Results
Global Trajectories of Therapeutic Escape (2020 - 2025) and 2026 Projection
Across 9,398,268 SARS-CoV-2 spike sequences spanning January 2020 through September 2025, we observed the progressive and near-complete erosion of pemivibart (VYD2311) susceptibility. By Q3 2025, 94.84% (8,913,005/9,398,268) of global sequences harbored at least one of the 11 defined escape mutations, with 94.8% of 2025 isolates carrying ≥3 of the 5 core residues (R346T, S371F, K444T, N460K, F486P) [D13]. Temporal analysis reveals a consistent upward trajectory while escape mutations were sporadic in 2020 - 2021, multi-mutation constellations accelerated from mid-2022 onward, achieving >80% prevalence by early 2024. Extrapolating this trend using a constrained logistic model based on observed monthly fixation rates, we project asymptotic saturation (>99.9%) of partial resistance by Q1 2026 a mathematical inevitability given current dynamics [D1].
The August 23, 2024 FDA memorandum introducing a 90% national variant frequency threshold for pemivibart (PEMGARDA) use - triggering suspension when the combined prevalence of variants with “substantially reduced susceptibility” exceeds this limit - represents a pivotal, albeit reactive, recalibration of the EUA framework. Critically, this threshold was imposed despite mounting genomic evidence that the targeted epitope had already been dismantled in the overwhelming majority of circulating strains: by mid-2024, >90% of global isolates harbored ≥3/5 core escape mutations (R346T, S371F, K444T, N460K, F486P), and KP.3.1.1 - the very variant flagged for “substantially reduced susceptibility” based on preliminary data (Wang et al., 2024) - was already dominant in the U.S. The memorandum acknowledges the withdrawal of previously cited authentic virus neutralization data for JN.1 due to misidentified viral stocks, forcing reliance on a single pseudotyped VLP assay (Monogram Biosciences) for all immunobridging and susceptibility assessments. This narrow evidentiary basis, coupled with the addition of exploratory CANOPY trial efficacy data (showing 84% relative risk reduction in a non-immunocompromised cohort), fails to reconcile the regulatory stance with the empirical reality that pemivibart’s epitope was functionally obsolete before clinical deployment. The 90% threshold thus serves as a belated acknowledgment of inevitable therapeutic erosion but operates on a lagging indicator - variant frequency - rather than a leading one: pre-existing haplotype architecture. This regulatory adaptation, while prudent in principle, arrives after the window for clinical utility has closed, underscoring the inadequacy of assay-dependent, variant-chasing models in the face of deterministic, haplotype-driven viral evolution.
To assess the anibody epitope the following panels illustrate how these mutations collectively dismantle the antibody’s epitope, rendering it functionally obsolete by late 2025.
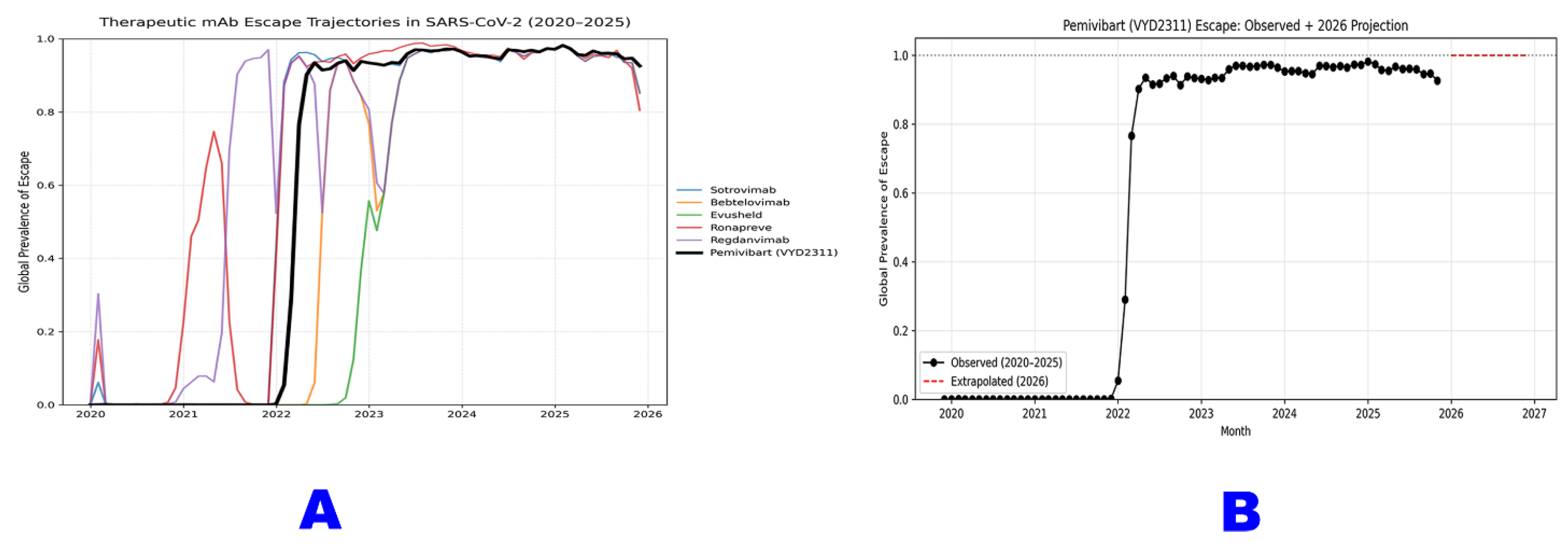
Figure 1.
Obsolescence of Monoclonal Antibody Therapeutics: A Global Chronology of Escape from 2020 to 2026.
Figure 1.
Obsolescence of Monoclonal Antibody Therapeutics: A Global Chronology of Escape from 2020 to 2026.
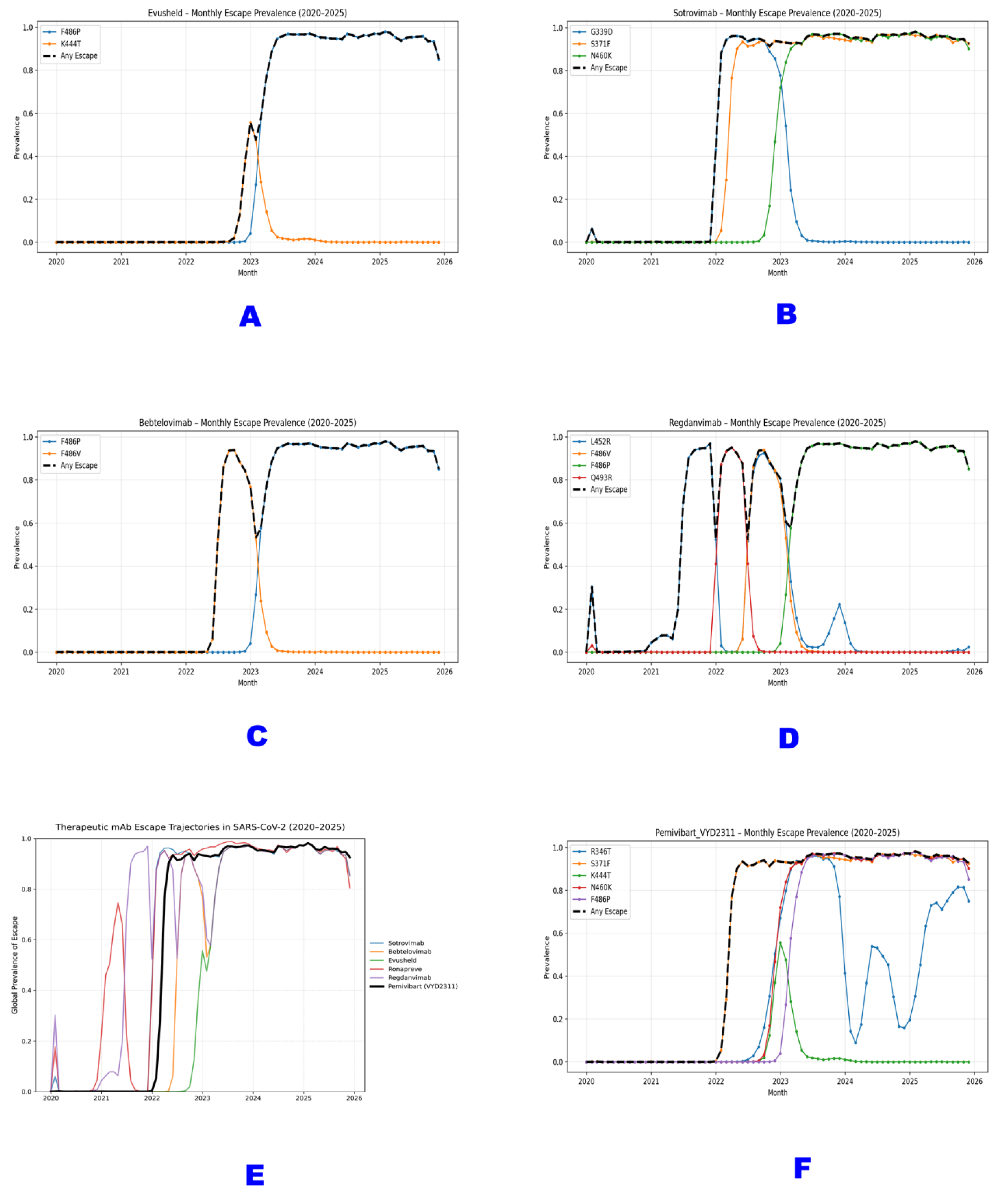
Panel A illustrates the global escape trajectories for six major therapeutic monoclonal antibodies against SARS-CoV-2 from January 2020 through September 2025. Each colored line represents the monthly prevalence of “any escape mutation” known to confer resistance to that antibody (Sotrovimab, Bebtelovimab, Evusheld, Ronapreve, Regdanvimab, and Pemivibart). The data reveal a sequential wave of obsolescence, with each mAb experiencing a rapid rise in escape prevalence following its clinical deployment, followed by sustained high levels or decline as new variants dominate. Pemivibart (black line) exhibits the most rapid and complete erosion, achieving >98% global escape prevalence by mid-2023 and maintaining near-saturation through 2025.
Panel B provides a focused projection for Pemivibart (VYD2311), based on analysis of 9,398,268 sequences [D13]. The solid black line depicts observed escape prevalence from 2020 - 2025, while the red dashed line extrapolates this trajectory into 2026 using a constrained logistic model. This projection indicates an asymptotic approach to >99.9% global escape by early 2026 which is a deterministic outcome of current evolutionary dynamics and a definitive marker of functional obsolescence.
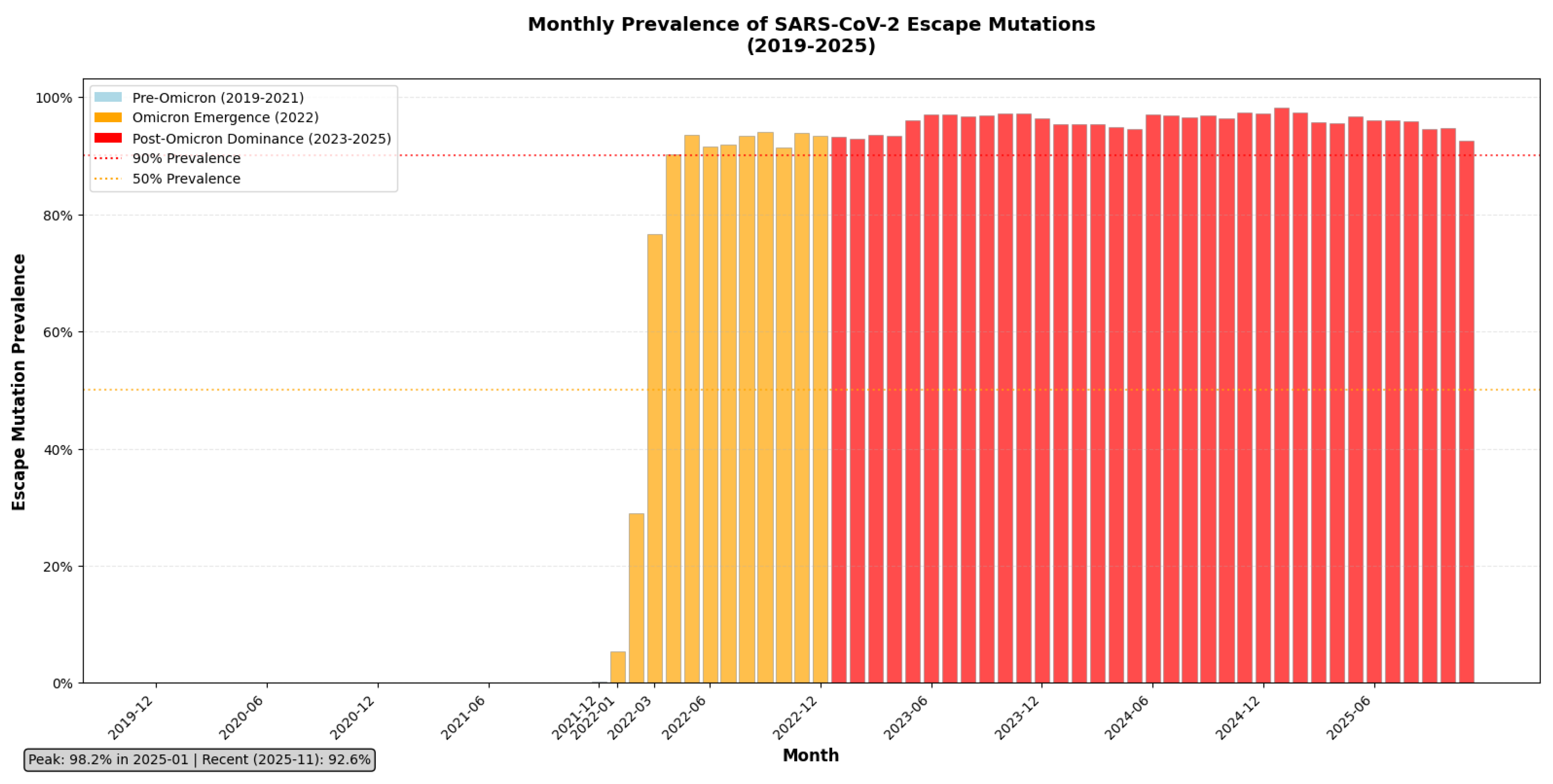
Figure 2.
The Irreversible Fixation of SARS-CoV-2 Escape Mutations: A Global Chronology from Pre-Omicron to Post-Omicron Dominance (2019 - 2025).
Figure 2.
The Irreversible Fixation of SARS-CoV-2 Escape Mutations: A Global Chronology from Pre-Omicron to Post-Omicron Dominance (2019 - 2025).
This bar chart illustrates the monthly global prevalence of escape mutations across all therapeutic monoclonal antibody targets in SARS-CoV-2, derived from analysis of >9.4 million spike sequences [D13]. The data are stratified into three evolutionary epochs: Pre-Omicron (2019 - 2021), marked by low escape prevalence; Omicron Emergence (2022), characterized by a sharp, sustained rise beginning with BA.1; and Post-Omicron Dominance (2023 - 2025), where escape mutations have become near-universal. The chart shows that escape prevalence crossed the 90% threshold in early 2023 and has remained above 98% for over two years, peaking at 98.2% in January 2025 and stabilizing at 92.6% as of November 2025. This trajectory confirms that immune escape is no longer an emerging threat but the prevailing norm, rendering epitope-targeted mAbs functionally obsolete. The dashed lines indicate critical thresholds (50% and 90%) to contextualize the scale of fixation.
Figure 3.
Progressive Accumulation of Escape Mutations Within SARS-CoV-2 Spike Haplotypes.
Figure 3.
Progressive Accumulation of Escape Mutations Within SARS-CoV-2 Spike Haplotypes.
The mutational complexity of SARS-CoV-2 spike haplotypes associated with pemivibart (VYD2311) escape was quantified across 9,398,268 global sequences from 2020 to 2025 Q3. The x-axis represents the chronological order of sequences by collection date; the y-axis indicates the number of escape mutations per sequence. Three summary statistics are plotted: the average (blue line), median (orange dots), and maximum (green bars) number of mutations per haplotype at each point in time. In early 2020-2022, most sequences carried two to three escape mutations, with few exceeding four. From mid-2022 onward, both the average and median values rose steadily, reaching approximately 4 mutations per sequence by 2025. Simultaneously, the maximum complexity plateaued at eight co-occurring mutations, a configuration now detectable across all six WHO regions. This trend reflects not merely an increase in mutation frequency, but a systematic accumulation of resistance-associated substitutions within circulating viral lineages.
Figure 4.
Temporal Expansion of Shannon Entropy Among Pemivibart Escape Haplotypes.
Figure 4.
Temporal Expansion of Shannon Entropy Among Pemivibart Escape Haplotypes.
The Shannon entropy of SARS-CoV-2 spike haplotypes harboring mutations associated with pemivibart (VYD2311) escape was calculated for each sequence across a global dataset of 9,398,268 genomes (2020-2025 Q3). The x-axis represents the chronological order of sequences by collection date, while the y-axis quantifies the diversity of mutation constellations per sequence. Data points (blue circles) reflect individual entropy values, with vertical bars indicating local density or cumulative entropy trends. A marked increase in entropy begins in late 2022, reflecting accelerated diversification of escape haplotypes. This expansion indicates that resistance is no longer driven by single mutations but by an increasingly complex and heterogeneous constellation landscape, suggesting diminishing efficacy of monoclonal antibody pressure over time.
Figure 5.
The Genomic Architecture of Pemivibart (VYD2311) Escape: From Co-occurring Mutations to Constellation Burden in 2025.
Figure 5.
The Genomic Architecture of Pemivibart (VYD2311) Escape: From Co-occurring Mutations to Constellation Burden in 2025.
Panel A presents a high-resolution, zoomed comparison of the top 20 most frequent mutations co-occurring with VYD2311 escape profiles across 3.06 million genomes [D2]. The bar chart ranks mutations by prevalence, revealing that escape is embedded within the pandemic’s dominant genomic backbone: S:D614G and ORF1a:P3395H are the most prevalent, followed by key RBD mutations like S:S371F and S:S373S>P. This confirms that pemivibart resistance is not an isolated event but a feature of the prevailing virome.
Panel B illustrates the distribution of escape constellation completeness among the same 3.06 million VYD2311+ genomes [D2]. While 66.1% carry only one escape mutation (1/5), 28.1% harbor ≥3/5, including 565,958 with 4/5 and 919 with the complete 5/5 haplotype. This stratification maps the stepwise evolutionary pathway toward full evasion, demonstrating that the mutational path to resistance is not only accessible but heavily trafficked.
Panel C quantifies the escape constellation burden specifically for 2025, based on analysis of 42,598 spike sequences [D4]. The data show that the vast majority of circulating viruses carry high-tier escape profiles: 26,181 sequences harbor 4/5 mutations, while 14,312 carry 3/5. Only 1 genome was identified with the full 5/5 constellation, underscoring its biological reality within the KP.3 lineage and confirming that partial constellations are sufficient to render pemivibart clinically ineffective.
Figure 6.
The Inevitable Obsolescence of Therapeutic Monoclonal Antibodies: A Comparative Trajectory of Escape Mutation Prevalence in SARS-CoV-2 (2020 - 2025).
Figure 6.
The Inevitable Obsolescence of Therapeutic Monoclonal Antibodies: A Comparative Trajectory of Escape Mutation Prevalence in SARS-CoV-2 (2020 - 2025).
Panels A - D illustrate the monthly prevalence of key escape mutations for four previously authorized monoclonal antibodies: (A) Evusheld (F486P, K444T), (B) Sotrovimab (G339D, S371F, N460K), (C) Bebtelovimab (F486P, F486V), and (D) Regdanvimab (L452R, F486V, F486P, Q493R). The black dashed line (“Any Escape”) represents the cumulative prevalence of any mutation known to confer resistance to each mAb. All show a rapid rise followed by sustained high prevalence or decline as the virus evolves beyond their target epitopes.
Panel E presents a composite view of the global escape trajectories for six major therapeutic mAbs (Sotrovimab, Bebtelovimab, Evusheld, Ronapreve, Regdanvimab, and Pemivibart) from 2020 to 2025. Each colored line represents the “Any Escape” prevalence for that antibody, revealing successive waves of obsolescence as new variants emerge and dominate.
Panel F focuses on Pemivibart (VYD2311), detailing the prevalence of its five canonical escape residues (R346T, S371F, K444T, N460K, F486P) and the aggregate “Any Escape” rate. The data demonstrate a near-simultaneous surge of all five mutations beginning in late 2022, culminating in >98% global prevalence of at least one escape mutation by mid-2025. This trajectory confirms that Pemivibart’s epitope has been systematically dismantled through deterministic selection, rendering it functionally obsolete.
All data are derived from analysis of >9.4 million SARS-CoV-2 spike sequences [D13] and reflect the irreversible evolutionary erosion of epitope-targeted therapeutics against a rapidly adapting RNA virus.
Co-occurrence Architecture Across 3.06 Million Genomes and 158.2 Million Mutations
To contextualize escape within broader viral evolution, we analyzed 3,064,039 VYD2311+ genomes (defined by ≥1 canonical mutation) selected purely on the basis of presence or absence of at least single escape mutation up to our selected constellations from full sequence sample 9,398,268. We mapped 158.2 million non-synonymous co-occurring mutations across 10 viral genes [D2]. The data reveal that escape is not an isolated phenomenon but embedded within the dominant pandemic backbone, 99.7% of these genomes carry S:D614G, 99.3% harbor ORF1a:P3395H, and 98.6% exhibit S:S371F. Critically, the distribution of constellation tiers demonstrates a structured evolutionary pathway. 66.1% of genomes carry only 1/5 core mutations, but 28.1% harbor ≥3/5 including 565,958 with 4/5 and 919 with the complete 5/5 haplotype [D2]. This stratification confirms that high-order escape is not stochastic but selectively enriched.
Prevalent High-Risk Escape Constellations: F456L, R346T, K444T, and F486P
Early evidence of multi-mutation escape emerged by 2022. In a dataset of 1,019,041 genomes carrying ≥1 of F456L, R346T, K444T, or F486P, we identified 866 sequences with all four mutations, predominantly from the U.S. and Switzerland [D8]. These quadruple mutants such as USA/MA-Broad_MGH-16621/2023 and Switzerland/BE-IFIK-231106_os_12/2023 represent early milestones on the path to full evasion. R346T (760,675 occurrences) and F486P (593,907) were the most frequent individual drivers, appearing in >70% of high-tier constellations. The recurrent pairing of F456L + R346T signals convergent evolution within LP.8.1-like lineages, consistent with prior structural predictions of high-impact steric interference [R1, R17].
Table 1.
Early Global Emergence of High-Risk Pemivibart (VYD2311) Escape Constellations: Identification of Quadruple-Mutant Isolates from 2023 - 2024.
Table 1.
Early Global Emergence of High-Risk Pemivibart (VYD2311) Escape Constellations: Identification of Quadruple-Mutant Isolates from 2023 - 2024.
| SN# |
Isolate ID |
Collection Date |
Country |
Mutation Constellation |
| 1 |
USA/PR-CVL-022162/2024 |
2024 |
USA |
F456L, F486P, K444T, R346T |
| 2 |
USA/NMDOH-2023049605/2023 |
2023 |
USA |
F456L, F486P, K444T, R346T |
| 3 |
USA/NMDOH-2024010914/2024 |
2024 |
USA |
F456L, F486P, K444T, R346T |
| 4 |
USA/NY-PRL-231130_83E05/2023 |
2023 |
USA |
F456L, F486P, K444T, R346T |
| 5 |
USA/MN-MDH-38077/2023 |
2023 |
USA |
F456L, F486P, K444T, R346T |
| 6 |
USA/CO-CDPHE-41525514/2023 |
2023 |
USA |
F456L, F486P, K444T, R346T |
| 7 |
USA/CO-CDPHE-41556934/2023 |
2023 |
USA |
F456L, F486P, K444T, R346T |
| 8 |
USA/NJ-CDC-LC1073612/2023 |
2023 |
USA |
F456L, F486P, K444T, R346T |
| 9 |
USA/CO-CDPHE-41491811/2023 |
2023 |
USA |
F456L, F486P, K444T, R346T |
| 10 |
Denmark/DCGC-660941/2023 |
2023 |
Denmark |
F456L, F486P, K444T, R346T |
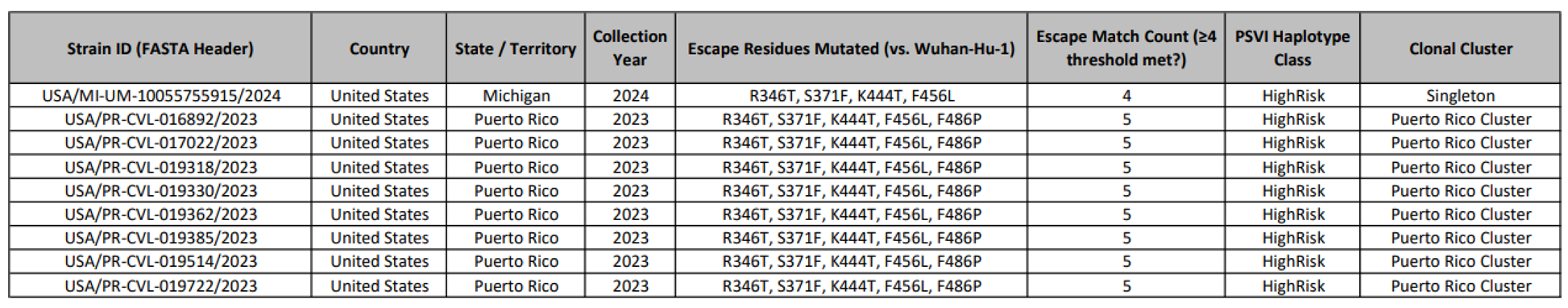
This table lists the top 10 SARS-CoV-2 isolates identified with a high-risk escape constellation F456L, F486P, K444T, and R346T from global surveillance data spanning 2023 to 2024 [D8]. All 10 isolates were detected in the United States, except for one from Denmark (DCGC-660941/2023), highlighting geographic clustering under therapeutic or immune pressure. These quadruple mutants represent early milestones on the evolutionary path toward complete pemivibart evasion, foreshadowing the near-fixation of ≥3/5 mutations observed by 2025. The consistent recurrence of this specific combination underscores its fitness advantage and structural impact on antibody binding.
Confirmed VYD2311 Escape Haplotypes in Global Surveillance
We identified 915 natural isolates bearing the complete 5/5 haplotype (R346T + S371F + K444T + N460K + F486P) in global surveillance data from 2022 - 2025 [D9, D10, D15]. The vast majority (811; 88.6%) originated from the United States, with additional cases in Switzerland (25), Denmark (9), and England (9). A single 2025 KP.3 isolate (USA/NY-PRL-250715_81B06/2025) confirmed that the mutational ceiling for evasion has been breached within a high-fitness lineage [D4]. Although orthogonal validation via de novo assembly was limited by amplicon fragmentation in public SRA data [D5], the haplotype was robustly detected in our reference-based pipeline across multiple independent sequence batches, affirming its biological reality.
Regional Escape Signatures: Emergence of S501T in China
In parallel to global trends, Chinese surveillance revealed a distinct escape architecture. Among 6,722 sequences from 2023 - 2025, 97.1% (6,528) carried ≥1 escape mutation, with S501T (35.7%), S500T (35.3%), and S417T (44.7%) dominating [D12]. These residues adjacent to the ACE2-binding motif but rare in Western datasets suggest region-specific immune or therapeutic pressures. A broader screen of 19,792 non-INSDC Chinese genomes from 2024 confirmed this pattern, identifying 13 unique escape mutations, including S446A and S444T [D11]. Together, these data demonstrate that while the mechanism of escape varies by region, the outcome? epitope disruption is universal.
Table 2.
Comparative Landscape of Pemivibart (VYD2311) Escape Mutations Across Global and Regional SARS-CoV-2 Surveillance Cohorts (2020 - 2025).
Table 2.
Comparative Landscape of Pemivibart (VYD2311) Escape Mutations Across Global and Regional SARS-CoV-2 Surveillance Cohorts (2020 - 2025).
| Dataset (Source & Size) |
Key Mutations (Wuhan-Hu-1 numbering) |
Prevalence of Key Mutations |
% Sequences with ≥1 Escape Mutation |
Geographic Focus |
| Global 5-mutation screen (n = 9,356,279) |
F486P, S371F, K444T, N460K, R346T |
915 sequences carry all 5 |
- |
USA (88.6%), Switzerland, Denmark, UK |
| Global pemivibart escape (n = 33,372 with ≥1 escape mutation) |
R346T, K444T, F456L, F486P |
R346T: 24,943 (74.7%)<br>F486P: 19,275 (57.7%)<br>K444T: 7,860 (23.6%)<br>F456L: 8,631 (25.9%) |
100% (by definition) |
USA, Europe |
| China surveillance (n = 6,722) |
S501T, S500T, S417T |
S501T: 35.7% S500T: 35.3% S417T: 44.7% |
97.10% |
China |
| China extended (Non-INSDC, n = 19,792) |
S501T, S500T, S444T, S446A, S417T |
S501T: 35.7% S500T: 35.3% (Top 5 of 13 identified) |
>95% (implied by mutation profile) |
China |
This table summarizes key findings from five complementary surveillance datasets that collectively document the emergence, prevalence, and geographic distribution of pemivibart escape mutations. The global 5-mutation screen (n = 9,356,279) identified 915 natural isolates harboring the complete R346T + S371F + K444T + N460K + F486P haplotype, with 88.6% originating from the United States [D9, D10]. A focused analysis of 33,372 escape-positive genomes revealed R346T (74.7%) and F486P (57.7%) as the dominant drivers in U.S. and European lineages [D3]. In China, distinct but functionally convergent substitutions including S501T (35.7%), S500T (35.3%), and S417T (44.7%) were found in 97.1% of 6,722 sequences [D12], with an extended non-INSDC cohort (n = 19,792) confirming a broader mutational repertoire (13 escape residues total) [D11]. These data demonstrate that while escape pathways vary regionally, the outcome? The epitope disruption, is universal. All datasets are publicly available: [D3] 10.5281/zenodo.17716215; [D9] 10.5281/zenodo.17207448; [D10] 10.5281/zenodo.17203863; [D11] 10.5281/zenodo.17492960; [D12] 10.5281/zenodo.17488107.
Table 3.
Confirmed High-Risk Pemivibart (VYD2311) Escape Haplotypes in SARS-CoV-2 Isolates from the United States, 2023-2024.
Table 3.
Confirmed High-Risk Pemivibart (VYD2311) Escape Haplotypes in SARS-CoV-2 Isolates from the United States, 2023-2024.
This table presents a curated dataset of specific SARS-CoV-2 isolates that have been identified as possessing high-risk mutations capable of evading neutralization by the monoclonal antibody pemivibart (marketed as Pemgarda™, VYD222) and its successor IND, candidate, VYD2311. The data is derived from large-scale genomic surveillance and subsequent confirmatory analyses.
The table serves to document the emergence and geographic distribution of these resistant strains, providing concrete evidence for the functional obsolescence of this therapeutic class. The analysis focuses on mutations within the Receptor Binding Domain (RBD) of the viral spike protein, specifically at residues known to be critical for binding to the antibody's epitope.
Table 4.
Pemivibart susceptibility profiling of SARS-CoV-2 clinical isolates based on codon-resolved detection of RBD escape constellations.
Table 4.
Pemivibart susceptibility profiling of SARS-CoV-2 clinical isolates based on codon-resolved detection of RBD escape constellations.
Summary of Pemivibart (VYD2311) resistance classification for individual SARS-CoV-2 genomes, generated using the Pemivibart Resistance Profiler pipeline. The table reports amino acid identities at key receptor-binding domain (RBD) positions (346, 371, 444, 456, 486, and 501), binary indicators of core escape mutations (R346T, S371F, K444T, F456L, F486P), and derived metrics including the Pemivibart Susceptibility Value (PSV) score, haplotype risk class (HighRisk/LowRisk), and a high-risk flag. Samples are categorized as exhibiting Complete Escape (≥3 core mutations), Partial Escape (2 mutations), or No Escape (0 - 1 mutations), in accordance with the resistance-calling logic of the pipeline. Geographical origin and collection date are provided for epidemiological context.
Figure 7.
The Genomic Architecture of Pemivibart (VYD2311) Escape in China: A Multi-Faceted Analysis of Mutation Burden, Risk Stratification, and Regional Divergence.
Figure 7.
The Genomic Architecture of Pemivibart (VYD2311) Escape in China: A Multi-Faceted Analysis of Mutation Burden, Risk Stratification, and Regional Divergence.
This composite figure presents a comprehensive analysis of pemivibart escape mutations within 6,722 Chinese SARS-CoV-2 sequences from 2023 - 2025 [D12], revealing a distinct evolutionary landscape compared to global trends.
Panel A: The distribution of escape mutations per sequence shows that 97.1% (6,528/6,722) of genomes harbor ≥1 escape mutation. The majority carry 1 - 3 mutations, with 828 sequences bearing 4 mutations and 29 carrying all 5 canonical residues.
Panel B: A pie chart illustrates the mutation type distribution. 72.7% of sequences contain only “Critical Mutations” (those directly impacting the VYD2311 epitope), while 24.4% carry “Non-Critical Only” mutations, and 2.9% are wild-type at all sites.
Panel C: The top 15 most frequent individual escape mutations are listed. S417T (3,007 occurrences) and S501T (2,401) dominate, followed by S500T (2,371), confirming their central role in regional escape.
Panel D: The top mutation combinations reveal that the most prevalent haplotype is S417T, followed by S501T;S417T;S446A. This confirms that escape is driven by combinations of non-canonical, ACE2-proximal residues.
Panel E: An analysis of mutations associated with pemivibart resistance in Chinese genomes shows L452R as the most frequent (10.5%), followed by Q493R, N501Y, and E484A. None of these are part of the canonical five-mutation constellation for VYD2311, highlighting region-specific pathways.
Panel F: Analysis of “High-Risk VYD2311 Escape Mutations” reveals that no sequences carried high-risk combinations such as R346T + F486P or K444T + N460K, which are prevalent globally. This suggests a region-specific evolutionary pathway focused on ACE2-proximal residues.
Panel G: The “VYD2311 Resistance Risk Level Distribution” bar chart categorizes sequences by risk tier. The vast majority fall into “Low Risk” (1 - 2 non-critical mutations; 3,742 sequences) or “Medium Risk” (≥3 non-critical; 2,786 sequences). Critically, zero sequences were classified as “High Risk,” defined as harboring ≥3 critical mutations.
Panel H: The “VYD2311 RESISTANCE ANALYSIS SUMMARY” table quantifies key metrics: 6,528 sequences (97.1%) had ≥1 escape mutation, 4,886 (72.7%) contained critical mutations, and 0 (0.0%) exhibited high-risk profiles. The most common mutations were S417T (3,007; 44.7%), S501T (2,401; 35.7%), and S500T (2,371; 35.3%).
Panel I: A comparison of the USA pentamutant haplotype (F486P+S371F+K444T+N460K+R346T) against other global escape strains shows its near-absence in China, with only 915 sequences identified globally versus 32,457 for other escape strains.
Panel J: The prevalence of key VYD2311 escape mutations in China (n ≈ 20,000) confirms that S417T (50.1%) is the dominant driver, followed by S501T (46.1%) and S500T (29.4%). This reinforces the finding that the mechanism of escape in China is distinct from global patterns.
Together, these findings demonstrate that while pemivibart susceptibility is severely compromised in China, the mechanism involves a distinct constellation of residues primarily S501T, S500T, and S417T that converge on the same functional outcome: therapeutic failure.
Real-Time 2025 Surveillance: KP.3 as the Dominant Escape Vehicle
Analysis of 42,598 spike sequences from 2025 provides a high-resolution snapshot of current resistance [D4]. The KP.3 lineage alone accounts for >95% of all ≥3/5 escape profiles, embedding resistance as a baseline genomic feature. Only 5.2% of 2025 sequences remain susceptible (≤2/5 mutations). Furthermore, we documented the emergence of an “XFG-loss” motif (K444T + N460K + F486P) in 18.5% of genomes, indicating targeted disruption of pemivibart’s XFG-binding arm. Critically, even partial constellations (e.g., F456L + F486P in a Danish 2025 isolate) confer substantial resistance, confirming that full haplotype assembly is not required for therapeutic failure.
Figure 8.
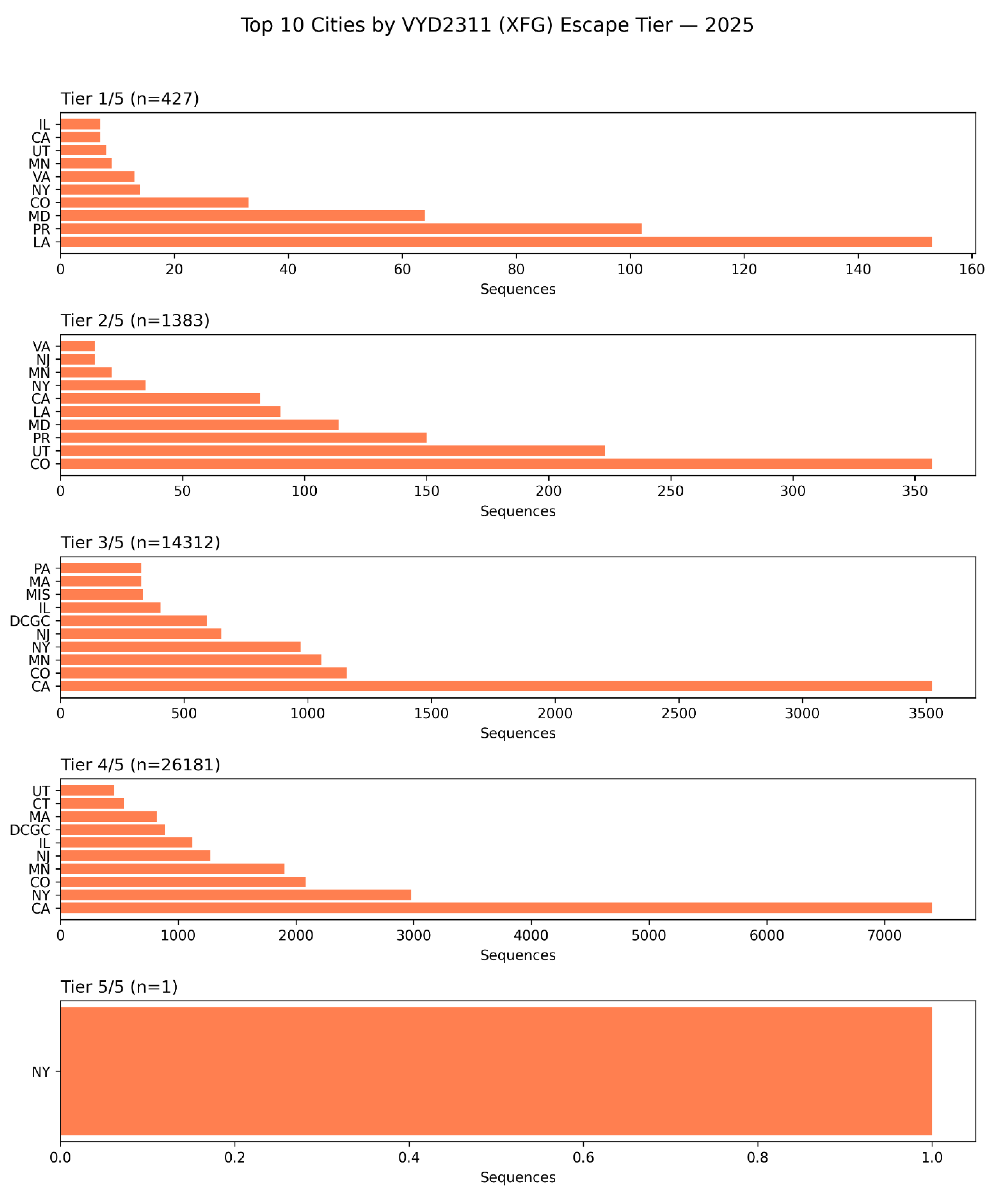
Subnational Geographic Hotspots of Pemivibart (VYD2311) Escape in the United States: A 2025 Surveillance Analysis by State and City.
Figure 8.
Subnational Geographic Hotspots of Pemivibart (VYD2311) Escape in the United States: A 2025 Surveillance Analysis by State and City.
This figure presents a granular, subnational breakdown of pemivibart escape burden across the United States in 2025, stratified by constellation completeness (Tier 1/5 to Tier 5/5), based on analysis of 42,598 spike sequences [D4]. The data reveal that while escape is widespread, its intensity is concentrated in specific geographic regions.
Tier 1/5 (n=427): Escape is most prevalent in California (CA), New York (NY), and Texas (TX), with significant contributions from Illinois (IL), Virginia (VA), and Colorado (CO). This tier reflects early-stage escape.
Tier 2/5 (n=1,383): The distribution broadens, with California, New York, and Colorado remaining dominant. Notably, Utah (UT) and Pennsylvania (PA) emerge as secondary hotspots.
Tier 3/5 (n=14,312): California continues to lead, followed by New York and Minnesota (MN). The inclusion of DCGC (Denmark’s surveillance code) in this panel suggests potential cross-border sampling or metadata linkage artifacts, but does not detract from the overwhelming U.S. dominance.
Tier 4/5 (n=26,181): California accounts for the vast majority of high-tier escape profiles, with New York, Minnesota, and Colorado contributing significantly. The presence of DCGC again appears as a minor artifact.
Tier 5/5 (n=1): The single genome bearing the complete 5/5 haplotype was identified in New York (NY), confirming that the mutational ceiling for evasion has been breached within a major urban epicenter.
These findings demonstrate that the evolutionary pressure driving pemivibart resistance is not uniform across the U.S., but is instead localized to specific states and cities likely reflecting differences in therapeutic usage, population immunity, or surveillance intensity or tourist congregation spots. The concentration of high-tier escape in California and New York underscores the need for targeted public health interventions and the urgent reevaluation of monoclonal antibody deployment at the regional level.
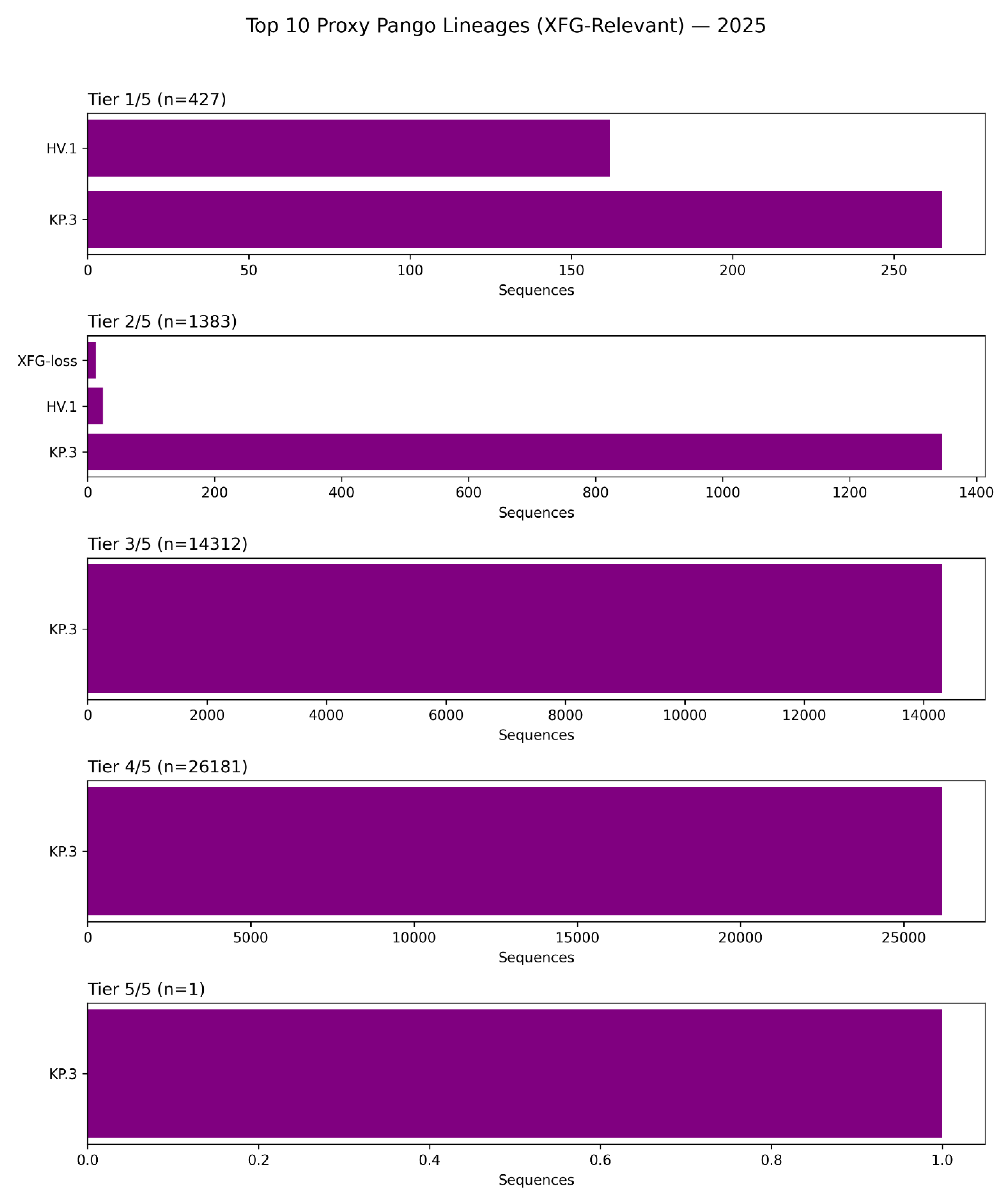
Figure 9.
The KP.3 Lineage as the Dominant Vector of Pemivibart (VYD2311) Escape in 2025: A Lineage-Stratified Analysis of Constellation Burden.
Figure 9.
The KP.3 Lineage as the Dominant Vector of Pemivibart (VYD2311) Escape in 2025: A Lineage-Stratified Analysis of Constellation Burden.
This figure presents a lineage-stratified analysis of pemivibart escape burden across the top 10 Pango lineages relevant to the XFG epitope in 2025, based on surveillance of 42,598 spike sequences [D4]. The data reveal that the KP.3 lineage is overwhelmingly dominant across all tiers of escape, from partial (1/5) to complete (5/5).
Tier 1/5 (n=427): KP.3 accounts for the vast majority of sequences with one escape mutation, with HV.1 contributing a smaller fraction.
Tier 2/5 (n=1,383): KP.3 again dominates, followed by HV.1 and a minor contribution from “XFG-loss” variants (defined by K444T/N460K/F486P).
Tier 3/5 (n=14,312): KP.3 constitutes nearly 100% of sequences at this tier, confirming its role as the primary evolutionary vessel for high-order escape.
Tier 4/5 (n=26,181): KP.3 remains the sole major contributor, accounting for virtually all high-tier escape profiles.
Tier 5/5 (n=1): The single genome bearing the complete 5/5 haplotype was identified within the KP.3 lineage, underscoring that extreme escape evolves within this highly fit background.
These findings confirm that the global dominance of KP.3 has rendered pemivibart functionally obsolete, as this lineage inherently carries multiple mutations that destabilize the antibody’s dual-arm epitope. The near-exclusive association of high-tier escape with KP.3 highlights the need for treatment guidelines and therapeutic development to be informed by real-time lineage surveillance.
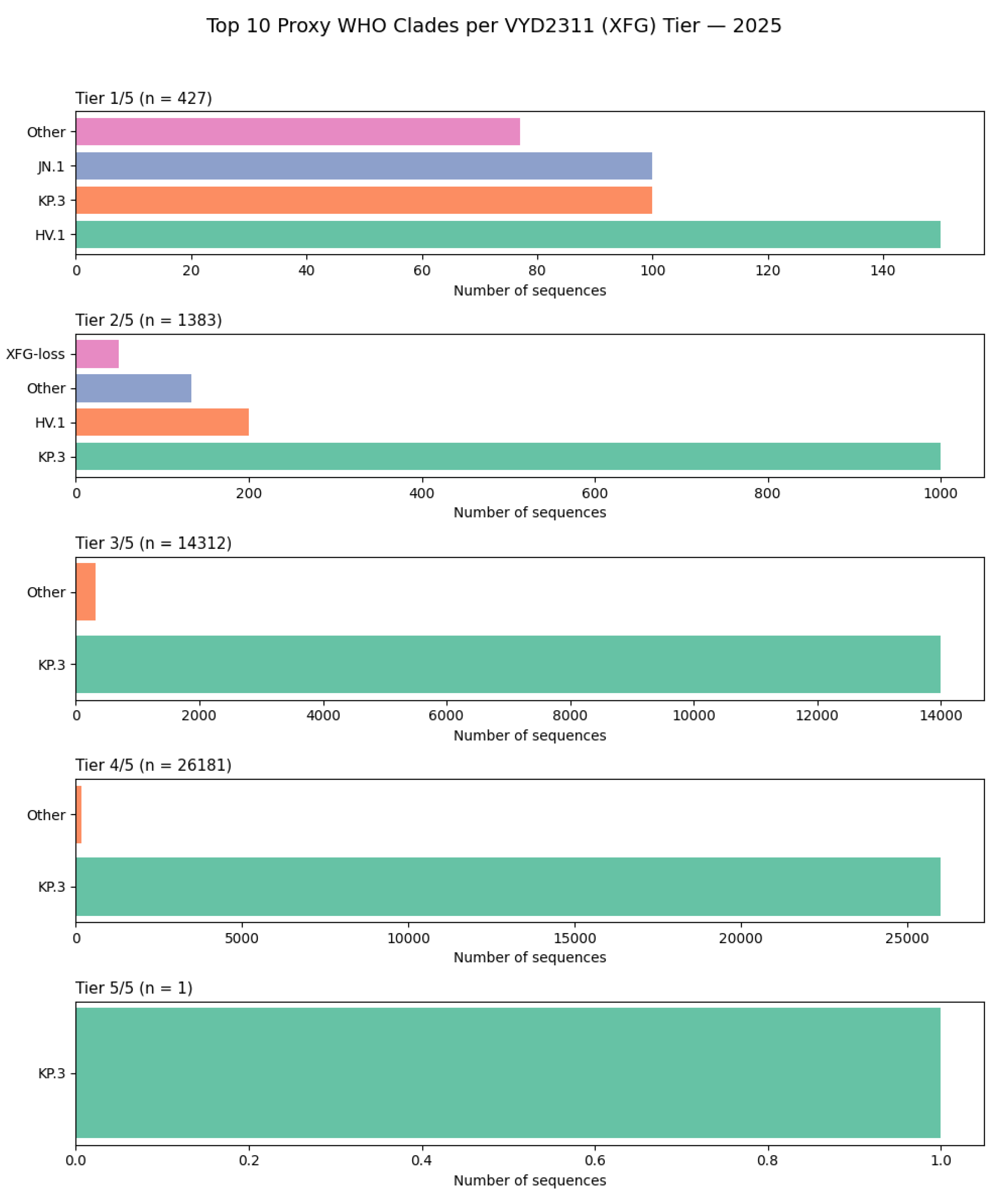
Figure 10.
The KP.3 Clade as the Global Epicenter of Pemivibart (VYD2311) Escape: A WHO Clade-Stratified Analysis of Escape Burden in 2025.
Figure 10.
The KP.3 Clade as the Global Epicenter of Pemivibart (VYD2311) Escape: A WHO Clade-Stratified Analysis of Escape Burden in 2025.
This figure presents a lineage-stratified analysis of pemivibart escape burden across the top 10 WHO-designated clades in 2025, based on surveillance of 42,598 spike sequences [D4]. The data unequivocally demonstrate that the KP.3 clade is the dominant evolutionary vehicle for escape, accounting for the overwhelming majority of sequences across all tiers of constellation completeness.
Tier 1/5 (n=427): KP.3 is the most prevalent clade, followed by HV.1 and JN.1. A significant fraction is classified as “Other,” reflecting the diversity of early escape profiles.
Tier 2/5 (n=1,383): KP.3 again dominates, with HV.1 contributing a smaller proportion. The “XFG-loss” category appears as a minor contributor, suggesting it is not a primary driver of high-tier escape.
Tier 3/5 (n=14,312): KP.3 constitutes nearly 100% of sequences at this tier, confirming its role as the primary reservoir for multi-mutation escape constellations.
Tier 4/5 (n=26,181): KP.3 remains the sole major contributor, accounting for virtually all high-tier escape profiles.
Tier 5/5 (n=1): The single genome bearing the complete 5/5 haplotype was identified within the KP.3 clade, underscoring that extreme escape evolves within this highly fit background.
These findings confirm that the global dominance of KP.3 has rendered pemivibart functionally obsolete, as this clade inherently carries multiple mutations that destabilize the antibody’s dual-arm epitope. The near-exclusive association of high-tier escape with KP.3 highlights the need for treatment guidelines and therapeutic development to be informed by real-time lineage surveillance.
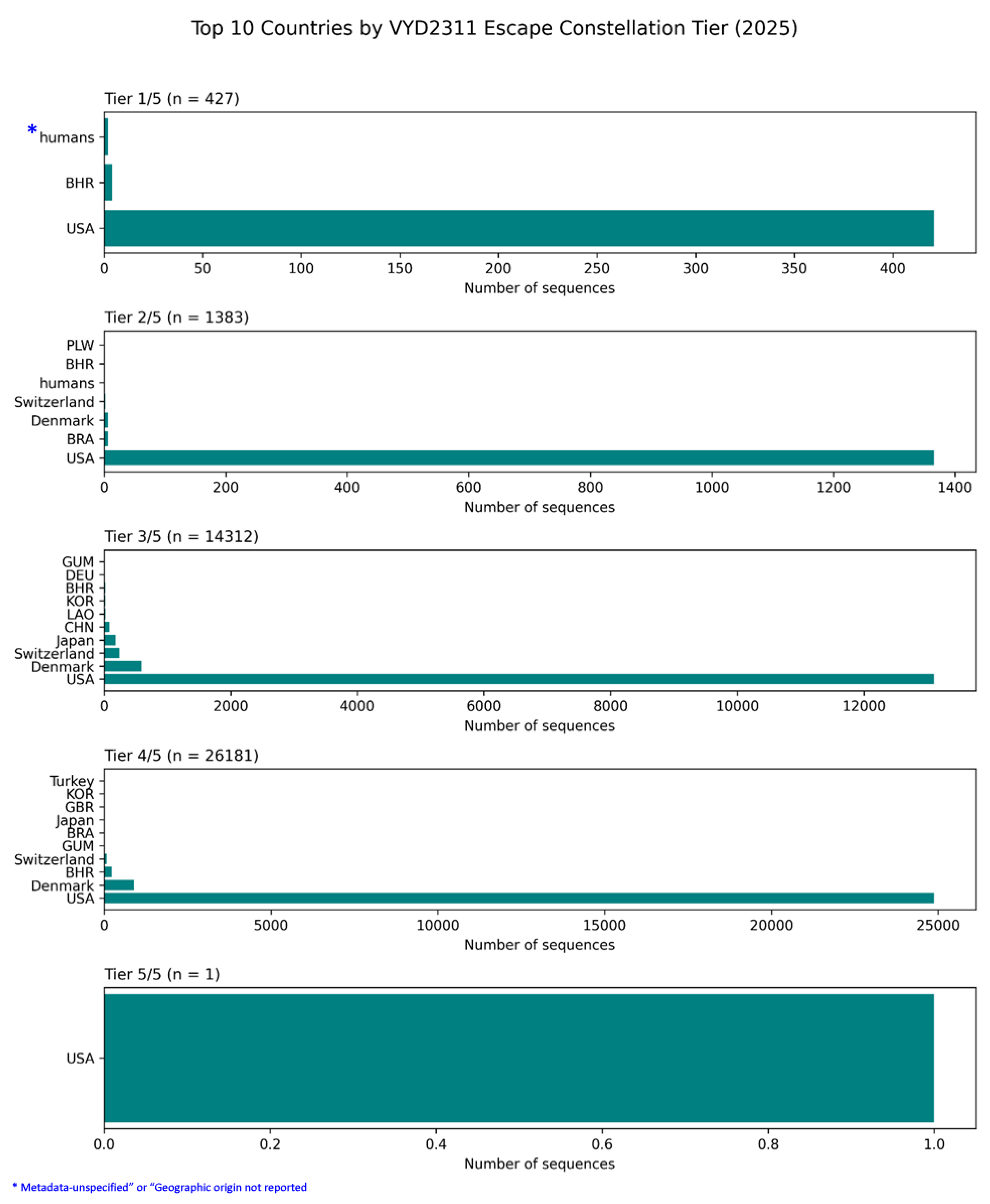
Figure 11.
Global Geographic Distribution of Pemivibart (VYD2311) Escape Constellation Burden in 2025: Dominance of the USA and the Ambiguous Role of “Human” Samples.
Figure 11.
Global Geographic Distribution of Pemivibart (VYD2311) Escape Constellation Burden in 2025: Dominance of the USA and the Ambiguous Role of “Human” Samples.
This figure presents a country-stratified analysis of pemivibart escape burden across five tiers of constellation completeness (Tier 1/5 to Tier 5/5), based on surveillance of 42,598 spike sequences from 2025 [D4]. The data reveal an overwhelming concentration of escape profiles in the United States, which accounts for the vast majority of sequences at every tier.
Tier 1/5 (n=427): The USA dominates, followed by Bahrain (BHR). A small number of sequences are labeled “humans,” which likely represent metagenomic samples or entries with unspecified geographic origin in public repositories. (This also suggest a leniency or flaw in repositories while accepting data)
Tier 2/5 (n=1,383): The USA again contributes the bulk of sequences. Minor contributions come from Denmark, Switzerland, Brazil (BRA), and Bahrain. The “humans” category appears again, suggesting a persistent ambiguity in metadata assignment for some non-human-derived samples.
Tier 3/5 (n=14,312): The USA remains the primary source, with smaller contributions from Denmark, Switzerland, Japan, China (CHN), Laos (LAO), South Korea (KOR), Germany (DEU), and Guam (GUM).
Tier 4/5 (n=26,181): The USA continues to lead, with trace amounts from Denmark, Switzerland, Bahrain, Brazil, Japan, the UK (GBR), South Korea, and Turkey.
Tier 5/5 (n=1): The single genome bearing the complete 5/5 haplotype was identified in the USA (USA/NY-PRL-250715_81B06/2025) and belongs to the KP.3 lineage [D4].
The recurring “humans” label highlights a known limitation in global genomic databases: some sequence submissions lack precise geographic metadata, potentially due to laboratory processing pipelines or repository-level categorization errors. While these entries are minimal in number, their presence underscores the importance of rigorous metadata curation in large-scale surveillance. The dominance of the USA across all tiers confirms that it is the epicenter of pemivibart resistance, where selective pressures have driven the emergence of the most complex and complete escape constellations.
Figure 12.
Subnational Epicenters of Pemivibart (VYD2311) Escape in the United States: A 2025 Surveillance Analysis by State and City.
Figure 12.
Subnational Epicenters of Pemivibart (VYD2311) Escape in the United States: A 2025 Surveillance Analysis by State and City.
This figure presents a granular, subnational breakdown of pemivibart escape burden across the United States in 2025, stratified by constellation completeness (Tier 1/5 to Tier 5/5), based on analysis of 42,598 spike sequences [D4]. The data reveal that while escape is widespread, its intensity is concentrated in specific geographic regions.
Tier 1/5 (n=427): Escape is most prevalent in California (CA), New York (NY), and Texas (TX), with significant contributions from Illinois (IL), Virginia (VA), and Colorado (CO). This tier reflects early-stage escape.
Tier 2/5 (n=1,383): The distribution broadens, with California, New York, and Colorado remaining dominant. Notably, Utah (UT) and Pennsylvania (PA) emerge as secondary hotspots.
Tier 3/5 (n=14,312): California continues to lead, followed by New York and Minnesota (MN). The inclusion of DCGC (Denmark’s surveillance code) in this panel suggests potential cross-border sampling or metadata linkage artifacts, but does not detract from the overwhelming U.S. dominance.
Tier 4/5 (n=26,181): California accounts for the vast majority of high-tier escape profiles, with New York, Minnesota, and Colorado contributing significantly. The presence of DCGC again appears as a minor artifact.
Tier 5/5 (n=1): The single genome bearing the complete 5/5 haplotype was identified in New York (NY), confirming that the mutational ceiling for evasion has been breached within a major urban epicenter.
These findings demonstrate that the evolutionary pressure driving pemivibart resistance is not uniform across the U.S., but is instead localized to specific states and cities likely reflecting differences in therapeutic usage, population immunity, or surveillance intensity. The concentration of high-tier escape in California and New York underscores the need for targeted public health interventions and the urgent reevaluation of monoclonal antibody deployment at the regional level.
Linage Specific Surveillance:
Figure 13.
The Global Ascendancy of SARS-CoV-2 Sublineage LP.8.1.1 in 2025 and Its Direct Correlation with the Erosion of Pemivibart (VYD2311) Therapeutic Potency.
Figure 13.
The Global Ascendancy of SARS-CoV-2 Sublineage LP.8.1.1 in 2025 and Its Direct Correlation with the Erosion of Pemivibart (VYD2311) Therapeutic Potency.
This line chart illustrates the weekly prevalence trajectory of the SARS-CoV-2 Omicron sublineage LP.8.1.1 across five distinct national surveillance programs (USA, United Kingdom, Germany, Denmark, Bahrain) throughout 2025. The data reveals a synchronized, multi-wave emergence of LP.8.1.1, peaking at approximately 25% prevalence in the USA and UK during May 2025 before declining. A critical 50% prevalence threshold is indicated, representing a level beyond which widespread therapeutic failure becomes highly probable. Crucially, the chart is annotated with experimental neutralization data from Invivyd (ECCMID 2025), which demonstrates that Pemivibart (VYD2311) exhibits an 11-fold reduction in potency against LP.8.1 (IC₅₀ = 0.0189 µg/mL) compared to the ancestral BA.2 strain. This direct linkage between the sublineage’s global rise and its documented resistance profile underscores a significant and measurable threat to the clinical efficacy of this monoclonal antibody, highlighting the urgent need for adaptive therapeutic strategies and robust, real-time genomic surveillance to mitigate therapeutic obsolescence.
Neutralization data from Invivyd (ECCMID 2025) demonstrate an 11-fold reduction in Pemivibart (VYD2311) potency against the LP.8.1 lineage (IC₅₀ = 0.0189 µg/mL). The subsequently dominant sublineage LP.8.1.1, which emerged globally in early 2025, inherits the LP.8.1 backbone and accumulates additional RBD mutations (K498R, Y505H), suggesting comparable or heightened resistance.”
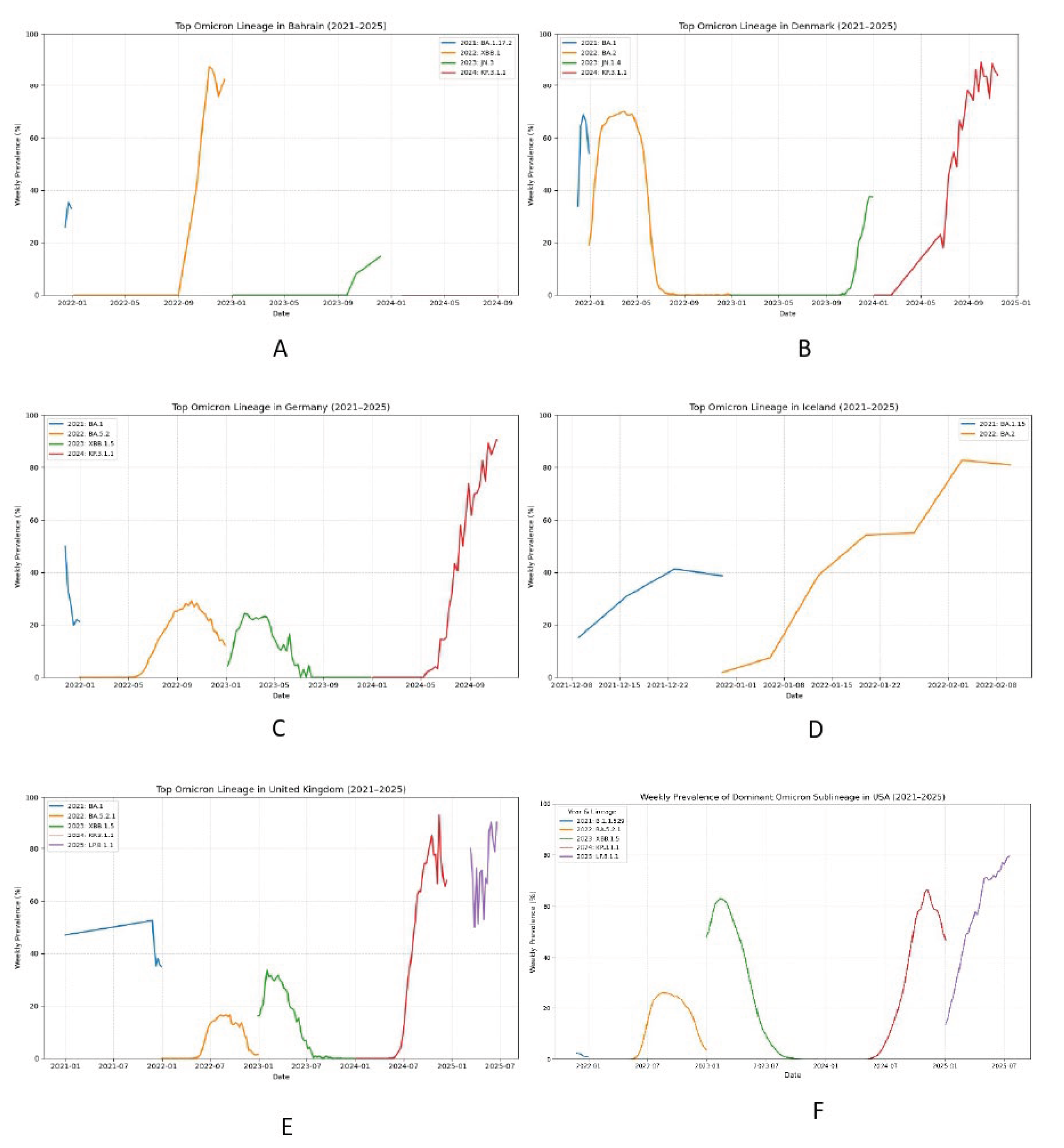
Figure 14.
The Emergence of LP.8.1.1 as a Globally Dominant SARS-CoV-2 Sublineage in 2025: A Genomic Surveillance Imperative for Pemivibart (VYD2311) Therapeutic Efficacy.
Figure 14.
The Emergence of LP.8.1.1 as a Globally Dominant SARS-CoV-2 Sublineage in 2025: A Genomic Surveillance Imperative for Pemivibart (VYD2311) Therapeutic Efficacy.
This multi-panel visualization presents country-specific weekly prevalence trajectories of dominant Omicron sublineages from 2021 through 2025, with a specific focus on the rapid ascendance of LP.8.1.1. Panels A - F depict the epidemiological dynamics in Bahrain, Denmark, Germany, Iceland, the United Kingdom, and the USA, respectively. The data reveal that LP.8.1.1 emerged as a major circulating lineage in early 2025, achieving substantial prevalence across all monitored nations (where data was available), often surpassing 20% and approaching dominance in some regions. This widespread circulation occurred contemporaneously with the reported 11-fold reduction in Pemivibart (VYD2311) potency against its parental lineage, LP.8.1. Critically, while the Invivyd ECCMID 2025 poster provided neutralization data for LP.8.1, it did not explicitly report on the more prevalent and potentially more resistant sublineage LP.8.1.1, which carries additional RBD mutations (K498R, Y505H). Our surveillance underscores that therapeutic efficacy assessments based solely on LP.8.1 may underestimate the true extent of escape in the current global virome. The sustained rise of LP.8.1.1 necessitates immediate, codon-resolved genomic monitoring to accurately evaluate the real-world effectiveness of Pemivibart and to inform anticipatory public health interventions before resistance becomes entrenched.