1. Introduction
Bacteriophages are the most abundant biological entities on Earth, with an estimated 10³¹ particles in the biosphere [
1]. Despite their ubiquity, our understanding of phage diversity remains limited. Phages have been called the “dark matter” of the biological world due to their enormous genetic diversity and many remaining secrets [
2]. Rapid sequencing advances have surged new phage genomes; complete ones in public databases doubled within three years [
2]. Yet, even with thousands available, they represent only a small fragment of total phage contigs diversity [
1,
3,
4]. Strikingly, many phage genes lack homologs in current databases [
5,
6]. These orphan genes show how each new phage broadens the known viral gene pool. Breadth-oriented studies continuously widen the phage pangenome, offering novel genes for future work [
7].
Phages also play key ecological and evolutionary roles. They drive bacterial dynamics and impact biogeochemical cycles by modulating microbial communities [
8,
9]. Via transduction and gene exchange, phages enable horizontal transfer among bacteria, e.g. affecting virulence and metabolism [
10,
11,
12]. In biotechnology and medicine, phages have been invaluable from early discoveries to modern phage therapy and molecular tools [
13,
14,
15,
16]. Yet, most characterized phages stem from a small set of model systems, leaving vast biodiversity unexplored.
Environmental surveys indicate every habitat—from oceans to soil to human gut—harbors unique phages adapted to local hosts. Wastewater is particularly rich in microbial diversity, hence likely in phages infecting those microbes [
9]. Urban sewage studies isolated distinct phages across host genera [
1]. Environments yield phages with varied genome sizes, morphologies, and specificities [
17,
18]. Thus, exploring under-sampled settings is crucial for new viral lineages and innovations.
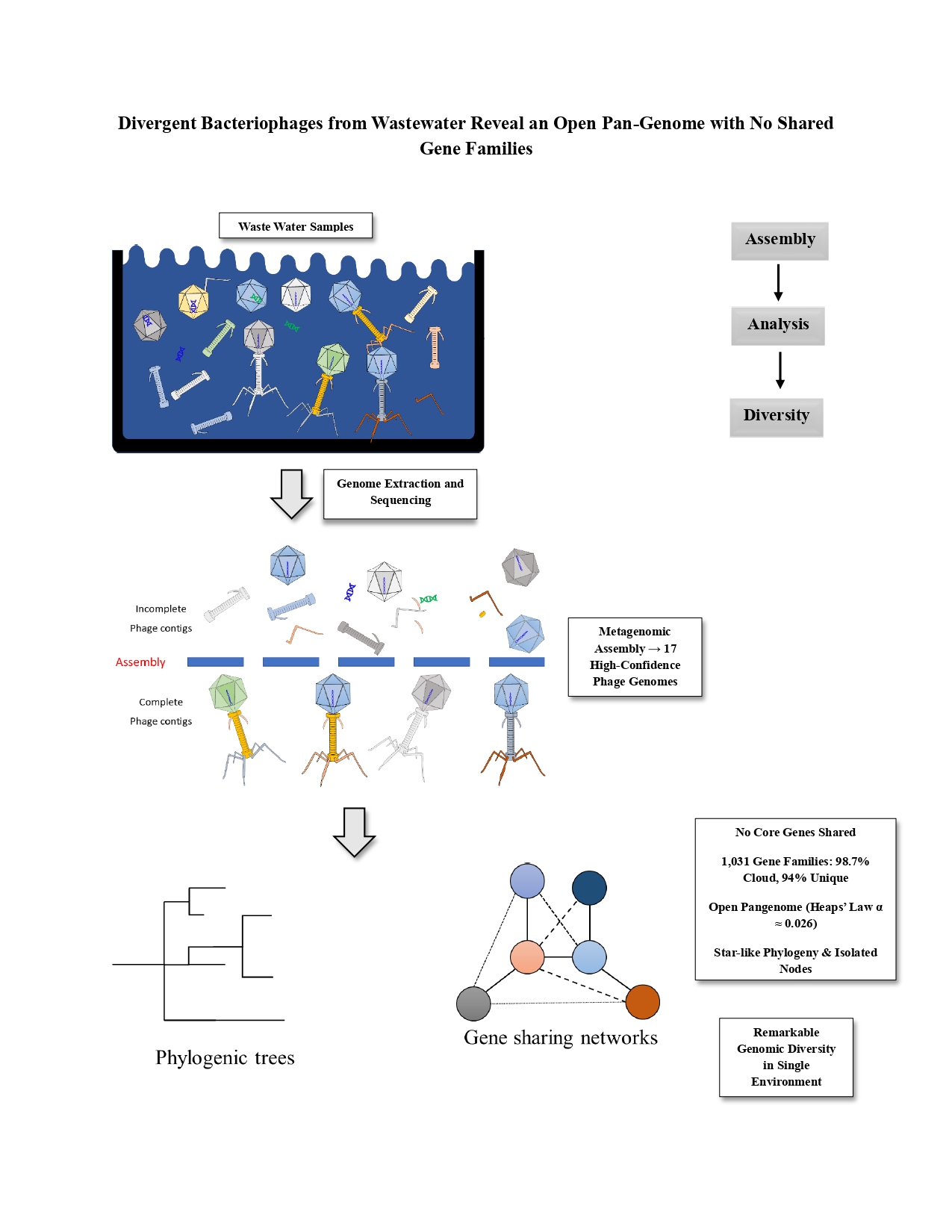
Here, metagenomic samples from rural wastewater facilities were analyzed for intact phage genomes. We hypothesized diverse microbes would yield high phage diversity, with largely unique genetic content per genome. Assembling viral contigs and identifying complete phages yielded a collection from these samples. Comprehensive comparative genomics characterized relatedness—or lack thereof—among them. This study expands environmental phage genomics, shows genomic distinctness in one setting, and assesses common/core genes. Such insights advance phage diversity and evolution, highlighting novelty from each new phage.
2. Materials and Methods
2.1. Data Collection
Metagenomic data were retrieved from BioProject PRJNA906478 (
https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA906478), encompassing rural decentralized wastewater treatment facilities in China, as part of a study conducted at Jiangnan University. These data, generated via Illumina MiSeq paired-end whole-genome sequencing (WGS) and published on November 30, 2022, were accessed from the NCBI Sequence Read Archive (SRA). The project included six SRA experiments with accession numbers SRX24547821, SRX18428665, SRX18428664, SRX18428663, SRX18428662, and SRX18428661. Datasets were imported into the UseGalaxy server for analysis. This resource provides substantial environmental data, indicating an abundance of bacteriophages with extensive genomic diversity.
2.2. Data Manipulation
FASTQ files from SRA entries were collected in UseGalaxy under unique identifiers. Trimmomatic package was applied to remove low-quality reads and adapter sequences. Contigs were assembled per sample using metaSPAdes tool [
19]. Taxonomic classification of reads was performed with Kraken tool, which assigns labels based on organism-based matches [
20,
21,
22]. Assembly outputs from metaSPAdes were subsequently classified against the viral_2020 database.
2.3. Phage Prediction and Annotation
Contigs were retained for intact bacteriophage identification. Each sample was screened for bacteriophages using the PHAge Search Tool with Enhanced Sequence Translation (PHASTEST) web tool under default settings [
23]. FASTA files per sample were preserved for annotation. Pharokka was employed for open reading frame (ORF) prediction, nucleotide translation, and annotation [
24]. GenBank (GBK) files were produced for each intact phage to facilitate downstream processing (see Additional file 1 for fully annotated phage genomes).
2.4. Computing Environment
Analyses were conducted on a Windows workstation utilizing Windows Subsystem for Linux (WSL) for MMseqs2-based sequence clustering [
25]. Python 3.10 with Biopython 1.85 handled genome parsing and protein extraction. R 4.5.1, incorporating base R and packages including micropan, ape, phangorn, vegan, ComplexHeatmap, ggplot2, ggtree, igraph, and ggraph, managed pangenome statistics, phylogenetics, and visualization. MMseqs2 (v13-45111+ds-2) was executed within WSL or via the wsl mmseqs command from Windows; all parameters listed represent values directly supplied to MMseqs2.
2.5. Genome Parsing and Protein Extraction
GenBank records were parsed using Biopython’s SeqIO module. For each coding sequence (CDS) feature, the canonical amino-acid sequence was extracted from the translation qualifier if available. Absent translations prompted in-frame translation of the nucleotide sequence via Biopython’s codon table, ensuring a consistent fallback. Proteins were output in FASTA format with headers encoding the genome identifier, a unique index, and genomic coordinates with strand where specified. This method maintains an annotation-consistent proteome for clustering, avoiding ORF re-prediction.
2.6. Clustering Proteins into Gene Families
Gene families resembling orthologs were inferred via MMseqs2’s greedy MEM clustering. All proteins across genomes were concatenated into a single FASTA file and converted to an MMseqs database (createdb). Clustering applied a minimum sequence identity of 0.30 and alignment coverage of 0.50 in coverage mode 1, enforcing the threshold on one sequence (pipeline standard). The default greedy algorithm with three refinement iterations was utilized; sensitivity matched MMseqs2’s clustering default, equivalent to -s 5. Each protein was assigned to at most one cluster, with a representative sequence retained. A membership table of cluster representatives versus member IDs was generated for presence–absence matrix construction.
2.7. Presence–Absence Matrix and Gene Partitions
A binary genome-by-family matrix "M"∈{0,1}^(G×F) was assembled from the cluster membership table by marking M_(g,f)=1 if any protein from genome g belonged to family f, and 0 otherwise. From the column sums of M, gene partitions were computed using widely adopted thresholds: core genes are present in 100% of genomes; soft-core in ≥95%; shell in 15–<95%; and cloud in <15% of genomes. These categories provide a succinct description of the pangenome’s structural composition and are used subsequently to summarize gene sharing.
2.8. Definition of Pangenome Metrics
Standard metrics were computed from the presence–absence matrix for clarity. Total predicted proteins (CDS) denote annotated coding sequences per genome. Total gene families (pangenome size) indicate unique orthologous clusters across genomes. Core gene families appear in 100% of genomes; soft-core in ≥95%. Shell gene families are present in 15–<95% of genomes, while cloud gene families occur in <15%. Singletons represent families unique to one genome. Heaps’ law α quantifies pangenome openness, with α < 1 signifying an open, expanding pangenome.
2.9. Pangenome Accumulation and Openness
Pangenome and core-genome growth were assessed through permutation rarefaction with micropan. For each k=1,…,G, distinct families after sampling k genomes were averaged over 500 permutations (seed = 42), yielding the mean pangenome curve and 95% interval. The core curve similarly reflected families present in all sampled genomes per permutation. Openness was evaluated by fitting Heaps’ law to the pangenome rarefaction: P(k)≈β_0+β_1 k^α,
where P(k) is the expected pangenome size at k genomes, α is Heaps’ α, and α < 1 is interpreted as an open pangenome (continued gene discovery with additional genomes), whereas α ≥ 1 indicates a more closed repertoire.
2.10. Gene-Content Distances, Clustering, and Heatmaps
Pairwise gene-content relatedness between genomes was measured using the Jaccard distance on the rows of M. For two genomes with presence vectors
, the similarity is
and the distance is
. A Neighbor-Joining (NJ) tree was inferred from this distance matrix; in parallel, an UPGMA tree was generated from hierarchical clustering using average linkage. To visualize global relatedness, a heatmap of Jaccard similarity was plotted with genomes ordered by the NJ tip order so that block structure in the matrix aligns with inferred relationships.
2.11. Intersections of Shared Gene Families
To summarize combinatorial sharing beyond the capacity of Venn diagrams, presence sets were constructed for each genome. The ComplexHeatmap implementation of UpSet was used to compute and visualize the largest intersections among (“distinct” mode). Empty sets (genomes without present families under the chosen filters) were excluded before counting. Displays featured bars for set and intersection cardinalities, illustrating genome combinations with maximal shared families.
2.12. Genome–Genome Gene-Sharing Network
Beyond pairwise distances, a weighted, undirected gene-sharing network was constructed whose nodes are genomes and whose edge weights are the Jaccard similarities . To avoid a saturated graph and emphasize non-trivial sharing, edges with weights below 0.10 were discarded. The resulting graph was simplified to merge parallel edges, and Louvain community detection was used to explore mesoscale structure. A force-directed layout (Fruchterman–Reingold) provided a stable visualization in which node sizes reflect each genome’s gene-family richness (row sums of M) and edge widths scale with Jaccard weight.
2.13. Ancestral Reconstruction of Presence/Absence and Branch Dynamics
Gene-family turnover was mapped phylogenetically by reconstructing ancestral states as absent/present characters. The NJ tree was midpoint-rooted, with multifurcations resolved to bifurcating topology. Maximum-likelihood states were estimated per family under an equal-rates (ER) model using ace from ape. Probable states at internal nodes were selected via argmax of likelihood vectors. Edge traversals counted 0-to-1 transitions as gains and 1-to-0 as losses; invariant families were omitted. Gain/loss tallies per node were aggregated and overlaid on the tree with ggtree to emphasize dynamic lineages.
2.14. Proteome-Level Relatedness by AAI
Average Amino-Acid Identity (AAI) provided an alternative proteome similarity metric via reciprocal best hits (RBH) in MMseqs2. Bidirectional searches per genome pair used minimum identity 0.30 and coverage 0.50 in mode 1. Best hits per query were chosen by composite score, favoring long, high-identity alignments. RBHs were mutual best hits. Pairwise AAI was the alignment-length-weighted mean percent identity across RBHs:
This approach mitigates bias from short HSPs. Thresholds were fixed as stated, though sensitivity adjustments were optional for divergent proteomes.
2.15. Visualization and Reporting
Figures were produced at 300 dpi using ggplot2, ComplexHeatmap, ggtree, and ggraph. The full analysis—encompassing pangenome curves with 95% envelopes, Heaps’ α estimation, Jaccard heatmaps, UpSet intersections, gene-sharing network, ancestral gain/loss overlays, and AAI heatmaps/histograms/networks—was documented in an R Markdown report rendered to HTML and PDF via xelatex for precise typesetting (see Additional file 2).
4. Discussion
The analysis of 17 bacteriophage genomes recovered from wastewater metagenomes revealed an extreme level of genetic diversity among viruses in environmental context. A key finding is that these phages share virtually no genes in common, in other words, there is no detectable core genome among them. This result aligns with the expectation that phages infecting different hosts or belonging to different taxonomic families are often highly divergent in gene content. Phage genomes evolve rapidly through mutation and horizontal gene transfer as part of their arms-race coevolution with hosts [
8], leading even related phages to accumulate distinct sets of genes. In the present dataset, this divergence is so pronounced that the pangenome is almost entirely cloud. Over 98% of the 1,031 gene families identified were found in only one or two phages, and 94% were singletons unique to a single phage genome. Such a skewed distribution of gene families is consistent with previous observations that many phage genes are unique or of unknown function [
6,
26]. It underscores the idea that each phage is genetically idiosyncratic, encoding a suite of ORFs not seen in others.
The lack of common genes across all phages is notable but not surprising. Unlike bacteria, which often share housekeeping genes, viruses have no universal marker gene present in all genomes. Even essential structural or replication genes can be so sequence-divergent or replaced by analogs that homology is unrecognizable across distant phages. The finding of zero core genes is therefore consistent with the understanding that the viral realm lacks a conserved backbone of genes connecting all members. In practical terms, by the current ICTV species demarcation criterion of ≤95% nucleotide identity across the genome [
2,
27,
28], each of these phages would be considered a separate species. Indeed, the pairwise AAI analysis showed that most phage pairs have essentially no identifiable protein similarity, reinforcing that they share no close evolutionary relationship. The two closest pairs, phage_16 and phage_17, and to a lesser extent phage_3 and phage_4, were the only cases with non-zero AAI and a modest number of shared genes. These pairs clustered together in gene-content-based phylogenetic trees and gene-sharing networks, indicating they represent small clades of related viruses amid an otherwise star-like array of unrelated genomes. Phage_16 and phage_17, shared on the order of a dozen gene families and showed ~46% AAI, suggesting they might belong to the same broader genus or have a recent common ancestor. Similarly, phage_3 and phage_4 form a duo with a handful of genes in common. Outside these, no other phage in the collection had more than a few genes overlapping with any other.
It is instructive to compare these results with prior studies of environmental phages. A large-scale isolation from sewage demonstrated that even within one environment, phages can be extremely diverse in terms of hosts and phenotypes [
1]. Similarly, a recent study by Wang et al. (2024) reconstructed over 18,000 viral operational taxonomic units (vOTUs) from wastewater treatment systems, revealing high novelty and a significant role of phages in nutrient cycling and host regulation, further supporting the extensive unexplored diversity and minimal gene sharing observed in our dataset [
29]. The genomic findings of our study add to this by showing that, at the DNA level, each phage can represent a distinct genetic lineage. In essence, the wastewater sample does not harbor a swarm of closely related phages infecting one type of bacterium, but rather a mélange of phages each potentially targeting different bacterial hosts. This likely reflects the complex microbial community in wastewater; phages tend to specialize on particular bacteria, so a diverse microbial ecosystem yields a correspondingly diverse set of phage genomes. The two small clusters we observed (phage_16–17 and phage_3–4), none of which were in one sample, might indicate instances where the same or closely related bacterial hosts were present in different samples, allowing related phages in different environments. Their limited gene sharing suggests they could be different strains of a phage lineage or have diverged from a common ancestor via acquisition of different accessory genes.
Pangenome openness is an important concept for contextualizing the results. It was found a Heaps’ law exponent α ≈ 0.026, which quantitatively signifies an extremely open pangenome. In practical terms, this means that every time a new phage genome is added to the analysis, a large number of brand-new gene families appear. The pangenome accumulation curve had not begun to plateau even by the 17th genome, and indeed the curve was climbing steeply. This is consistent with Flores et al. (2024), who identified over 1,700 novel phage genomes from urban wastewater, with 80% exhibiting no shared core genes with reference databases, reinforcing that each new phage contributes unique gene families to the pangenome [
30]. This trajectory indicates that the gene repertoire of phages in this environment is effectively inexhaustible, sampling additional phages would likely continue to reveal many novel genes. Open pangenomes are common in broad collections of bacteria or viruses that have high diversity. In bacteriophages, this is often taken to the extreme. In this regard, even phages infecting the same bacterial species can differ markedly in gene content, let alone phages from different families. The findings of this study exemplifies this phenomenon; the collective genome of these 17 phages is vast and mosaic, while the intersection of all 17 is essentially empty. From an evolutionary perspective, such openness reflects how phage genomes are constantly gaining and losing genes, often through horizontal gene transfer, recombination, and modular shuffling of functional gene cassettes. Prior research has shown that phages maintain diversity by exchanging flexible gene modules while sometimes preserving a core genome within a lineage [
8]. Here, however, since our phages do not share a core lineage, we are observing diversity at the highest level, essentially, phages of disparate origins, rather than variations on one node.
Interestingly, the constructed gene-sharing network distilled the same insight in graphical form. With a threshold requiring at least 10% Jaccard similarity for an edge, only two edges remained. All other phages stood as solitary nodes, isolated due to negligible overlap with any other genome. This star-like network topology is characteristic of systems where most entities have unique content and few commonalities. It highlights that no larger communities or subgroups of phages exist in the dataset beyond the two small pairs, there is no evidence of a big cluster of virions sharing a host or evolutionary lineage. The network also highlighted phage_10 as a standout node due to its size and contributed the highest number of unique gene families. Phage_10’s outsized gene repertoire suggests it could be a jumbo phage or at least a relatively large-genome phage carrying many auxiliary genes. In phage ecology, larger genomes often encode additional metabolic or defense genes that can enhance the phage’s lifestyle or broaden its host range. The uniqueness of phage_10’s gene content might imply novel functions or adaptations worthy of further study. Particularly, despite its complexity, phage_10 still did not share any detectable gene with the other phages.
While sequence-based analysis shows no direct gene homology, the functionally analogous genes for capsid assembly, DNA packaging, and lysis were present, but were so sequence-divergent that they fall into different “gene families” in the clustering. Bacteriophages often achieve similar ends using proteins that, over evolutionary time, share little sequence identity. Thus, the data do not imply that these phages lack fundamental structural genes, but rather that those genes are not conserved at the sequence level across the group. From a broader standpoint, the genomic uniqueness of environmental phages suggests that every new phage genome can potentially encode novel biochemical capabilities. This is consistent with suggestions by Hatfull and others that exploring phage diversity may lead to discovery of new molecular mechanisms and tools [
15]. Accordingly, the high-level divergence we observed hints that these phages likely employ different strategies to infect hosts or evade host defenses. Some may carry unique tail fiber genes for host recognition, others unusual nucleases or replication proteins. Indeed, phages are known to occasionally carry host-like genes, and in a diverse set like this, one might find evidence of genes borrowed from various bacterial pathways. Identifying and characterizing those unique genes was beyond the scope of our pangenome-focused analysis, but it remains an exciting avenue for future research.
The ancestral gene gain/loss analysis further provided insight into how phage genomes evolved in this collection. Although broad inference is difficult given the lack of close relationships, the analysis suggested an episodic pattern. In this regard, most lineages accumulated few or no new genes along their branches, but a couple of lineages underwent “burst” events of gene acquisition. Accordingly, the branch leading to the phage_16–17 clade showed a concentrated gain of around 13 gene families, possibly marking a major innovation. These bursts stand out against a background of relative stasis, where other phages changed little in gene content from their inferred ancestors. This scenario is consistent with the idea that phage genomes can undergo rare but significant exchanges, such as acquiring a plasmid or another phage’s genes during co-infection, resulting in a sudden increase in genome size and coding potential. Over longer timescales, these rare events contribute disproportionately to the expansion of the overall phage gene pool. Meanwhile, lineages without such exchanges mainly diversify via mutation in their existing genes, which would not create new gene families. The open pangenome nature observed is thus driven by those occasional leaps in gene content. It also mirrors patterns seen in some bacterial open pangenomes, where the cumulative diversity arises from a minority of lineages acquiring most of the novel genes, while others remain relatively stable.
Overall, the results support the initial hypothesis that a diverse environment like a wastewater system harbors an diverse set of bacteriophages. We see direct evidence that each phage is unique, confirming that phages from complex microbial communities overlap in genomic content unless they are very closely related. This highlights how rich the unexplored phage space is. The findings resonate with the broader literature, phage diversity research is expanding the boundaries of the viral pangenome, uncovering myriad new genes and gene combinations . Additionally, the study underscores the challenge in phage genomics of making generalizations. Each phage genome must be examined on its own merits. From an applied perspective, the novel genes within these phages could include useful enzymes like novel lysins, polymerases, or CRISPR inhibitors that might have biotechnological or therapeutic value. Expanding the sampling would also help determine whether certain gene modules recur in specific niches or whether they remain entirely unique. Additionally, we did not investigate the taxonomic family of each detected phage, which could be addressed in future studies.
There are some limitations for this study. First, the analysis was restricted to 17 phage genomes in six different bio-samples, which represents only a small subset of the wastewater virome and may not capture the full diversity present. Second, the genomes were derived from publicly available metagenomic assemblies and onlu complete genomes retained, and contigs with incomplete or chimeric sequences were discarded. Third, no host information was available, which limits the ecological interpretation of these phages, which could be addressed by determining the bacteriophages’ families. Future studies combining broader sampling, and host prediction will provide a more comprehensive picture of wastewater phage diversity.
Figure 1.
Genome Summary. A. Predicted protein counts per phage genome. A lollipop chart showing the number of protein-coding genes in each of the 17 phage genomes (points), sorted from smallest to largest genome. The dashed line indicates the mean genome size (66 proteins) with the 95% confidence interval. Genome sizes vary over a 6-fold range (30 to 172 proteins), highlighting considerable heterogeneity in coding capacity among the phages. This variation provides the context for pan-genome analyses, as larger genomes contribute many unique gene families. B. Genome features of the analyzed phages. (Top left) Distribution of genome sizes (kb), shown as a boxplot with individual genomes overlaid. (Top right) Distribution of predicted open reading frames (ORFs) across genomes. (Bottom left) Correlation between genome size and predicted ORFs, showing a strong positive linear relationship. (Bottom right) Relationship between genome size and GC content (%) modeled with a smooth curve, highlighting variability in GC% across different genome sizes.
Figure 1.
Genome Summary. A. Predicted protein counts per phage genome. A lollipop chart showing the number of protein-coding genes in each of the 17 phage genomes (points), sorted from smallest to largest genome. The dashed line indicates the mean genome size (66 proteins) with the 95% confidence interval. Genome sizes vary over a 6-fold range (30 to 172 proteins), highlighting considerable heterogeneity in coding capacity among the phages. This variation provides the context for pan-genome analyses, as larger genomes contribute many unique gene families. B. Genome features of the analyzed phages. (Top left) Distribution of genome sizes (kb), shown as a boxplot with individual genomes overlaid. (Top right) Distribution of predicted open reading frames (ORFs) across genomes. (Bottom left) Correlation between genome size and predicted ORFs, showing a strong positive linear relationship. (Bottom right) Relationship between genome size and GC content (%) modeled with a smooth curve, highlighting variability in GC% across different genome sizes.


Figure 2.
Gene-content phylogeny (Neighbor-Joining at top and UPGMA bottom)and Clustering. A. An unrooted NJ tree based on Jaccard distances between phage gene repertoires. Branch lengths reflect gene content dissimilarity. Phages that cluster together (adjacent in the tree) share more gene families. Notably, small-genome phages 16 and 17 branch as closest neighbors, indicating their gene sets overlap substantially. Phages 3 and 4 also cluster, consistent with their moderate gene sharing. In contrast, phage_10 is isolated on a long branch, highlighting its high divergence (it shares virtually no genes with others). The NJ analysis thus groups phages by gene content similarity, revealing discrete clades of more closely related genomes. B. A UPGMA dendrogram built from the same Jaccard distance matrix of gene presence/absence. The overall clustering pattern mirrors that of the NJ tree, with the same major groupings observed. Phages 16 and 17 again form a distinct cluster, and phages 3 and 4 group together similarly. The consistency between UPGMA and NJ trees suggests that these relationships are robust. Minor differences in branch lengths or order are due to the clustering algorithm, but no fundamentally new groupings appear. Both methods underscore the high level of gene content divergence among the phages, as evidenced by the long branches separating most clusters. C. Heatmap of pairwise gene-content distances among the 17 phage genomes. Jaccard distance (1 − Jaccard similarity) is shown for each genome pair. Genomes are ordered according to a neighbor-joining tree derived from these distances, so that closely related phages appear adjacent. The matrix is shows minimal shared gene content). Notably, two darker off-diagonal cells are visible, one corresponds to the phage_16–phage_17 comparison, and another to phage_3–phage_4, indicating these pairs have the highest gene content. D. Ancestral gene-family gain/loss reconstruction on the phage phylogeny. A phylogenetic tree of the 17 phages is shown with internal nodes annotated by the number of gene families gained (blue) or lost (red) along each branch (inferred by maximum likelihood). Circle sizes are proportional to the total number of changes (gains + losses) at that node. Most branches have zero or few changes (no circle or very small), indicating relative gene content stasis. However, a few ancestral branches stand out with large circles – for example, one node (marked with an arrow) shows 13 gene gains, and another shows ~9 gains. These results suggest that while gene content evolution is generally conservative, there have been episodic bursts of gene acquisition in certain lineages, which contributed significantly to the divergence in phage gene content.
Figure 2.
Gene-content phylogeny (Neighbor-Joining at top and UPGMA bottom)and Clustering. A. An unrooted NJ tree based on Jaccard distances between phage gene repertoires. Branch lengths reflect gene content dissimilarity. Phages that cluster together (adjacent in the tree) share more gene families. Notably, small-genome phages 16 and 17 branch as closest neighbors, indicating their gene sets overlap substantially. Phages 3 and 4 also cluster, consistent with their moderate gene sharing. In contrast, phage_10 is isolated on a long branch, highlighting its high divergence (it shares virtually no genes with others). The NJ analysis thus groups phages by gene content similarity, revealing discrete clades of more closely related genomes. B. A UPGMA dendrogram built from the same Jaccard distance matrix of gene presence/absence. The overall clustering pattern mirrors that of the NJ tree, with the same major groupings observed. Phages 16 and 17 again form a distinct cluster, and phages 3 and 4 group together similarly. The consistency between UPGMA and NJ trees suggests that these relationships are robust. Minor differences in branch lengths or order are due to the clustering algorithm, but no fundamentally new groupings appear. Both methods underscore the high level of gene content divergence among the phages, as evidenced by the long branches separating most clusters. C. Heatmap of pairwise gene-content distances among the 17 phage genomes. Jaccard distance (1 − Jaccard similarity) is shown for each genome pair. Genomes are ordered according to a neighbor-joining tree derived from these distances, so that closely related phages appear adjacent. The matrix is shows minimal shared gene content). Notably, two darker off-diagonal cells are visible, one corresponds to the phage_16–phage_17 comparison, and another to phage_3–phage_4, indicating these pairs have the highest gene content. D. Ancestral gene-family gain/loss reconstruction on the phage phylogeny. A phylogenetic tree of the 17 phages is shown with internal nodes annotated by the number of gene families gained (blue) or lost (red) along each branch (inferred by maximum likelihood). Circle sizes are proportional to the total number of changes (gains + losses) at that node. Most branches have zero or few changes (no circle or very small), indicating relative gene content stasis. However, a few ancestral branches stand out with large circles – for example, one node (marked with an arrow) shows 13 gene gains, and another shows ~9 gains. These results suggest that while gene content evolution is generally conservative, there have been episodic bursts of gene acquisition in certain lineages, which contributed significantly to the divergence in phage gene content.


Figure 3.
Pangenomic Openness. A. Pangenome accumulation curves for the intact phages. The total number of gene families (pangenome, teal line) and the number of core families (present in all genomes, red line) are plotted as a function of genomes sampled, with shading indicating the 95% confidence interval across 500 random genome addition permutations. The pangenome size shows a steady increase, reaching 1030 families after all genomes are included, while the core genome size rapidly declines toward zero and remains essentially zero beyond 5 genomes. The non-saturating pangenome curve and vanishing core indicate an open pangenome (consistent with Heaps’ law exponent α ≈ 0.026). B. Composition of the phage pangenome by gene frequency category. A pie chart illustrates the proportion of gene families that are core (present in all genomes, 0%), soft-core (present in ≥95% of genomes, 0%), shell (present in 15–<95% of genomes, 1.3%), and cloud (present in <15% of genomes, 98.7%). The cloud segment dominates, reflecting that almost all gene families are found in only one or a few phages. Shell families constitute a negligible fraction, and no universal core genes were identified among these phages.
Figure 3.
Pangenomic Openness. A. Pangenome accumulation curves for the intact phages. The total number of gene families (pangenome, teal line) and the number of core families (present in all genomes, red line) are plotted as a function of genomes sampled, with shading indicating the 95% confidence interval across 500 random genome addition permutations. The pangenome size shows a steady increase, reaching 1030 families after all genomes are included, while the core genome size rapidly declines toward zero and remains essentially zero beyond 5 genomes. The non-saturating pangenome curve and vanishing core indicate an open pangenome (consistent with Heaps’ law exponent α ≈ 0.026). B. Composition of the phage pangenome by gene frequency category. A pie chart illustrates the proportion of gene families that are core (present in all genomes, 0%), soft-core (present in ≥95% of genomes, 0%), shell (present in 15–<95% of genomes, 1.3%), and cloud (present in <15% of genomes, 98.7%). The cloud segment dominates, reflecting that almost all gene families are found in only one or a few phages. Shell families constitute a negligible fraction, and no universal core genes were identified among these phages.

Figure 5.
Pairwise AAI Analysis of 17 Phage Genomes. A. Network of AAI Similarities. A network visualization where nodes represent phages and edges indicate AAI values above a threshold, emphasizing connected components. The network highlights sparse connections, with notable edges between phage_16–phage_17 and phage_3–phage_4, consistent with their gene content similarities. Node sizes reflect the number of gene families per phage, and edge thickness corresponds to AAI similarity strength. The sparsity of edges reinforces the limited protein-level commonality among the phages. B. Heatmap of Pairwise AAI. Each cell shows the average amino-acid identity (AAI) as a percentage between proteins of two genomes, calculated from reciprocal best-hit pairs. Dark purple/black indicates no significant shared proteins (AAI ~0%), while brighter colors (yellow/green) indicate higher identity levels. Most genome pairs have AAI = 0%. A few pairs show low-to-moderate AAI: for instance, phage_16 vs phage_17 (≈46% AAI) and phage_11 vs phage_14 (≈70% AAI) appear as the brightest cells off the diagonal. These correspond to the same genome pairs that showed the closest relationships in gene content. The heatmap underscores that protein-level similarity between these phages is minimal in most cases, consistent with their divergent gene sets.
Figure 5.
Pairwise AAI Analysis of 17 Phage Genomes. A. Network of AAI Similarities. A network visualization where nodes represent phages and edges indicate AAI values above a threshold, emphasizing connected components. The network highlights sparse connections, with notable edges between phage_16–phage_17 and phage_3–phage_4, consistent with their gene content similarities. Node sizes reflect the number of gene families per phage, and edge thickness corresponds to AAI similarity strength. The sparsity of edges reinforces the limited protein-level commonality among the phages. B. Heatmap of Pairwise AAI. Each cell shows the average amino-acid identity (AAI) as a percentage between proteins of two genomes, calculated from reciprocal best-hit pairs. Dark purple/black indicates no significant shared proteins (AAI ~0%), while brighter colors (yellow/green) indicate higher identity levels. Most genome pairs have AAI = 0%. A few pairs show low-to-moderate AAI: for instance, phage_16 vs phage_17 (≈46% AAI) and phage_11 vs phage_14 (≈70% AAI) appear as the brightest cells off the diagonal. These correspond to the same genome pairs that showed the closest relationships in gene content. The heatmap underscores that protein-level similarity between these phages is minimal in most cases, consistent with their divergent gene sets.


Figure 6.
UpSet plot of shared gene families among the phages. The matrix at bottom indicates specific combinations of genomes (black dots connected by lines), and the vertical bars above show the number of gene families in the intersection of those genomes’ gene sets. Only the 25 largest intersections are shown. The tallest bars correspond to gene families unique to individual phages, single-dot intersections, with phage_10, phage_2, and phage_3 having the largest unique sets (bar heights 168, 112, 76 respectively). Far fewer gene families are shared by multiple phages. Notable smaller intersections include the set of families common to phage_16 and phage_17 (bar ≈16) and a set shared by phage_15–phage_16–phage_17. The horizontal bar chart on the right shows the total number of gene families present in each phage, which corresponds to each phage’s genome gene count. This plot highlights that most gene families are genome-specific, and only very limited overlaps exist between even the most closely related phages.
Figure 6.
UpSet plot of shared gene families among the phages. The matrix at bottom indicates specific combinations of genomes (black dots connected by lines), and the vertical bars above show the number of gene families in the intersection of those genomes’ gene sets. Only the 25 largest intersections are shown. The tallest bars correspond to gene families unique to individual phages, single-dot intersections, with phage_10, phage_2, and phage_3 having the largest unique sets (bar heights 168, 112, 76 respectively). Far fewer gene families are shared by multiple phages. Notable smaller intersections include the set of families common to phage_16 and phage_17 (bar ≈16) and a set shared by phage_15–phage_16–phage_17. The horizontal bar chart on the right shows the total number of gene families present in each phage, which corresponds to each phage’s genome gene count. This plot highlights that most gene families are genome-specific, and only very limited overlaps exist between even the most closely related phages.

Table 1.
Summary of pangenome statistics for the 17 bacteriophage genomes analyzed. Percentages for gene family categories are relative to the total number of gene families. The Heaps’ law α parameter (with α < 1 indicating an open pangenome) is shown to quantify pangenome openness.
Table 1.
Summary of pangenome statistics for the 17 bacteriophage genomes analyzed. Percentages for gene family categories are relative to the total number of gene families. The Heaps’ law α parameter (with α < 1 indicating an open pangenome) is shown to quantify pangenome openness.
| Metric |
Value |
| Number of phage genomes |
17 |
| Total predicted proteins (CDS) |
1122 |
| Total gene families (pangenome size) |
1031 |
| Core gene families (present in 100% genomes) |
0 (0%) |
| Soft-core gene families (present in ≥95% genomes) |
0 (0%) |
| Shell gene families (present in 15–<95% genomes) |
13 (1.3%) |
| Cloud gene families (present in <15% genomes) |
1018 (98.7%) |
| Singleton gene families (unique to 1 genome) |
966 (93.7%) |
| Heaps’ law α (pangenome openness) |
0.026 (Open) |