was then centrifuged at 16,000 × g for 30 min at 4 °C. The supernatant was collected for subsequent use. The protein concentration was determined using the Coomassie Brilliant Blue method at 595 nm. All determinations were repeated three times, and the average value was calculated.
2.3.2.6. Extraction of Hazelnut Proteins
According to the extraction method of Fang et al. [
13], 30 g of hazelnut powder was mixed with n-hexane at a ratio of 1:15 (w/v) for defatting. The mixture was shaken for extraction for 6 hours, then centrifuged twice at 8000 r/min, each time for 10 min. The supernatant was removed to obtain the precipitate. After removing n-hexane from the precipitate, ultrapure water was added at a ratio of 1:6 (w/v). The mixture was homogenized at high speed (12,000 r/min) for 2 min using a magnetic stirrer. After homogenization, the pH was adjusted to 8.0 with dilute alkali (1% NaOH), followed by water bath extraction at 40 °C with shaking for 4 hours. The mixture was then centrifuged (6000 r/min, 10 min) to separate the phases, and the supernatant was collected for subsequent use. The protein concentration was determined using the Coomassie Brilliant Blue method at 595 nm.
2.3.2.7. Extraction of Pistachio Proteins
According to the extraction method of Noorbakhsh et al. [
14], 10 g of pistachio powder was mixed with n-hexane at a ratio of 1:15 (w/v) for defatting. The mixture was shaken for extraction for 6 hours, then centrifuged (8000 r/min for 20 min). The supernatant was removed to obtain the precipitate. After removing n-hexane from the precipitate, it was mixed with phosphate buffer (20 mmol/L, pH 7.4) at a ratio of 1:10 (w/v). The mixture was stirred uniformly and shaken for extraction for 6 h, then centrifuged at 9000 × g for 30 min. The supernatant was collected for subsequent use. The protein concentration was determined using the Coomassie Brilliant Blue method at 595 nm.
2.3.2.8. Extraction of Cashew Proteins
According to the extraction method of Zhong et al. [
15], plump cashew nuts uniform in texture and free from disease and insect damage were selected prior to the experiment. A certain amount of cashew nuts was crushed in a grinder for subsequent use. The powder was defatted by shaking with n-hexane at a ratio of 1:15 (w/v) for 6 hours, followed by centrifugation (8000 r/min; 20 min). The supernatant was removed to obtain the precipitate. After removing n-hexane from the precipitate, 1 g was taken and 40 mL of distilled water was added. The pH was adjusted to 9 using 0.01 mol/L NaOH solution, and the mixture was stirred to dissolve. Extraction was performed at 35 °C for 1.5 h, followed by centrifugation at 4000 r/min for 15 min. The supernatant was collected for protein content determination.
2.3.2.9. Extraction of Carrot Proteins
According to the extraction method of Guo [
16], vigorously growing carrots were selected and homogenized using a blender. The homogenate was mixed with acetone at a ratio of 1:4 (w/v) and magnetically stirred for 3 h, followed by centrifugation at 12,000 r/min for 10 min at 4 °C. The supernatant was discarded. After removing acetone from the precipitate, phosphate buffer (0.02 mol/L, pH 7.2) was added at a ratio of 1:10 (w/v) for extraction for 2 h. The mixture was then centrifuged at 15,000 r/min for 20 min, and the supernatant was collected for subsequent use. The protein concentration was determined using the Coomassie Brilliant Blue method at 595 nm.
2.3.2.10. Extraction of Peach Proteins
According to the extraction method of Pasini et al. [
17], peach pulp homogenate was mixed with acetone at a ratio of 1:3 (w/v) for defatting for 5 h. The defatted sample was then dissolved in 0.01 mol/L phosphate buffer (0.015 mol/L sodium chloride, pH 7.2) at a ratio of 1:10 (w/v), followed by shaking extraction for 2 h. The mixture was centrifuged (10,000 × g, 30 min, 4 °C), and the supernatant was collected and stored at -20 °C for subsequent use. The protein concentration was determined using the Coomassie Brilliant Blue method at 595 nm.
2.3.2.11. Extraction of Lychee Proteins
According to the extraction method of Song et al. [
18], 3 g of lychee pulp homogenate was mixed with 15 mL of cold acetone and subjected to shaking extraction for 2 h, followed by centrifugation at 15,000 × g for 10 min at 4 °C. This step was repeated twice. After removing acetone from the precipitate, it was mixed with phosphate buffer (0.01 M, pH 7.4, 0.15 M NaCl) at a ratio of 1:15 (w/v) and extracted with shaking at 4 °C. After centrifugation (20,000 × g, 45 min), the supernatant was collected and stored frozen for subsequent use. The protein concentration was determined using the Coomassie Brilliant Blue method at 595 nm.
2.3.2.12. Extraction of Pear Proteins
According to the extraction method of Zhang et al. [
19], fresh fragrant pear pulp homogenate was mixed with solvent at a solid-to-liquid ratio of 1:10 (w/v). The pH was adjusted to 10 using 0.1 mol/L NaOH. Extraction was performed at 40 °C with constant-temperature shaking in a water bath for 2 h. The mixture was centrifuged at 8000 r/min for 15 min, and the supernatant was collected for subsequent use. The protein concentration was determined using the Coomassie Brilliant Blue method at 595 nm.
2.3.2.13. Extraction of Apple Proteins
According to the extraction method of Deng et al. [
20], 1 g of apple pulp homogenate was mixed with 25 mL of 100 mmol/L Tris-HCl buffer (pH 9.0) and magnetically stirred for 2 h. The mixture was then centrifuged at 12,000 × g for 20 min. The supernatant was collected and stored at -20 °C for subsequent use. The protein concentration was determined using the Coomassie Brilliant Blue method at 595 nm.
2.3.2.14. Extraction of Banana Proteins
According to the method of Nikolic et al. [
21], peeled banana pulp (160 g) was homogenized in 300 mL of 50 mM NH₄HCO₃ buffer (pH 8.5) and extracted for 2 h. Clarification of the protein extract was achieved by centrifugation at 3000 × g for 15 min at 4 °C. The supernatant was collected, dialyzed against ammonium bicarbonate buffer (50 mM NH₄HCO₃, pH 8.5) for 48 h, aliquoted, and stored frozen for subsequent use. The protein concentration was determined using the Coomassie Brilliant Blue method at 595 nm.
2.3.3. Identification of Mango Cross-Reactive Allergens by Immunoblotting Using Sera from Mango-Allergic Patients
The protein extracts from mango, wheat, peanut, shrimp, almond, hazelnut, pistachio, cashew, carrot, peach, lychee, pear, apple, and banana were subjected to SDS-PAGE separation and subsequently electrotransferred onto a polyvinylidene fluoride (PVDF) membrane. The immunoblotting procedure [
22] was performed on the membrane. The transferred PVDF membrane was washed with distilled water for 5 min, followed by rinsing four times with TBST, each for 15 min at 37 °C. Blocking buffer was added and incubated at 37 °C for 1 h, then washed with water for 5 min. 1:10 diluted sera from patients with mango allergy (the hospital of Zhejiang Gongshang University) was added to ELISA plate and incubated overnight at 4 °C, followed by a 5-min water wash, each for 10 min. 1:1000 diluted HRP-conjugated mouse anti-human IgE secondary antibody was added and incubated at 37 °C for 2 h, followed by a 5-min water wash and then three times washes with TBST (10 mM Tris-HCl buffer, pH 7.4, containing 0.8% NaCl and 0.1% Tween 20), each for 10 min. The PVDF membrane was immersed in a freshly prepared 4-chloro-1-naphthol solution (St. Louis, MO, USA) for color development at 37 °C for 35 min. After color development, the reaction was terminated by rinsing with high-purity water, and the membrane was air-dried between two layers of filter paper for storage. Western blotting was studied using control pooled sera from several non-allergic individuals.
2.3.4. Prediction of Similar Sequences of Cross-Reactive Allergens
Based on the identification results from immunoblotting, BLAST analysis was employed to predict similar sequences with potential cross-reactive allergenic properties.
2.3.5. Synthesis, Purification, and Characterization of Linear Epitopes
The similar sequences (peptides) were synthesized using solid-phase synthesis [
23,
24,
25], purified by liquid chromatography, and the molecular weights of the synthesized sequences were determined by mass spectrometry to verify their accuracy.
2.3.6. Identification of Linear Epitopes of Mango Cross-Reactive Allergens by Serum IgE from Mango-Allergic Patients
The synthesized peptides were screened using enzyme-linked immunosorbent assay (ELISA) [
26] with serum IgE from mango-allergic patients to identify potential cross-reactive linear epitopes. The synthesized similar sequences were assayed in a 96-well microplate, each well of which was first coated with streptavidin, then sequentially received the sequences to be subjected to epitope mapping, and finally underwent antibody detection.
Procedure:
The lyophilized peptides were dissolved in 100% dimethyl sulfoxide (DMSO), the stock solution was 10 mg/mL, the working solution was 1 mg/mL, stored at -70 °C.
Microplates were coated with 50 μL of streptavidin (5 μg/mL diluted in deionized water) per well.
Plates were washed four times with PBST, and nonspecific binding sites were blocked with 2% BSA/PBS for 2 h at room temperature.
Peptides were diluted to a final concentration of 20 μg/mL in 0.1% (w/v) BSA/PBS, and 50 μL of the peptide solution was added to each well. Plates were incubated overnight at 4 °C. Wells without peptide solution served as controls.
Primary antibody diluted in 0.1% BSA/PBS was added and incubated for 2 h.
After four washes, horseradish peroxidase (HRP)-conjugated secondary antibody (diluted 1:800 in BSA/PBS) was added and incubated for 2 h.
Plates were washed four times, followed by addition of 50 μL TMB substrate per well. The reaction was terminated with 50 μL of 100 mmol/L sulfuric acid when blue color developed. Absorbance was measured at 450 nm.
3. Results
3.1. Protein Concentration Results of Mango and Other Foods
The protein concentrations of mango, wheat, peanut, shrimp, almond, hazelnut, pistachio, cashew, carrot, peach, lychee, crystal pear, fragrant pear, apple, and banana were determined using the Coomassie Brilliant Blue method [
27], with the results shown in
Table 1. Among these, the hazelnut extract exhibited the highest protein concentration at 37.372 mg/mL, followed by peanut (17.607 mg/mL), shrimp (15.059 mg/mL), almond (12.247 mg/mL), pistachio (7.875 mg/mL), and cashew (5.498 mg/mL). Wheat protein concentration was 1.495 mg/mL. These protein concentrations were relatively high compared to those of fruits and vegetables, where crystal pear contained 0.415 mg/mL, mango 0.389 mg/mL, fragrant pear 0.251 mg/mL, lychee 0.180 mg/mL, banana 0.088 mg/mL, apple 0.072 mg/mL, peach 0.065 mg/mL, and carrot 0.056 mg/mL.
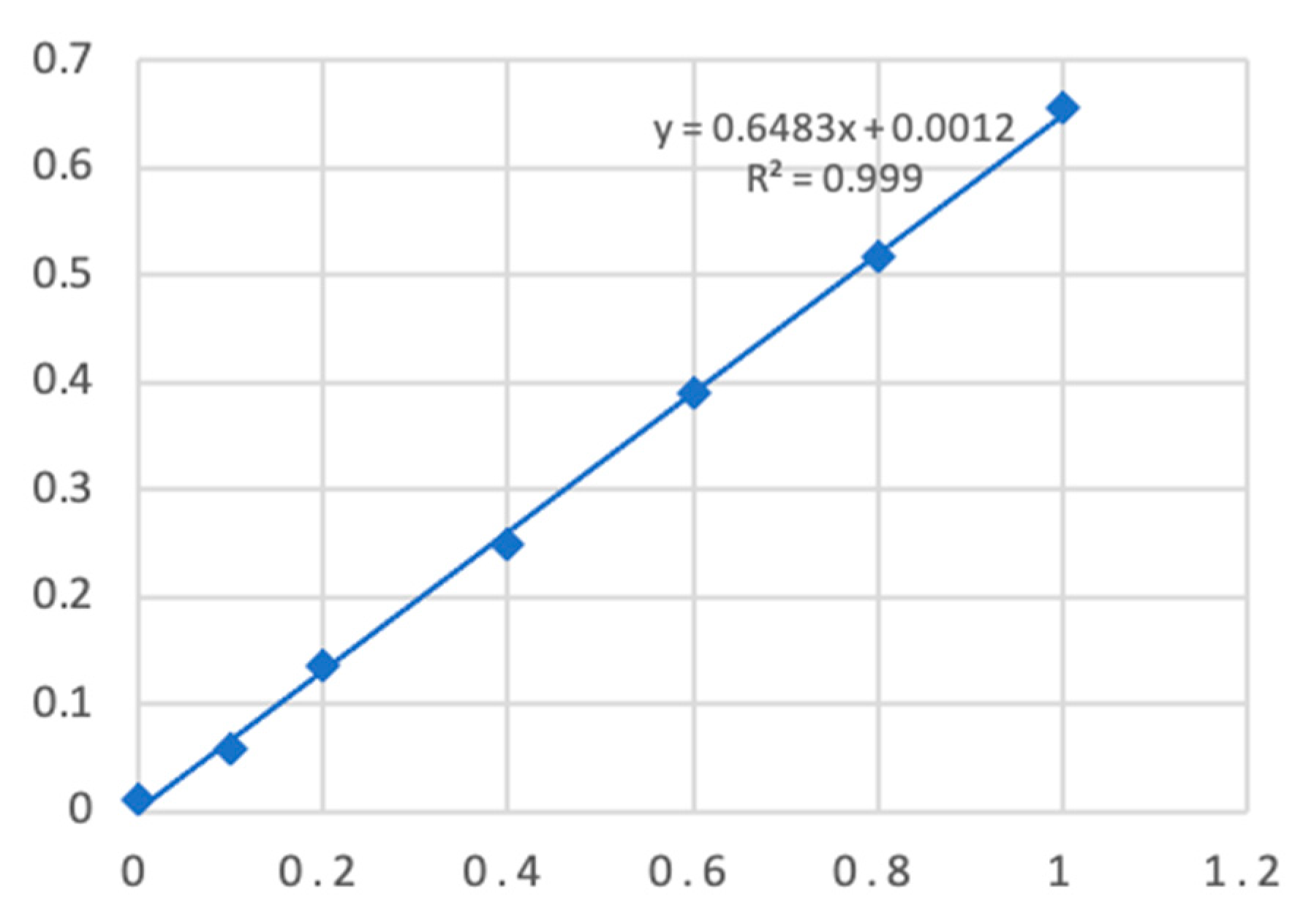
Figure 1 illustrates the standard curve for protein concentration determination by the Coomassie Brilliant Blue method, with R² > 0.999.
3.2. Prediction Results of Mango Cross-Reactive Allergens
According to the Codex Alimentarius Commission (CAC), potential cross-sensitization should be considered—indicating the presence of potential cross-reactivity—if any sequence exhibits greater than 35% similarity over 80 amino acids or contains eight consecutive identical amino acids [
28]. To enhance the accuracy of subsequent serological validation, sequences with similarity exceeding 30% were also classified as potential cross-reactive allergens. The prediction results of mango cross-reactive allergens via BLAST are presented in
Table 2. Mango glyceraldehyde-3-phosphate dehydrogenase showed >30% similarity with wheat Tri a 34. Mango chitinase demonstrated >30% similarity with wheat Tri a 37, hazelnut Cor a 1, Cor a 12, and banana Mus a 2. Mango profilin exhibited >30% similarity with wheat Tri a 12, Tri a 29, Tri a 30, and Tri a 37; peanut Ara h 5; almond Pru du 4; shrimp Cra c 6; hazelnut Cor a 2 and Cor a 13; pistachio Pis v 1; peach Pru p 4; lychee Lit c 1; banana Mus a 1; apple Mal d 4; pear Pyr c 4; carrot Dau c 4; and banana Mus a 5. Mango β-1,3-glucanase showed >30% similarity with banana Mus a 5.
3.3. Immunoblot Results of Mango Cross-Reactive Allergens Identified by Sera from Allergic Patients
3.3.1. Immunoblot Analysis of Mango Allergens Recognized by Sera from Mango-Allergic Patients
Mango exhibited a reaction with antibodies in the serum of mango-allergic patients, as shown in
Figure 2a. One band was detected in mango at approximately 63 kDa. To exclude false-positive reactions, immunoblotting was performed using negative control sera from healthy individuals, as presented in
Figure 2b. No reaction was observed between mango and the negative control sera. Therefore, it can be concluded that mango displayed one specific band at 63 kDa in the immunoblot, indicating a specific immune reaction with sera from mango-allergic patients.
3.3.2. Immunoblot Analysis of Wheat and Peanut Allergens Recognized by Sera from Mango-Allergic Patients
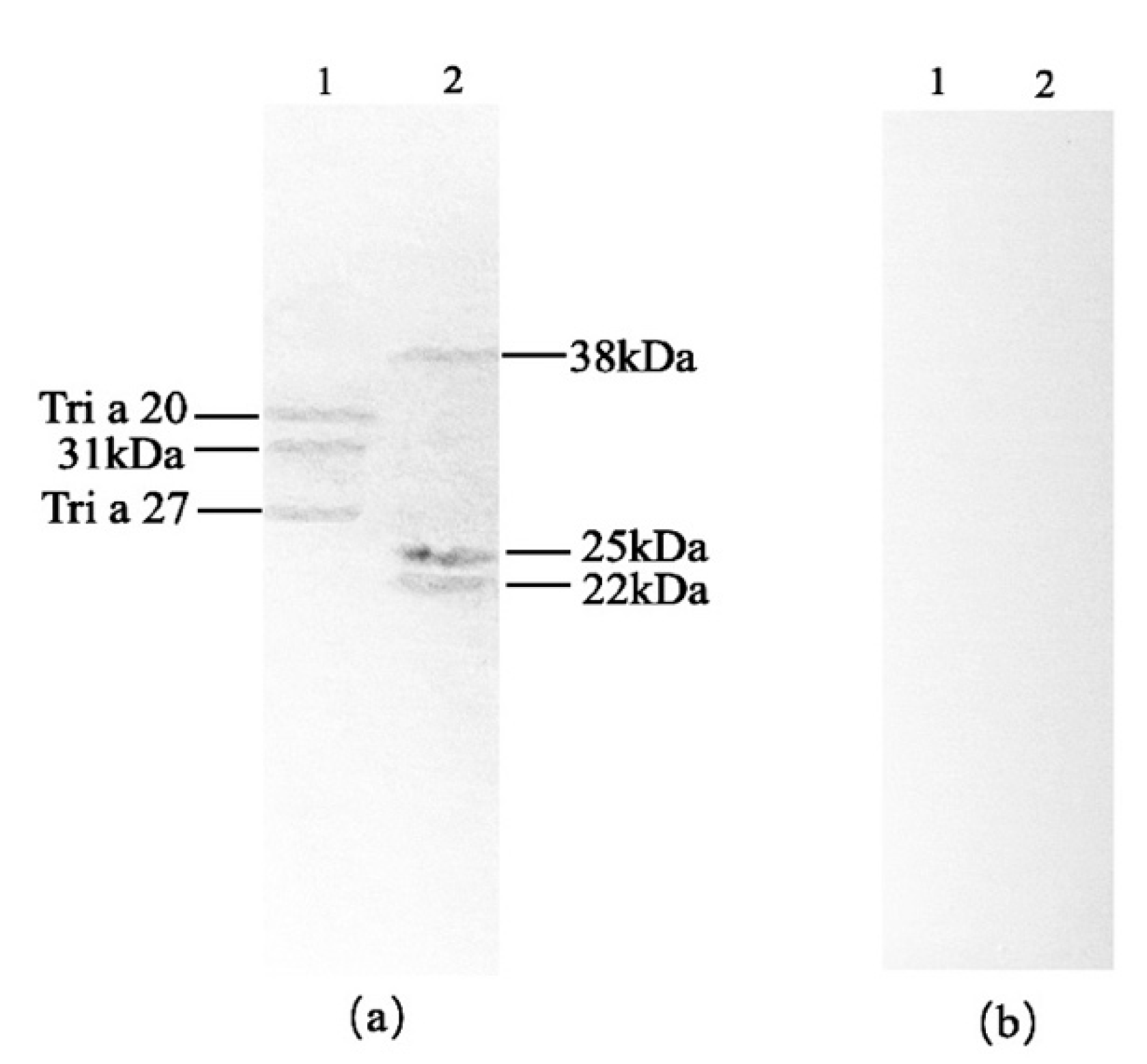
Both wheat and peanut exhibited reactions with serum antibodies from mango-allergic patients, as shown in
Figure 3a. Wheat displayed three bands corresponding to Tri a 20 (35 kDa), 31 kDa, and Tri a 27 (27 kDa). Peanut showed three bands at 38 kDa, 25 kDa, and 22 kDa. To exclude false-positive reactions, immunoblot analysis was performed using serum from healthy individuals as a negative control, as presented in
Figure 3b. Neither wheat nor peanut reacted with the negative control serum. Therefore, it can be concluded that wheat specifically reacted with sera from mango-allergic patients, showing three cross-reactive allergen bands: Tri a 20 (35 kDa), 31 kDa, and Tri a 27 (27 kDa). Similarly, peanut specifically reacted with the sera from mango-allergic patients, exhibiting three cross-reactive allergen bands at 38 kDa, 25 kDa, and 22 kDa. Song et al. [
29] previously predicted high sequence similarity between mango profilin and peanut Ara h 5 profilin, though this was not experimentally validated.
3.3.3. Immunoblot Analysis of Cashew and Pistachio Allergens Recognized by Sera from Mango-Allergic Patients
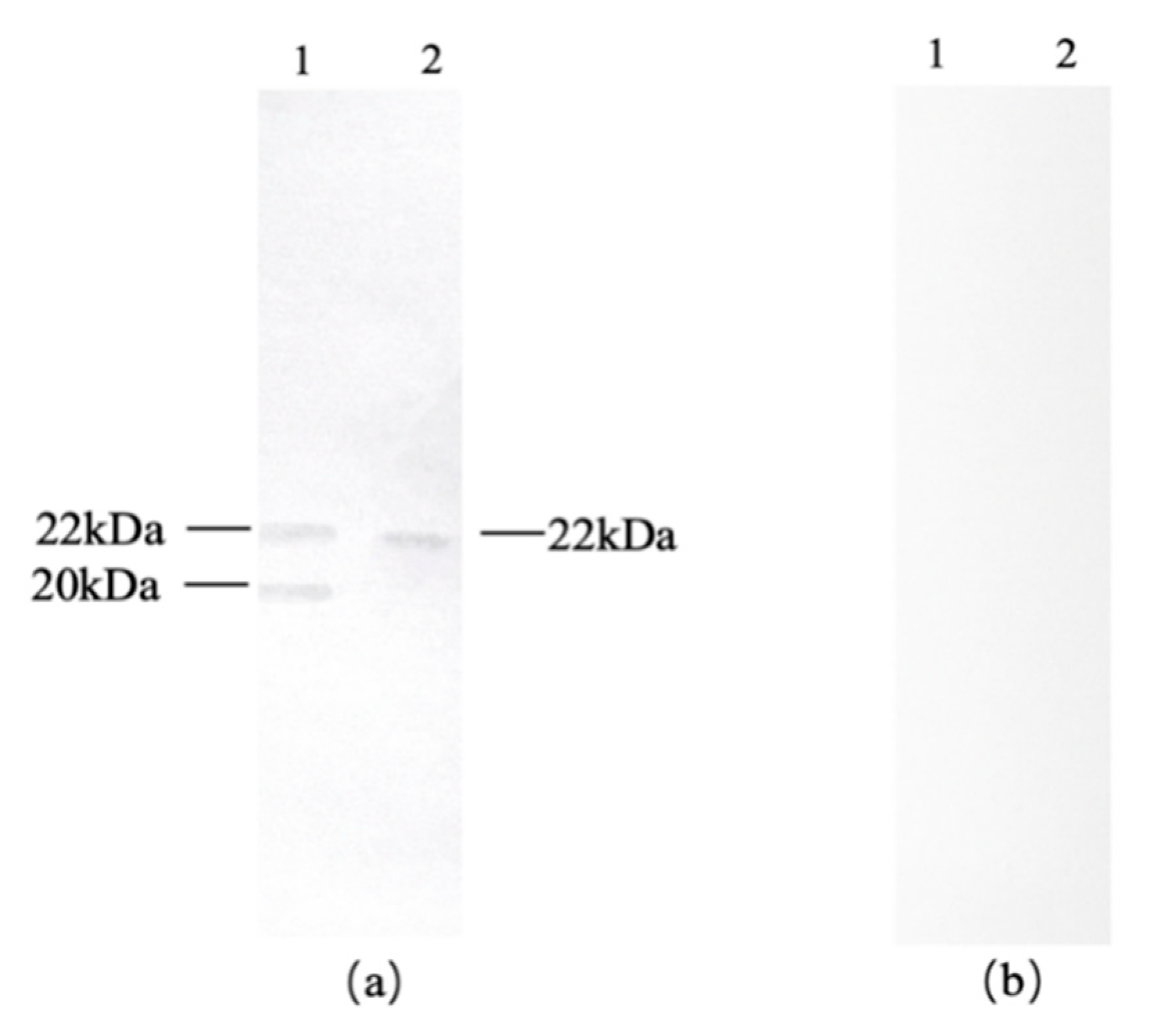
Both cashew and pistachio exhibited immunoreactivity with serum antibodies from mango-allergic patients, as shown in
Figure 4a. Cashew displayed two bands at approximately 22 kDa and 20 kDa, while pistachio showed one band at 22 kDa. To exclude false-positive reactions, immunoblot analysis was performed using serum from healthy individuals as a negative control, as presented in
Figure 4b. Neither cashew nor pistachio reacted with the negative control serum. Therefore, it can be concluded that cashew specifically reacted with sera from mango-allergic patients, showing two cross-reactive allergen bands at 22 kDa and 20 kDa, while pistachio exhibited one cross-reactive allergen band at 22 kDa. Bastiaan-Net et al. [
30] reported substantial cross-reactivity between mango and cashew via inhibitory immunoblotting using sera from cashew-allergic patients, suggesting that chitinase and β-1,3-glucanase may be involved in this cross-reactivity.
3.3.4. Immunoblot Analysis of Hazelnut and Almond Allergens Recognized by Sera from Mango-Allergic Patients
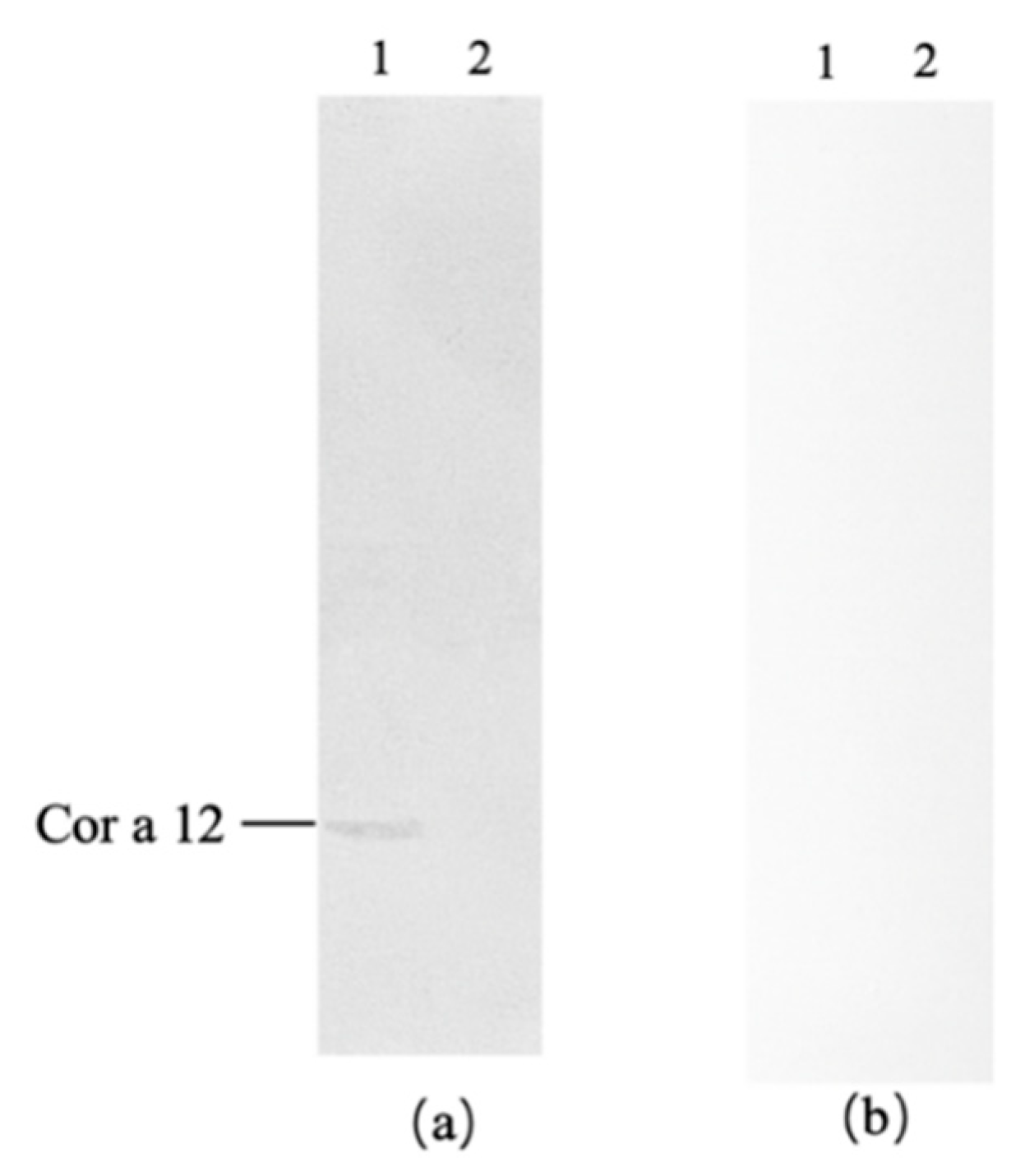
Almond did not react with antibodies from mango-allergic patients, while hazelnut exhibited specific immunoreactivity, as shown in
Figure 5a. Hazelnut displayed one band corresponding to Cor a 12 (17 kDa). To exclude false-positive reactions, immunoblot analysis was performed using serum from healthy individuals as a negative control, as presented in
Figure 5b. Neither hazelnut nor almond reacted with the negative control serum. Therefore, it can be concluded that hazelnut specifically reacted with sera from mango-allergic patients, showing one cross-reactive allergen band: Cor a 12 (17 kDa), indicating a cross-reactive allergen between hazelnut and mango.
3.3.5. Immunoblot Analysis of Lychee, Banana and Apple Allergens Recognized by Sera from Mango-Allergic Patients
Lychee, banana, and apple exhibited no immunoreactivity with either serum antibodies from mango-allergic patients or negative control sera, as shown in
Figure 6. Song et al. [
29] predicted high sequence similarity between mango profilin and apple Mal d 4 profilin, though this was not experimentally validated. Yan et al. [
31] demonstrated high homology between mango and lychee profilins through amino acid sequence alignment, suggesting a potential basis for cross-reactivity, but no experimental confirmation was performed.
3.3.6. Immunoblot Analysis of Carrot, Fragrant Pear, Crystal Pear, Peach and Shrimp Allergens Recognized by Sera from Mango-Allergic Patients
Carrot, fragrant pear, crystal pear, peach, and shrimp showed no immunoreactivity with serum antibodies from either mango-allergic patients or negative controls, as shown in
Figure 7. Song et al. [
29] predicted high sequence similarity between mango profilin and profilins from pear (Pyr c 4), peach (Pru p 4), apple (Mal d 4), peanut (Ara h 5), and carrot (Dau c 4), though these predictions lack experimental validation.
3.4. Prediction Results of Similar Sequences of Mango Cross-Reactive Allergens
The similar sequences between mango and its cross-reactive allergens (peanut, wheat, pistachio, hazelnut, and cashew) were predicted, and the results are presented in
Table 3. The predicted potential functional epitopes include: mango chitinase with wheat Tri a 27; mango profilin (Man i 3) with peanut Ara h 5; and mango profilin with pistachio Pis v 1.
3.5. Identification of Cross-Reactive Linear Epitopes in Mango and Cross-Reactive Allergens by Sera from Mango-Allergic Patients
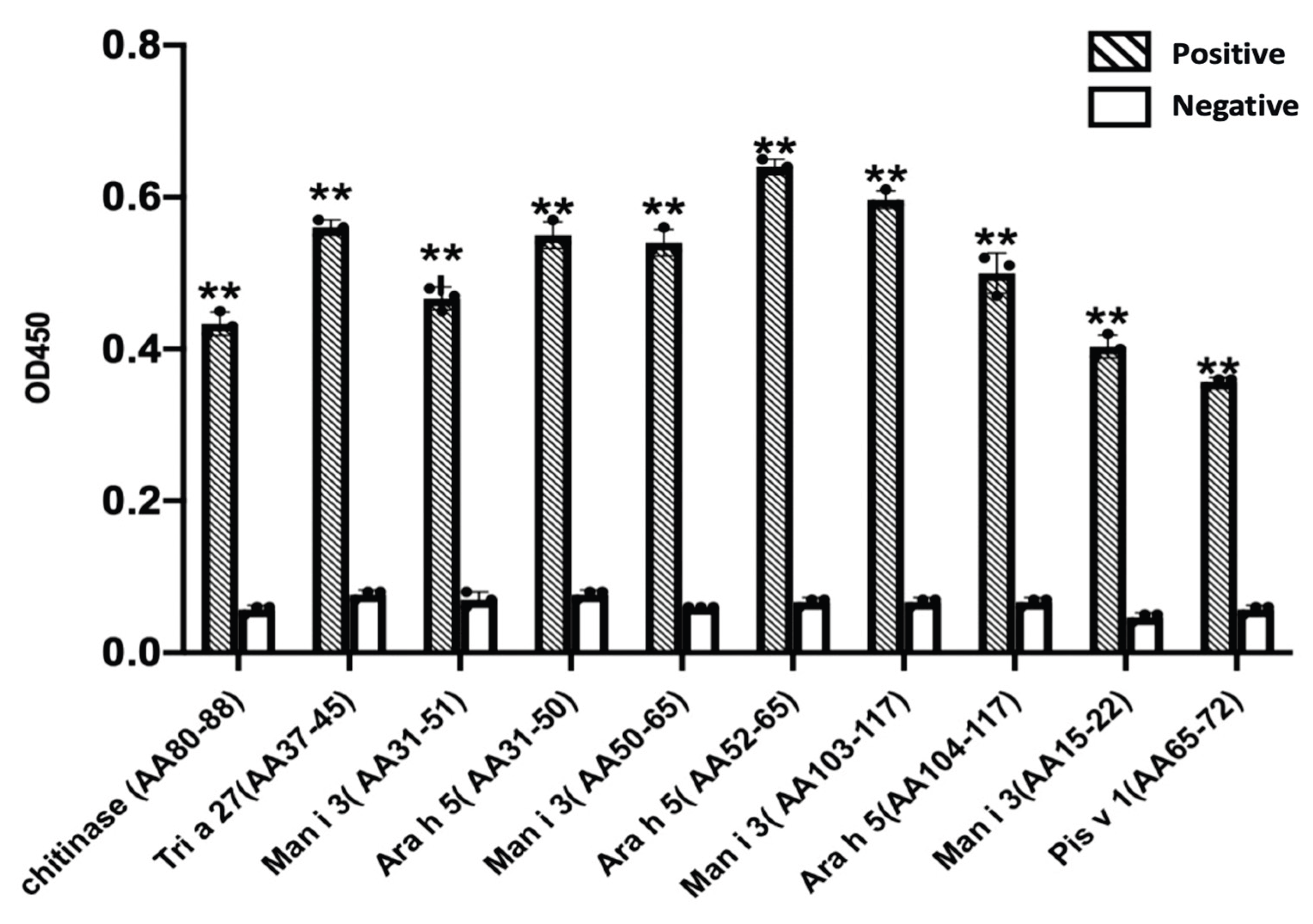
Significance analysis of the results obtained by using enzyme-linked immunosorbent assay (ELISA) with serum IgE from mango-allergic patients to identify synthetic peptides is shown in
Figure 8. The absorbance values of the similar sequences with sera from mango-allergic patients were significantly higher than those with negative control sera (P < 0.05), indicating that all predicted similar sequences are cross-reactive linear epitopes. The specific results are as follows: the AA80–88 sequence of mango chitinase and the AA37–45 sequence of wheat Tri a 27 constitute a cross-reactive epitope; the AA15–22 sequence of mango profilin and the AA65–72 sequence of pistachio Pis v 1 constitute a cross-reactive epitope; the AA31–51 sequence of mango profilin and the AA31–50 sequence of peanut Ara h 5 constitute a cross-reactive epitope; the AA50–65 sequence of mango profilin and the AA52–65 sequence of peanut Ara h 5 constitute a cross-reactive epitope; the AA76–96 sequence of mango profilin and the AA76–96 sequence of peanut Ara h 5 constitute a cross-reactive epitope; the AA103–117 sequence of mango profilin and the AA104–117 sequence of peanut Ara h 5 constitute a cross-reactive epitope.
Tsai et al. [
32] analyzed the B-cell epitopes of mango glyceraldehyde-3-phosphate dehydrogenase and suggested that the sequences 154–164, 215–222, and 318–326 are potential B-cell epitopes. Yan et al. [
31] analyzed the structural domains of mango profilin and chitinase using the SMART software and proposed that the structural domains may represent functional epitopes of profilin or chitinase. Cao et al. [
33] validated through serological experiments and identified the 8–28 aa sequence (chitin-binding domain) as the major linear IgE-binding epitope of chitinase. Zhang et al. [
34] analyzed the B-cell epitopes of mango chitinase and identified potential regions as follows: 38–46, 64, 67, 75–76, 89, 97–101, 115, 122, 124–126, 128–129, 133–138, 144–150, 152, 176, 178–181, and 222–225.
4. Discussion
Numerous studies have reported on the sensitization characteristics of individual mango allergens, whereas research on mango cross-reactive allergens remains relatively limited. This area represents a challenging and active research focus, with current investigations relying primarily on bioinformatic predictions. Paschke et al. [
35] identified the 14 kDa protein in mango exhibiting cross-reactivity with birch pollen Bet v 1 through inhibitory immunoblotting. Furthermore, using sera from mango-allergic patients, they demonstrated that proteins of approximately 40, 43, and 67 kDa in mango were primarily responsible for cross-reactivity with carrot. Song et al. [
29] predicted high sequence similarity between mango profilin and profilins from pear (Pyr c 4), peach (Pru p 4), apple (Mal d 4), peanut (Ara h 5), and carrot (Dau c 4), although experimental validation was not conducted. Yan et al. [
31] performed homology alignment of profilin amino acid sequences from various fruits and revealed high homology among mango, grape, peach, citrus, and lychee, suggesting a potential basis for cross-reactivity, yet without experimental confirmation. Funes et al. [
36] demonstrated cross-reactivity among pistachio, cashew, and mango within the Anacardiaceae family using RAST inhibition. In 2018, Cardona et al. [
37] utilized immunoblotting with sera from mango-allergic patients and suggested potential cross-reactivity between mango and three banana components: Mus a 1, Mus a 2, and Mus a 5. Similarly, Bastiaan-Net et al. [
30] employed immunoblotting with cashew-allergic sera and revealed substantial cross-reactivity between mango and cashew, potentially associated with chitinase and β-1,3-glucanase.
In this study, using serum IgE from mango-allergic patients as a probe, we systematically and scientifically verified mango cross-reactive allergens through immunoblotting. It was found that sera from mango-allergic patients exhibited cross-reactivity with wheat, peanut, cashew, pistachio, and hazelnut. This study innovatively identified wheat and hazelnut as cross-reactive allergens of mango. Furthermore, previously unreported cross-reactive allergen bands were detected in immunoblotting. The proteins that showed cross-reactivity with serum antibodies from mango-allergic patients included: wheat proteins with molecular weights of Tri a 20 (35 kDa), 31 kDa, and Tri a 27 (27 kDa); peanut proteins of 38 kDa, 25 kDa, and 22 kDa; cashew proteins of 22 kDa and 20 kDa; pistachio protein of 22 kDa; and hazelnut protein Cor a 12 (17 kDa). For future research, it is recommended to isolate and purify individual mango allergens and prepare their monoclonal antibodies to further characterize mango cross-reactive allergens.
Currently, there are limited reports on B-cell epitopes of individual mango allergens, and no studies have been reported on cross-reactive linear epitopes. Yan et al. [
31] analyzed the structural domains of mango profilin and chitinase using SMART software and proposed that these domains may represent linear epitopes of profilin or chitinase. Tsai et al. [
32] analyzed B-cell epitopes of mango glyceraldehyde-3-phosphate dehydrogenase and suggested that the sequences 154–164, 215–222, and 318–326 may serve as potential B-cell epitopes. Cao et al. [
33] experimentally confirmed the aa8–28 sequence as the major linear IgE-binding epitope of chitinase. Zhang et al. [
34] analyzed B-cell epitopes of mango chitinase and identified potential regions as follows: 38–46, 64, 67, 75–76, 89, 97–101, 115, 122, 124–126, 128–129, 133–138, 144–150, 152, 176, 178–181, and 222–225. This study, for the first time, identified cross-reactive linear epitopes of mango: the AA80–88 sequence of mango chitinase with the AA37–45 sequence of wheat Tri a 27; the AA15–22 sequence of mango profilin with the AA65–72 sequence of pistachio Pis v 1; the AA31–51 sequence of mango profilin with the AA31–50 sequence of peanut Ara h 5; the AA50–65 sequence of mango profilin with the AA52–65 sequence of peanut Ara h 5; the AA76–96 sequence of mango profilin with the AA76–96 sequence of peanut Ara h 5; and the AA103–117 sequence of mango profilin with the AA104–117 sequence of peanut Ara h 5.
Due to the limitations of bioinformatics, the prediction of cross-reactive allergens carries inherent uncertainty. In vitro serological identification using IgE antibodies is a commonly employed method to validate these predictions. As polyclonal IgE antibody reactions often yield false positives, the inclusion of negative serum controls is crucial to ensure result accuracy. Currently, the IgE antibodies used in experiments are either prepared based on allergen-specific epitopes or epitopes capable of inducing cross-reactivity. The proportion of cross-reactive IgE produced depends on the patient’s genetic factors [
38]. Some studies suggest that immunological cross-reactivity is specific, occurring primarily when serum IgE antibodies from allergic patients bind to conserved epitopes of allergens. The strength of cross-reactivity is associated with the affinity of IgE antibodies; however, not all IgE cross-reactions translate to clinical cross-reactivity, making in vivo validation essential. The mango cross-reactive allergens and cross-reactive linear epitopes identified in this study via immunoblotting should be further validated through future animal experiments. In the context of food allergy, linear IgE-binding epitopes appear more relevant since conformational epitopes are easily degraded by gastrointestinal digestion [
39]. Therefore, this study primarily focused on identifying mango cross-reactive linear epitopes.
5. Conclusions
In summary, mango exhibits cross-reactivity with peanut, wheat, cashew, pistachio, and hazelnut, among which wheat and hazelnut are novel findings. Furthermore, this study innovatively identified the following cross-reactive linear epitopes: the AA80–88 sequence of mango chitinase with the AA37–45 sequence of wheat Tri a 27; the AA15–22 sequence of mango profilin with the AA65–72 sequence of pistachio Pis v 1; the AA31–51 sequence of mango profilin with the AA31–50 sequence of peanut Ara h 5; the AA50–65 sequence of mango profilin with the AA52–65 sequence of peanut Ara h 5; the AA76–96 sequence of mango profilin with the AA76–96 sequence of peanut Ara h 5; and the AA103–117 sequence of mango profilin with the AA104–117 sequence of peanut Ara h 5.
Author Contributions
Conceptualization: Wenxuan Zhao and YanYun Cong; resources: Honglei Guo and YanYun Cong; writing—original draft preparation: Wenxuan Zhao and Honglei Guo; review and editing: YanYun Cong; funding acquisition: YanYun Cong. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Dairy Science and Technology Innovation Fund of China Dairy Industry Association (CDIAKCJJ-TP-2024-007) and National Science and Technology Major Project of China (2019YFC1605002).
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| FAO |
Food and Agriculture Organization |
| BLAST |
Basic Local Alignment Search Tool |
| BLASTP |
protein-protein BLAST |
| CAC |
Codex Alimentarius Commission |
| AA |
amino acid |
| SDS-PAGE |
sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| PVDF |
polyvinylidene fluoride |
| TBST |
Tris-buffered saline with Tween |
| HRP |
horseradish peroxidase |
| TMB |
3,3′,5,5′-tetramethylbenzidine |
| ELISA |
enzyme-linked immunosorbent assay |
| DMSO |
dimethyl sulfoxide |
| BSA |
bovine serum albumin |
| PBS |
phosphate-buffered saline |
| PBST |
PBS with Tween-20 |
| IgE |
immunoglobulin E |
References
- Miyazawa, H.; Nishie, W.; Hata, H.; Shimizu, H.; Matsumura, K. A severe case of mango dermatitis. J. Eur. Acad. Dermatol. Venereol. 2018, 32, e160. [Google Scholar] [CrossRef] [PubMed]
- O’Hern, K.; Zhang, F.; Zug, K.A.; Pace, N.C.; Hamann, C.R. “Mango Slice” dermatitis: Pediatric allergic contact dermatitis to mango pulp and skin. Dermatitis 2022, 33, e46–e47. [Google Scholar] [CrossRef]
- Pesqué, D.; Canal Garcia, E.; Rozas-Muñoz, E.; Pujol Vallverdú, R.M.; Giménez Arnau, A.M. Non-occupational protein contact dermatitis induced by mango fruit. Contact Dermatitis 2021, 458–460. [Google Scholar] [CrossRef] [PubMed]
- Raison-Peyron, N.; Aljaber, F.; Al Ali, O.A.; Dereure, O. Mango dermatitis: An unusual cause of eyelid dermatitis in France. Contact Dermatitis 2021, 85, 599–600. [Google Scholar] [CrossRef]
- Messina, M.; Venter, C. Recent surveys on food allergy prevalence. Nutr. Today 2020, 55, 22–29. [Google Scholar] [CrossRef]
- Kwon, J.; Lee, Y.M.; Wen, H. Knowledge, attitudes, and behaviors about dining out with food allergies: A cross-sectional survey of restaurant customers in the United States. Food Control 2020, 107, 106776. [Google Scholar] [CrossRef]
- Dube, M.; Zunker, K.; Neidhart, S.; Carle, R.; Steinhart, H.; Paschke, A. Effect of technological processing on the allergenicity of mangoes (Mangifera indica L.). J. Agric. Food Chem. 2004, 52, 3938–3945. [Google Scholar] [CrossRef]
- Hou, L.Y. Mango allergen components analysis by three different extracting methods. Chin. J. Food Hyg. 2014, 417–421. [Google Scholar]
- Zhao, J.; Li, Z.; Khan, M.U.; Gao, X.; Yu, M.; Gao, H.; Li, Y.; Zhang, H.; Dasanayaka, B.P.; Lin, H. Extraction of total wheat (Triticum aestivum) protein fractions and cross-reactivity of wheat allergens with other cereals. Food Chem. 2021, 347, 129064. [Google Scholar] [CrossRef]
- Zhang, Y. Evaluation of changes in protein allergenicity in fresh peanuts during thermal processing. Master’s thesis, Nanchang University, Nanchang, China, 2019. [Google Scholar]
- Yin, L.a.; Jiang, X.; Fan, Y.; Wang, J.; Xue, C.; Xue, Y. Preparation, Gel electrophoresis analysis, and nutritional evaluation of a functional krill protein concentrate with low fluoride level from Antarctic krill (Euphausia superba). J. Aquat. Food Prod. Technol. 2017, 26, 958–968. [Google Scholar] [CrossRef]
- L’Hocine, L.; Pitre, M. Quantitative and qualitative optimization of allergen extraction from peanut and selected tree nuts. Part 1. Screening of optimal extraction conditions using a D-optimal experimental design. Food Chem. 2016, 194, 780–786. [Google Scholar] [CrossRef]
- Fang, Y.; Li, J.; Zhu, X.; Li, X.; Xu, S.; Wu, H.; Zhang, H.; Luo, Y. Optimization and purification of natural protein extract from hazelnut press cake and its antioxidant activity. Front. Nutr. 2025, 12, 1636534. [Google Scholar] [CrossRef]
- Noorbakhsh, R.; Mortazavi, S.A.; Sankian, M.; Shahidi, F.; Maleki, S.J.; Nasiraii, L.R.; Falak, R.; Sima, H.R.; Varasteh, A. Influence of processing on the allergenic properties of pistachio nut assessed in vitro. J. Agric. Food Chem. 2010, 58, 10231–10235. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.Z.; Dun, R.Y.; Huang, Z.L.; Liu, W.; Liu, C.M. Optimizing the extraction of protein from cashew nuts. Food Sci. 2014, 35, 18–22. [Google Scholar]
- Guo, Z.B. Somatic embryogenesis and protein expression in maize and carrot. J. Anhui Agric. Sci. 2010, 38, 9979–9980. [Google Scholar]
- Pasini, G.; Curioni, A.; Vegro, M.; Pagani, M.; Masi, A.; Schievano, E.; Antico, A. Extraction and mass spectrometry identification of a major peach allergen Pru p 1. J. Sci. Food Agric. 2012, 92, 570–576. [Google Scholar] [CrossRef] [PubMed]
- Song, J.J.; Zhang, H.Y.; Liu, Z.G.; Ran, P.X. Cloning of the panallergen profilin from lychee fruit and its cross-reactivity with birch pollen profilin Bet v 2. Food Agric. Immunol. 2007, 18, 129–138. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, L.; Li, Y.T.; Wang, X.J.; Hailiqian, T.E.D.H.; Yang, F. Study on the optimum extraction technology of protein from korla pear. J. Food Saf. Qual. 2020, 11, 4643–4648. [Google Scholar]
- Deng, L.L.; Pan, X.Q.; Sheng, J.P.; Shen, L. Optimization of conditions for determining micro-soluble protein content in apple tissue by Coomassie brilliant blue method. Food Sci. 2012, 33, 185–189. [Google Scholar]
- Nikolić, J.; Nešić, A.; Kull, S.; Schocker, F.; Jappe, U.; Gavrović-Jankulović, M. Employment of proteomic and immunological based methods for the identification of catalase as novel allergen from banana. J. Proteomics 2018, 175, 87–94. [Google Scholar] [CrossRef]
- Paschke, A.; Kinder, H.; Zunker, K.; Wigotzki, M.; Weßbecher, R.; Vieluf, D.; Steinhart, H. Characterization of allergens in mango fruit and ripening dependence of the allergenic potency. Food Agric. Immunol. 2001, 13, 51–61. [Google Scholar] [CrossRef]
- Harris, P.W.R.; Lee, D.J.; Brimble, M.A. A slow gradient approach for the purification of synthetic polypeptides by reversed phase high performance liquid chromatography. J. Pept. Sci. 2012, 18, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Mottola, S.; Del Bene, A.; Mazzarella, V.; Cutolo, R.; Boccino, I.; Merlino, F.; Cosconati, S.; Di Maro, S.; Messere, A. Sustainable Ultrasound-Assisted Solid-Phase peptide synthesis (SUS-SPPS): Less Waste, more efficiency. Ultrason. Sonochem 2025, 114, 107257. [Google Scholar] [PubMed]
- Karas, M.; Hillenkamp, F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 1988, 60, 2299–2301. [Google Scholar] [CrossRef]
- Zhao, J.; Camus-Ela, M.; Zhang, L.; Wang, Y.; Rennie, G.H.; Wang, J.; Raghavan, V. A comprehensive review on mango allergy: clinical relevance, causative allergens, cross-reactivity, influence of processing techniques, and management strategies. Compr. Rev. Food Sci. Food Saf. 2024, 23, e13304. [Google Scholar] [CrossRef]
- Kielkopf, C.L.; Bauer, W.; Urbatsch, I.L. Bradford assay for determining protein concentration. Cold Spring Harb. Protoc. 2020, 2020, pdb-prot102269. [Google Scholar] [CrossRef]
- Herman, R.A.; Song, P.; Mirsky, H.P.; Roper, J.M. Evidence-based regulations for bioinformatic prediction of allergen cross-reactivity are needed. Regul. Toxicol. Pharmacol. 2021, 120, 104841. [Google Scholar] [CrossRef]
- Song, J.; Zhang, H.; Liu, Z.; Ran, P. Mango profilin: cloning, expression and cross-reactivity with birch pollen profilin Bet v 2. Mol. Biol. Rep. 2008, 35, 231–237. [Google Scholar] [CrossRef]
- Bastiaan-Net, S.; Reitsma, M.; Cordewener, J.H.G.; van der Valk, J.P.M.; America, T.A.H.P.; Dubois, A.E.J.; Gerth van Wijk, R.; Savelkoul, H.F.J.; de Jong, N.W.; Wichers, H.J. IgE cross-reactivity of cashew nut allergens. Int. Arch. Allergy Immunol. 2019, 178, 19–32. [Google Scholar]
- Yan, H.; Huang, X.; Ma, Z. Progress in understanding hypersensitivity reaction after ingestion of mango fruits. Food Sci. 2017, 38, 305–309. [Google Scholar]
- Tsai, W.C.; Wu, T.C.; Chiang, B.L.; Wen, H.W. Cloning, expression, and purification of recombinant major mango allergen Man i 1 in Escherichia coli. Protein Expr. Purif. 2017, 130, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Cao, H. Establishment of a mouse model for mango allergy and identification of a novel major allergen chitinase. Master’s thesis, Shenzhen University, Shenzhen, China, 2018. [Google Scholar]
- Zhang, S.; Liu, Z.B.; Wu, Z.M.; Yan, H.Q. Sequence and B-cell epitope analysis of mango allergen chitinase. J. Guizhou Norm. Univ. (Nat. Sci.) 2018, 36, 40–44. [Google Scholar]
- Paschke, A.; Kinder, H.; Zunker, K.; Wigotzki, M.; Steinhart, H.; Wessbecher, R.; Vieluf, I. Characterization of cross-reacting allergens in mango fruit. Allergy 2001, 56, 237–242. [Google Scholar] [CrossRef]
- Funes, E.; Millan, J.M.; Pagán, J.A. Allergy to anarcadiaceae. Ident. Allerg. 1999, 14, 82–89. [Google Scholar]
- Cardona, E.E.G.; Heathcote, K.; Teran, L.M.; Righetti, P.G.; Boschetti, E.; D’Amato, A. Novel low-abundance allergens from mango via combinatorial peptide libraries treatment: A proteomics study. Food Chem. 2018, 269, 652–660. [Google Scholar] [CrossRef] [PubMed]
- Aalberse, R.C. Assessment of allergen cross-reactivity. Clin. Mol. Allergy 2007, 5, 2. [Google Scholar] [CrossRef]
- Pekar, J.; Ret, D.; Untersmayr, E. Stability of allergens. Mol. Immunol. 2018, 100, 14–20. [Google Scholar] [CrossRef] [PubMed]
Figure 1.
Standard curve ofmango and cross allergen protein. Note: The abscissa represents protein concentration (mg/mL), and the ordinate represents OD value.
Figure 1.
Standard curve ofmango and cross allergen protein. Note: The abscissa represents protein concentration (mg/mL), and the ordinate represents OD value.
Figure 2.
Western blot of mango with mango allergic patients serum antibody. (a) Immunoblot with positive serum; (b) Immunoblot with negative control serum.
Figure 2.
Western blot of mango with mango allergic patients serum antibody. (a) Immunoblot with positive serum; (b) Immunoblot with negative control serum.
Figure 3.
Western blot of wheat and peanut with mango allergic patients serum antibody. (a) Immunoblot with positive serum; (b) Immunoblot with negative control serum; 1: wheat, 2: peanut.
Figure 3.
Western blot of wheat and peanut with mango allergic patients serum antibody. (a) Immunoblot with positive serum; (b) Immunoblot with negative control serum; 1: wheat, 2: peanut.
Figure 4.
Western blot of cashew and pistachio with mango allergic patients’ serum antibody. (a) Immunoblot with positive serum; (b) Immunoblot with negative control serum; 1: cashew, 2: pistachio.
Figure 4.
Western blot of cashew and pistachio with mango allergic patients’ serum antibody. (a) Immunoblot with positive serum; (b) Immunoblot with negative control serum; 1: cashew, 2: pistachio.
Figure 5.
Western blot of hazelnut and almond with mango allergic patients serum antibody. (a) Immunoblot with positive serum; (b) Immunoblot with negative control serum; 1: hazelnut, 2: almond.
Figure 5.
Western blot of hazelnut and almond with mango allergic patients serum antibody. (a) Immunoblot with positive serum; (b) Immunoblot with negative control serum; 1: hazelnut, 2: almond.
Figure 6.
Western blot of litchi, bananas and apples with mango allergic patients serum antibody. (a) Immunoblot with positive serum; (b) Immunoblot with negative control serum; 1: lychee, 2: banana, 3: apple.
Figure 6.
Western blot of litchi, bananas and apples with mango allergic patients serum antibody. (a) Immunoblot with positive serum; (b) Immunoblot with negative control serum; 1: lychee, 2: banana, 3: apple.
Figure 7.
Western blot of carrots, pears, crystal pears, peaches, and shrimps with mango allergic patients’ serum antibody. (a) Immunoblot with positive serum; (b) Immunoblot with negative control serum; 1: carrot, 2: fragrant pear, 3: crystal pear, 4: peach, 5: shrimp.
Figure 7.
Western blot of carrots, pears, crystal pears, peaches, and shrimps with mango allergic patients’ serum antibody. (a) Immunoblot with positive serum; (b) Immunoblot with negative control serum; 1: carrot, 2: fragrant pear, 3: crystal pear, 4: peach, 5: shrimp.
Figure 8.
Results of serum recognition epitopes in mango allergy patients. Note: Chitinase AA80–88 represents the AA80–88 sequence of mango chitinase; Tri a 27 AA37–45 represents the AA37–45 sequence of wheat Tri a 27; Man i 3 AA31–51 represents the AA31–51 sequence of mango profilin; Ara h 5 AA31–50 represents the AA31–50 sequence of peanut Ara h 5; Man i 3 AA50–65 represents the AA50–65 sequence of mango profilin; Ara h 5 AA52–65 represents the AA52–65 sequence of peanut Ara h 5; Man i 3 AA103–117 represents the AA103–117 sequence of mango profilin; Ara h 5 AA104–117 represents the AA104–117 sequence of peanut Ara h 5; Man i 3 AA15–22 represents the AA15–22 sequence of mango profilin; Pis v 1 AA65–72 represents the AA65–72 sequence of pistachio Pis v 1.
Figure 8.
Results of serum recognition epitopes in mango allergy patients. Note: Chitinase AA80–88 represents the AA80–88 sequence of mango chitinase; Tri a 27 AA37–45 represents the AA37–45 sequence of wheat Tri a 27; Man i 3 AA31–51 represents the AA31–51 sequence of mango profilin; Ara h 5 AA31–50 represents the AA31–50 sequence of peanut Ara h 5; Man i 3 AA50–65 represents the AA50–65 sequence of mango profilin; Ara h 5 AA52–65 represents the AA52–65 sequence of peanut Ara h 5; Man i 3 AA103–117 represents the AA103–117 sequence of mango profilin; Ara h 5 AA104–117 represents the AA104–117 sequence of peanut Ara h 5; Man i 3 AA15–22 represents the AA15–22 sequence of mango profilin; Pis v 1 AA65–72 represents the AA65–72 sequence of pistachio Pis v 1.
Table 1.
Protein concentration results of mango and mango cross allergen protein.
Table 1.
Protein concentration results of mango and mango cross allergen protein.
| |
OD595nm |
Protein Concentrati (mg/mL) |
| Mango |
0.253±0.004 |
0.389±0.004 |
| Wheat |
0.098±0.018 |
1.495±0.129 |
| Peanut |
0.255±0.033 |
17.607±1.017 |
| Shrimp |
0.245±0.012 |
15.059±0.359 |
| Almond |
0.200±0.006 |
12.247±0.204 |
| Hazelnut |
0.486±0.008 |
37.372±0.319 |
| Pistachio |
0.342±0.033 |
7.875±0.339 |
| Cashew |
0.358±0.009 |
5.498±0.070 |
| Carrot |
0.038±0.007 |
0.056±0.006 |
| Peach |
0.043±0.002 |
0.065±0.002 |
| Lychee |
0.118±0.009 |
0.180±0.007 |
| Crystal Pear |
0.270±0.008 |
0.415±0.006 |
| Fragrant Pear |
0.164±0.009 |
0.251±0.007 |
| Apple |
0.048±0.017 |
0.072±0.012 |
| Banana |
0.058±0.008 |
0.088±0.006 |
Table 2.
Prediction results of mango cross-reactive allergens.
Table 2.
Prediction results of mango cross-reactive allergens.
| Mango Allergen |
Other Allergen |
Max score |
Total score |
Query cover |
E value |
Per ident |
AA length |
| Glyceraldehyde-3-phosphate dehydrogenase |
Wheat Tri a 34 |
602 |
602 |
98% |
0.0 |
86.23% |
337 |
| Chitinase |
Wheat Tri a 37 |
18.5 |
18.5 |
9% |
0.042 |
54.55% |
240 |
| Chitinase |
Wheat Tri a 27 |
20.4 |
20.4 |
17% |
0.023 |
32.56% |
202 |
| Chitinase |
Hazelnut Cor a 12 |
21.2 |
21.2 |
13% |
0.009 |
33.33% |
159 |
| Chitinase |
Banana Mus a 2 |
172 |
172 |
96% |
4e-57
|
41.42% |
318 |
| Profilin |
Wheat Tri a 12 |
216 |
216 |
99% |
7e-79
|
75.38% |
131 |
| Profilin |
Wheat Tri a 29 |
18.1 |
18.1 |
13% |
0.031 |
33.33% |
120 |
| Profilin |
Wheat Tri a 37 |
18.1 |
18.1 |
16% |
0.026 |
40.91% |
111 |
| Profilin |
Peanut Ara h 5 |
219 |
219 |
100% |
2e-80 |
77.1% |
131 |
| Profilin |
Almond Pru du 4 |
228 |
228 |
100% |
7e-84 |
80.92% |
131 |
| Profilin |
Shrimp Cra c 6 |
18.9 |
18.9 |
10% |
0.024 |
50.00% |
150 |
| Profilin |
Hazelnut Cor a 2 |
227 |
227 |
100% |
3e-83 |
80.92% |
131 |
| Profilin |
Hazelnut Cor a 13 |
19.2 |
19.2 |
32% |
0.013 |
34.09% |
140 |
| Profilin |
Pistachio Pis v 1 |
18.9 |
18.9 |
14% |
0.019 |
42.11% |
149 |
| Mango |
Cashew |
—— |
|
|
|
|
|
| Profilin |
Peach Pru p 4 |
230 |
230 |
100% |
1e-84 |
81.68% |
131 |
| Profilin |
Lychee Lit c 1 |
237 |
237 |
100% |
2e-87 |
83.97% |
131 |
| Profilin |
Apple Mal d 4 |
227 |
227 |
100% |
1e-83 |
79.39% |
131 |
| Profilin |
Banana Mus a 1 |
228 |
228 |
100% |
1e-83 |
81.68% |
131 |
| Profilin |
Pear Pyr c 4 |
237 |
237 |
100% |
2e-87 |
85.50% |
131 |
| Profilin |
Carrot Dau c 4 |
213 |
213 |
100% |
6e-78 |
75.37% |
134 |
| β-1,3- Glucanase |
Banana Mus a 5 |
201 |
201 |
93% |
5e-69 |
57.65% |
340 |
Table 3.
Prediction results of similar series.
Table 3.
Prediction results of similar series.
| Mango Chitinase Antigenic Sequence |
Wheat Tri a 27 |
Score |
Expect |
Identities |
Positives |
Gaps |
| RDGFLNAAN(AA80-88) |
RDGLLDAAN(AA37-45) |
23.5 bits (48) |
2e-07 |
7/9(78%) |
8/9(88%) |
0/9(0%) |
| Mango Man i 3 Antigenic Sequence
|
Peanut Ara h 5
|
Score |
Expect |
Identities |
Positives |
Gaps |
| SVWAQSANFPKLNPEEITAIN(AA31-51) |
SVWTESPNFPKFKPEEIAGI(AA31-50) |
43.5 bits(95) |
6e-12 |
13/20(65%) |
14/20(70%) |
0/20(0%) |
| INKDFDEPGSLAPTGL(AA50-65) |
KDFEEPGHLAPTGL(AA52-65) |
38.4 bits(83) |
2e-10 |
12/14(86%) |
13/14(92%) |
0/14(0%) |
| QGEPGAVIRGKKGPGGVTVKK(AA76-96) |
QGEPGVVIRGKKGTGGITIKK(AA76-96) |
52.4 bits(116) |
4e-15 |
17/21(81%) |
19/21(90%) |
0/21(0%) |
| IGIYDEPMTPGQCNM(AA103-117) |
IYDEPMTPGQCNL(AA104-117) |
48.6 bits(107) |
3e-14 |
13/14(93%) |
14/14(100%) |
0/14(0%) |
| Mango Man i 3 Antigenic Sequence |
Pistachio Pis v 1 |
Score |
Expect |
Identities |
Positives |
Gaps |
| IEGHHLTA(AA15-22) |
QDGHSLTA(AA65-72)
|
16.3 bits(31)
|
1e-04
|
5/7(71%)
|
6/7(85%)
|
0/7(0%)
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |