Submitted:
24 December 2025
Posted:
26 December 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Reagents and Instruments
2.2. Extraction of Genomic DNA and Sample Preparation for ARMS-PCR
2.3. Preparation of AuNPs and AuNP Labeled Anti-FITC Antibody Conjugates for LFS
2.4. Assembly of LFS
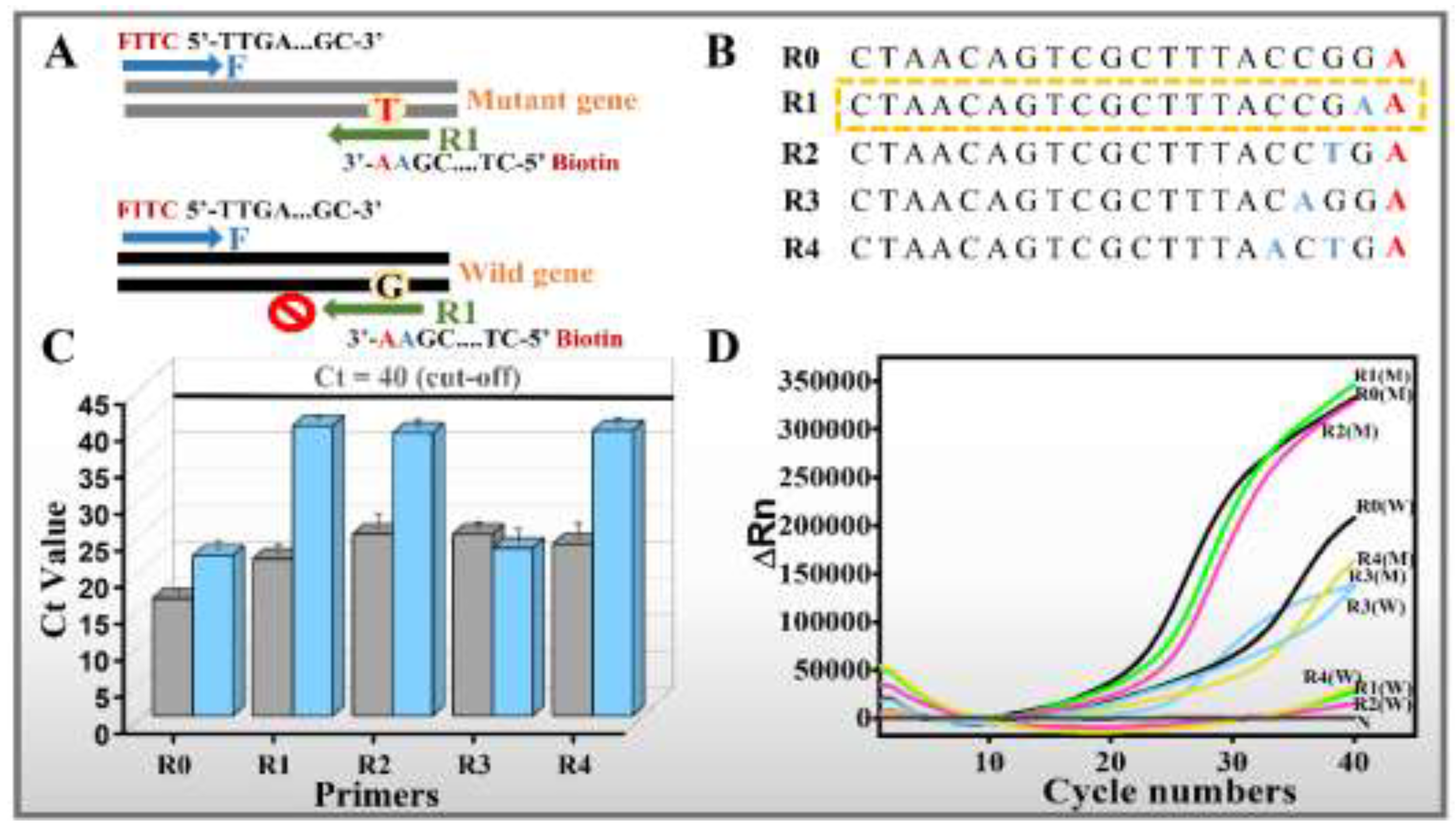
2.5. Primer Design for the Accurate Identification of SNP in the Target Eae Gene
2.6. ARMS-qPCR for Primer Design Evaluation
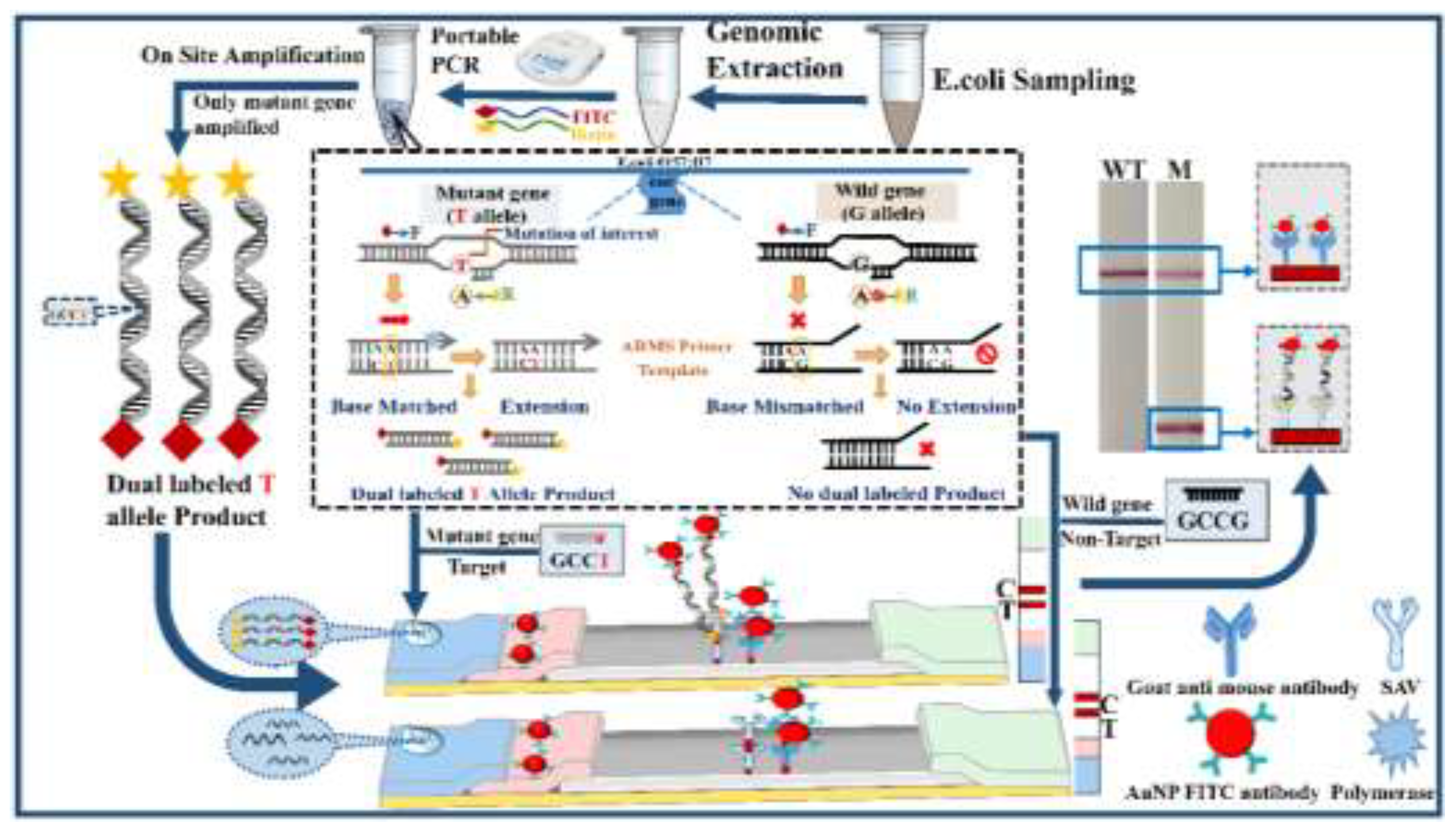
2.7. ARMS-PCR Assisted Rapid and Visual Identification Of SNP in Eae Gene of E. coli with the Lateral Flow Strip
3. Results
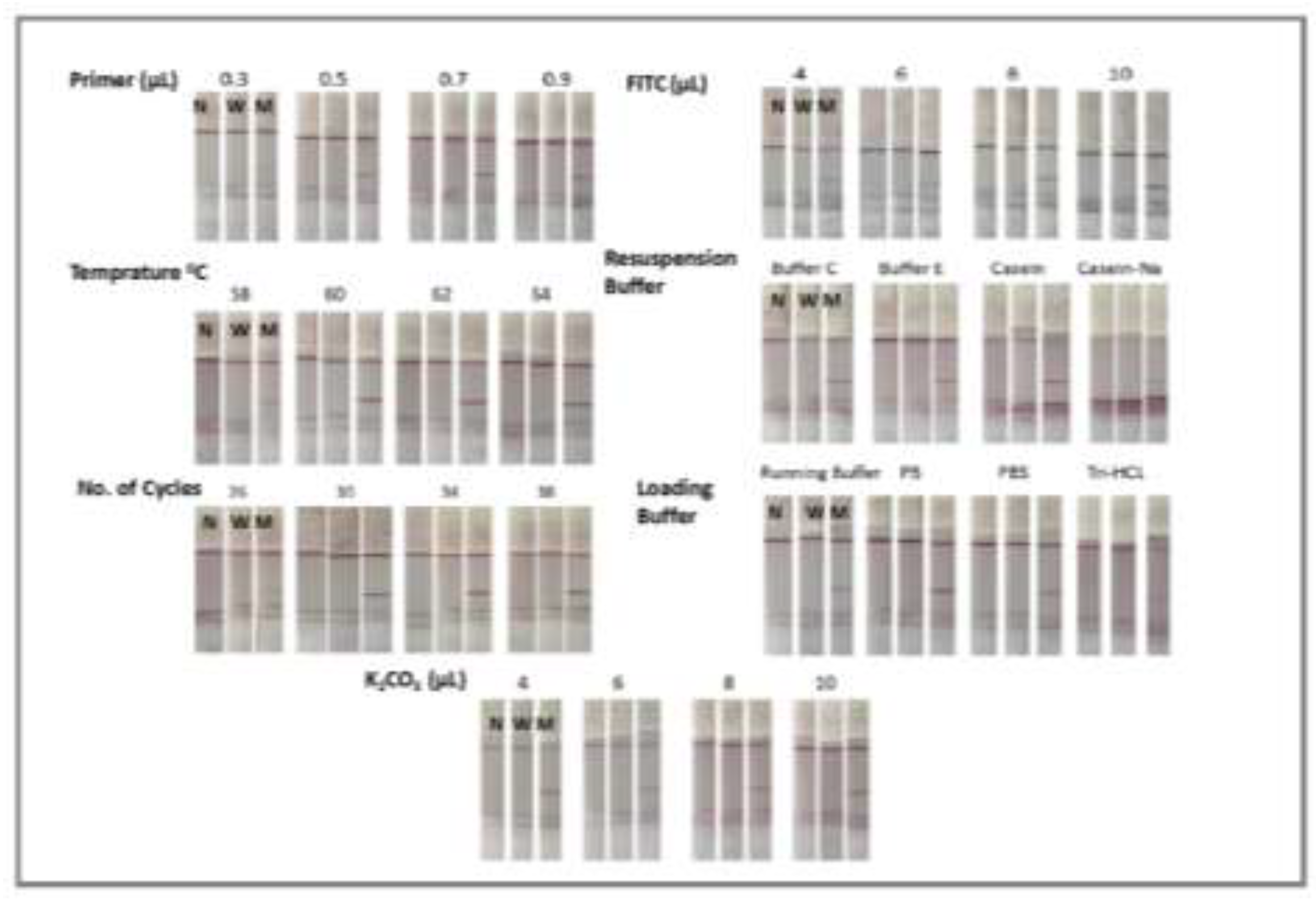
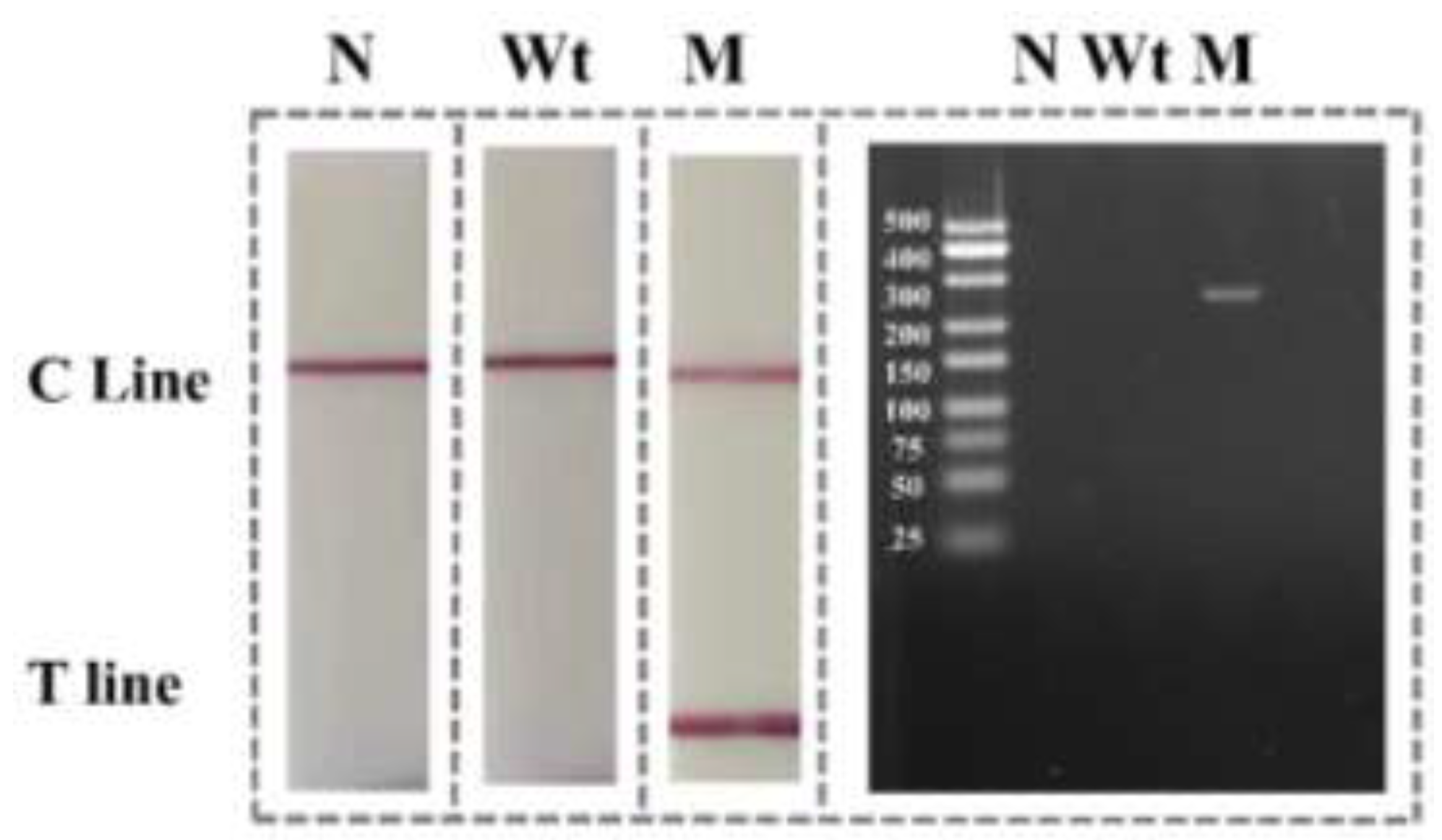
3.1. Detection of Mutation in Eae Gene of E. coli by the Designed ARMS-PCR Mediated Lateral Flow Strip in the Visual Mode
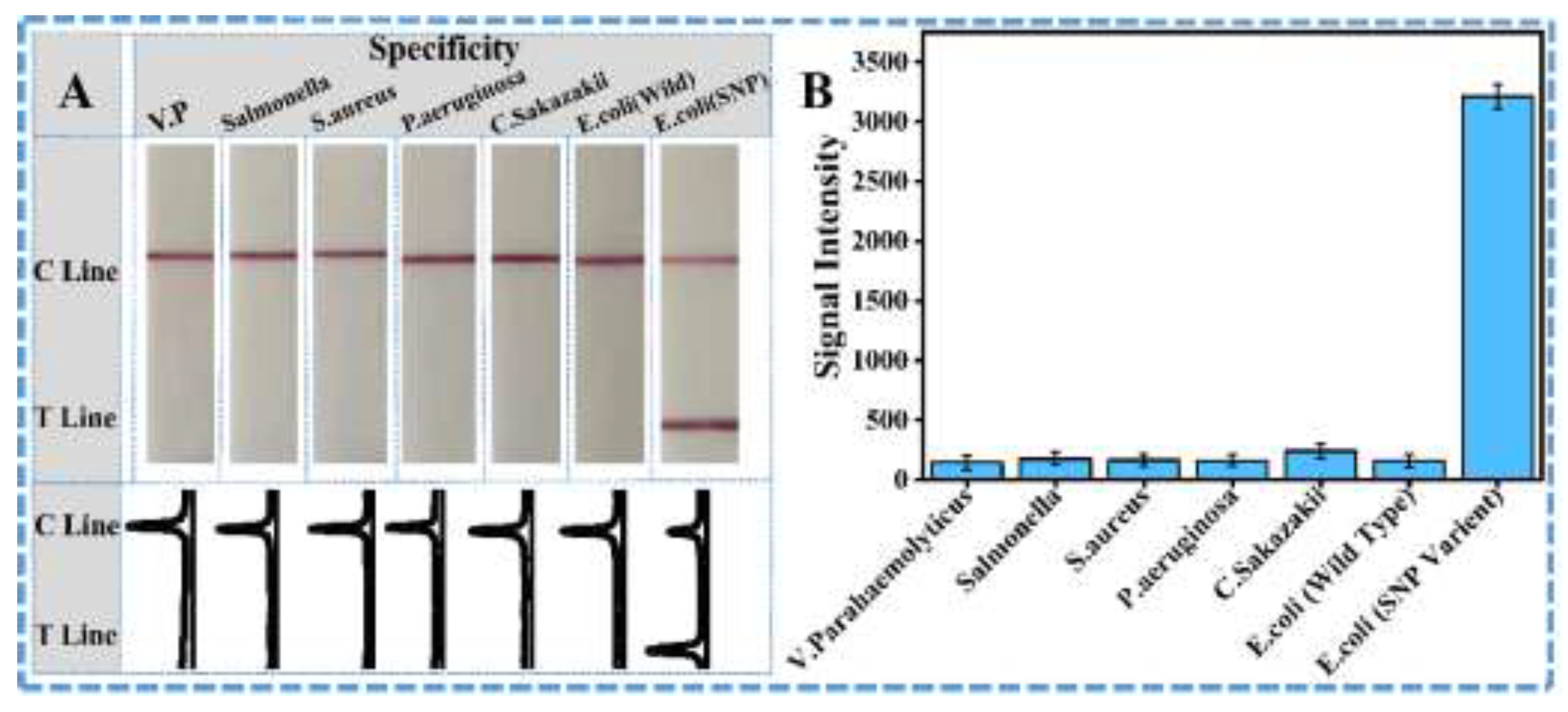
3.1. Possibility Verification of the Designed ARMS-PCR Mediated LFS Assay for Precise Visual SNP Identification
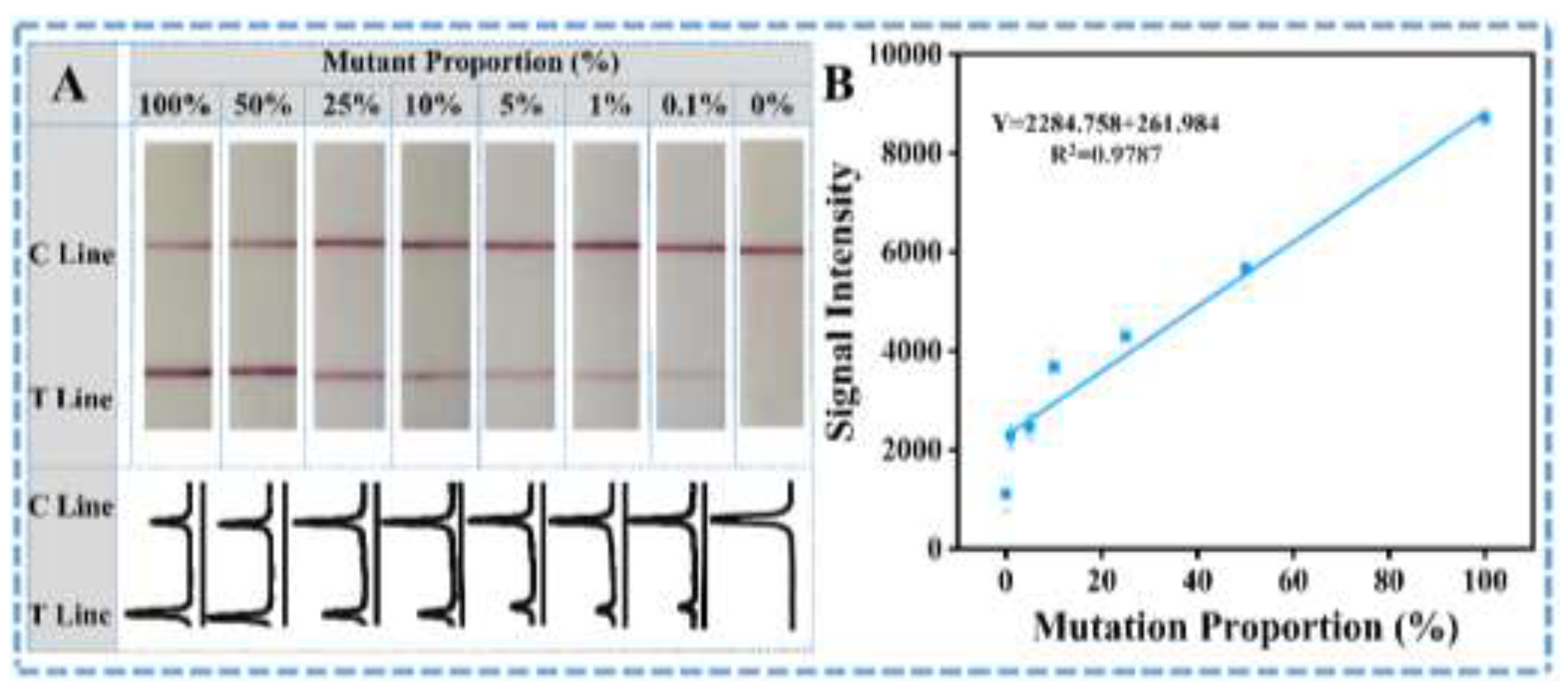
3.3. Visual-Identification Performance of the SNP in the Mutant Genes with the Designed ARMS-PCR-LFS
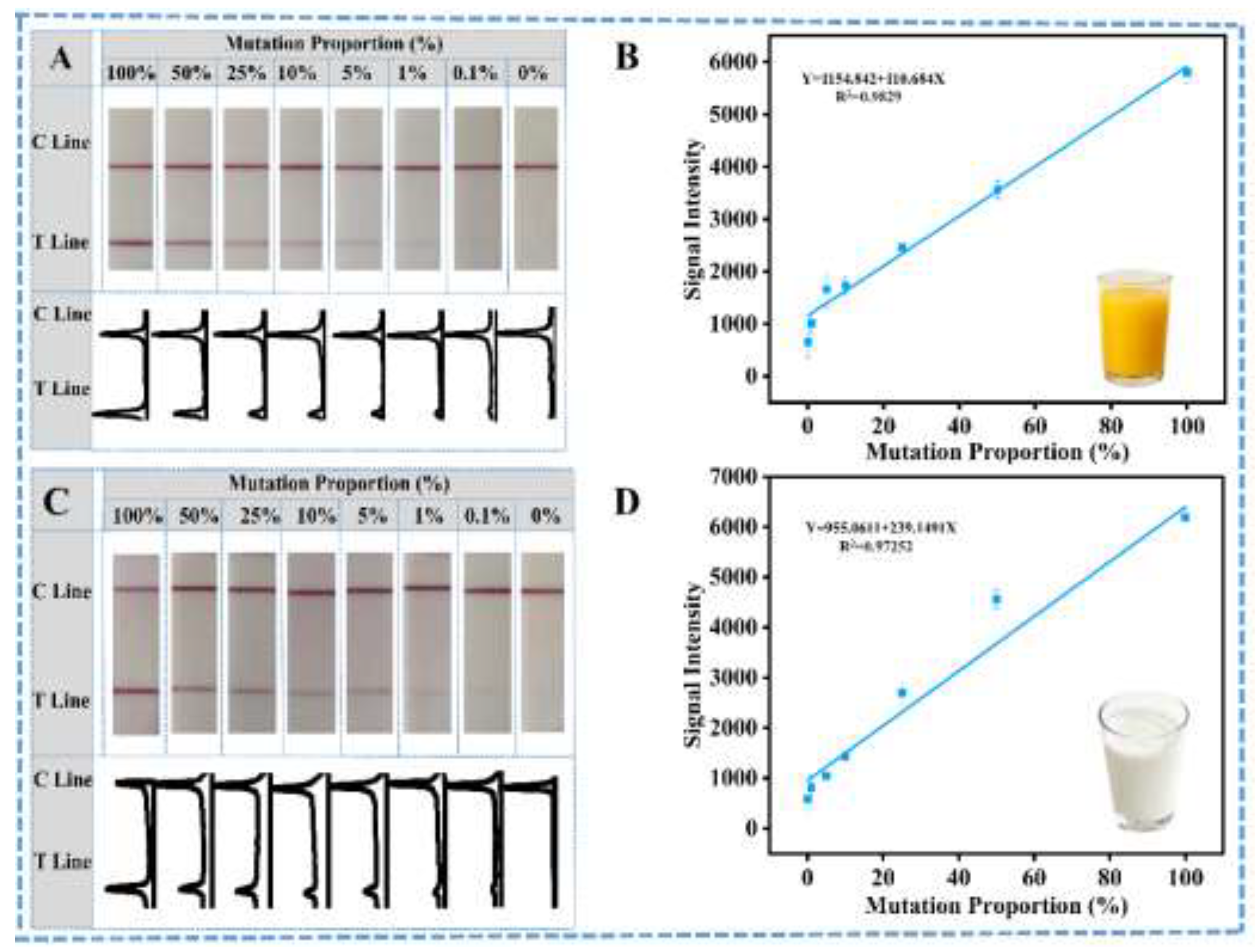
3.4. Practical Detection Research of SNP Mutated Eae Genes of E. coli in Spiked Wild-Type E.coli Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
Appendix A.1
| Primers and Probe | Primer and probe Sequence | Size(bp) | Method |
| F PCR R1 |
TTGATCAAACCAAGGCCAGC CTAACAGTCGCTTTACCGAA |
250 | ARMS-PCR |
| F LFS R1 |
FITC-TTGATCAAACCAAGGCCAGC Biotin-CTAACAGTCGCTTTACCGAA |
250 | ARMS PCR-LFS |
| F qPCR R1 |
TTGATCAAACCAAGGCCAGC CTAACAGTCGCTTTACCGAA |
250 | ARMS-qPCR |
| P- qPCR | 5’-FAMTCCCGTGGTTGCTTGCGTTTGAGACT-BHQ1 |
References
- Lee, H.; Yoon, Y. Etiological agents implicated in foodborne illness world wide. Food science of animal resources 2021, 41, 1. [Google Scholar] [CrossRef] [PubMed]
- Kariuki, S. Global burden of antimicrobial resistance and forecasts to 2050. The Lancet 2024, 404, 1172–1173. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Alsayeqh, A.F. Review of major meat-borne zoonotic bacterial pathogens. Frontiers in Public Health 2022, 10, 1045599. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO bacterial priority pathogens list, 2024: Bacterial pathogens of public health importance, to guide research, development, and strategies to prevent and control antimicrobial resistance. 2024: World Health Organization.
- Odo, S.E.; Uchechukwu, C.F.; Ezemadu, U.R. Foodborne diseases and intoxication in Nigeria: Prevalence of Escherichia coli 0157: H7, Salmonella, Shigella and Staphylococcus aureus. J Adv Microbiol 2021, 20, 84–94. [Google Scholar] [CrossRef]
- Bonten, M.; et al. Epidemiology of Escherichia coli bacteremia: A systematic literature review. Clinical Infectious Diseases 2021, 72, 1211–1219. [Google Scholar] [CrossRef]
- Mueller, M.; Tainter, C.R. Escherichia coli infection, in StatPearls [Internet]. 2023, StatPearls Publishing.
- Pokharel, P.; Dhakal, S.; Dozois, C.M. The diversity of Escherichia coli pathotypes and vaccination strategies against this versatile bacterial pathogen. Microorganisms 2023, 11, 344. [Google Scholar] [CrossRef]
- Hasan, J.M.; Najim, S.S. A review of the Prevalence of Enterohemorrhagic E. coli in Iraq. Journal of Biotechnology Research Center 2024, 18, 33–39. [Google Scholar] [CrossRef]
- Heo, N.; et al. The epidemiological and clinical characteristics of the largest outbreak of enterohemorrhagic Escherichia coli in Korea. J. Korean Med. Sci. 2023, 38. [Google Scholar] [CrossRef]
- Shuvra, S. Isolation of Shiga Toxin Producing Escherichia coli 0157: H7 from Environmental and Clinical Samples in Dhaka City—A Review. J Immuno Allerg 2023, 4, 40–62. [Google Scholar] [CrossRef]
- Jiang, L.; et al. Virulence-related O islands in enterohemorrhagic Escherichia coli O157: H7. Gut microbes 2021, 13, 1992237. [Google Scholar] [CrossRef]
- Liu, Q.; et al. Genetic diversity and expression of Intimin in Escherichia albertii isolated from humans, animals, and food. Microorganisms 2023, 11, 2843. [Google Scholar] [CrossRef]
- Perraud, Q.; Sperandio, V. Enterohemorrhagic E. coli (EHEC) and the microbiome. PLoS pathogens 2025, 21, e1013224. [Google Scholar] [CrossRef] [PubMed]
- Rosauer, M.L.; et al. Validation of the 3M™ Molecular Detection Assay 2-STEC Gene Screen (stx and eae) for the Detection of Shiga Toxin Gene (stx and eae) in Fresh Raw Beef Trim, Fresh Raw Ground Beef and Fresh Spinach: AOAC Performance Tested Method SM 071902. J. AOAC Int. 2022, 105, 902–914. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; et al. Molecular characteristics of eae-positive clinical Shiga toxin-producing Escherichia coli in Sweden. Emerging microbes & infections 2020, 9, 2562–2570. [Google Scholar] [CrossRef]
- Rahman, M.-M.; Lim, S.-J.; Park, Y.-C. Development of single nucleotide polymorphism (SNP)-based triplex PCR marker for serotype-specific Escherichia coli detection. Pathogens 2022, 11, 115. [Google Scholar] [CrossRef]
- Cheng, C.; Fei, Z.; Xiao, P. Methods to improve the accuracy of next-generation sequencing. Frontiers in bioengineering and biotechnology 2023, 11, 982111. [Google Scholar] [CrossRef]
- Yang, H.; et al. Sensitive detection of a single-nucleotide polymorphism in foodborne pathogens using CRISPR/Cas12a-signaling ARMS-PCR. Journal of Agricultural and Food Chemistry 2022, 70, 8451–8457. [Google Scholar] [CrossRef]
- Han, Y.; Wu, H. Fluorescent primers amplification refractory mutation system qPCR (FP ARMS-qPCR) for MTHFR C677T SNP genotyping. Molecular Biology Reports 2024, 51, 1122. [Google Scholar] [CrossRef]
- Muneeswaran, K.; et al. Genotyping SNPs and indels: A method to improve the scope and sensitivity of high-resolution melt (HRM) analysis based applications. Clinica Chimica Acta 2024, 562, 119897. [Google Scholar] [CrossRef]
- Kishi, J.Y.; et al. SABER amplifies FISH: Enhanced multiplexed imaging of RNA and DNA in cells and tissues. Nature methods 2019, 16, 533–544. [Google Scholar] [CrossRef]
- Dardani, I.; et al. ClampFISH 2.0 enables rapid, scalable amplified RNA detection in situ. Nature methods 2022, 19, 1403–1410. [Google Scholar] [CrossRef]
- Liu, X.; et al. In situ Cas12a-based allele-specific PCR for imaging single-nucleotide variations in foodborne pathogenic bacteria. Analytical Chemistry 2024, 96, 2032–2040. [Google Scholar] [CrossRef]
- Islam, M.T.; et al. A rapid and cost-effective multiplex ARMS-PCR method for the simultaneous genotyping of the circulating SARS-CoV-2 phylogenetic clades. Journal of medical virology 2021, 93, 2962–2970. [Google Scholar] [CrossRef]
- Mesrian Tanha, H.; et al. Modified tetra-primer ARMS PCR as a single-nucleotide polymorphism genotyping tool. Genetic testing and molecular biomarkers 2015, 19, 156–161. [Google Scholar] [CrossRef]
- Khehra, N.; Padda, I.S.; Swift, C.J. Polymerase chain reaction (PCR). 2023.
- Kinyua, D.M.; et al. Advancements and Applications of Lateral Flow Assays (LFAs): A Comprehensive Review. Sensors 2025, 25, 5414. [Google Scholar] [CrossRef]
- Zhao, L.; et al. A PCR-based lateral flow assay for the detection of Turkey ingredient in food products. Food Control 2020, 107, 106774. [Google Scholar] [CrossRef]
- Li, B.; et al. Filtration assisted pretreatment for rapid enrichment and accurate detection of Salmonella in vegetables. Food Science and Human Wellness 2023, 12, 1167–1173. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





