Submitted:
20 November 2025
Posted:
20 November 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methods
2.1. Transformer
2.2. Workflow
2.3. Datasets
2.4. Evaluation of TE Models
2.5. Software and Hardware Specifications
3. Results
3.1. Parameters Used for Training TE Models
3.2. Training of TE Models
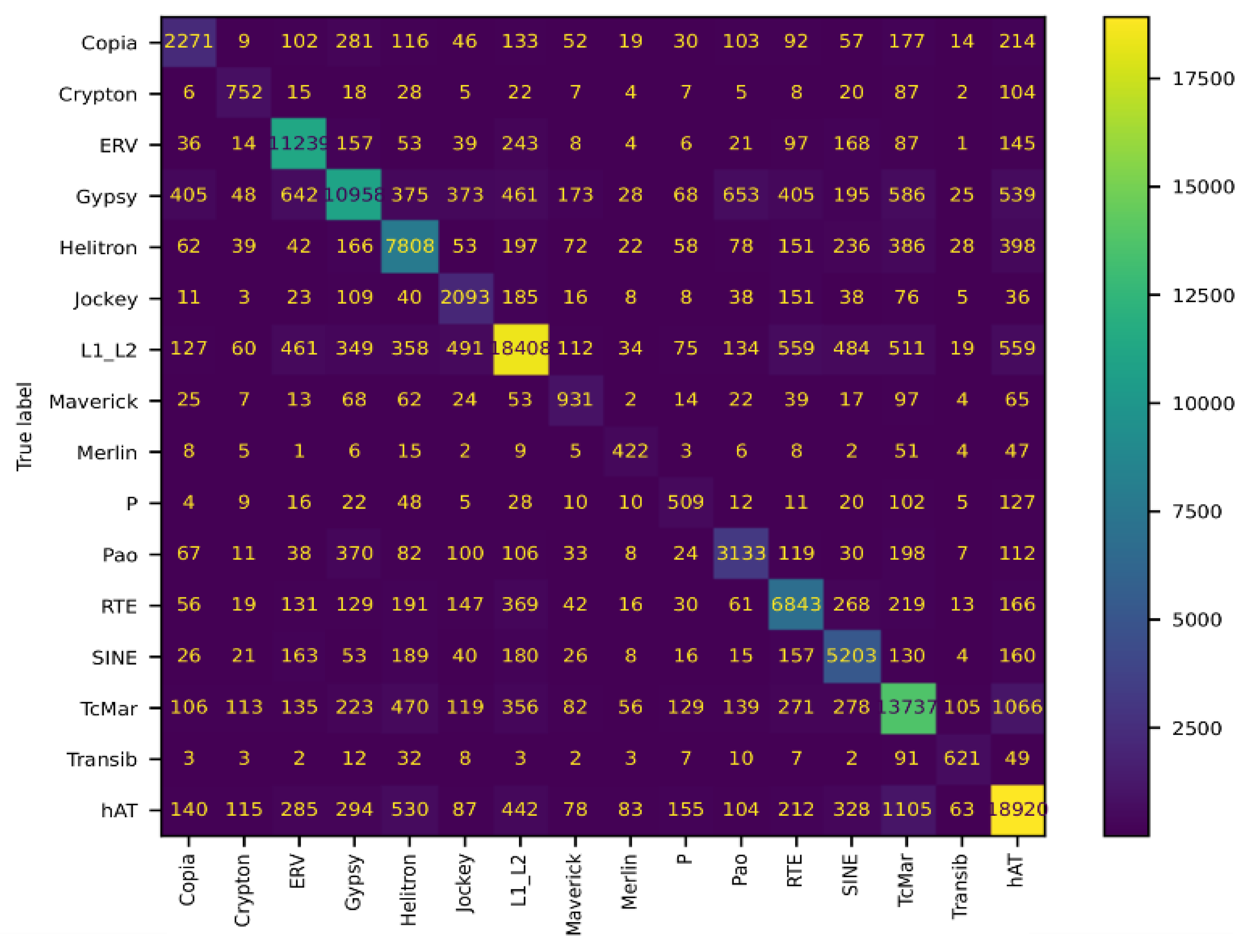
3.3. Comparison with Other Machine Learning Tools
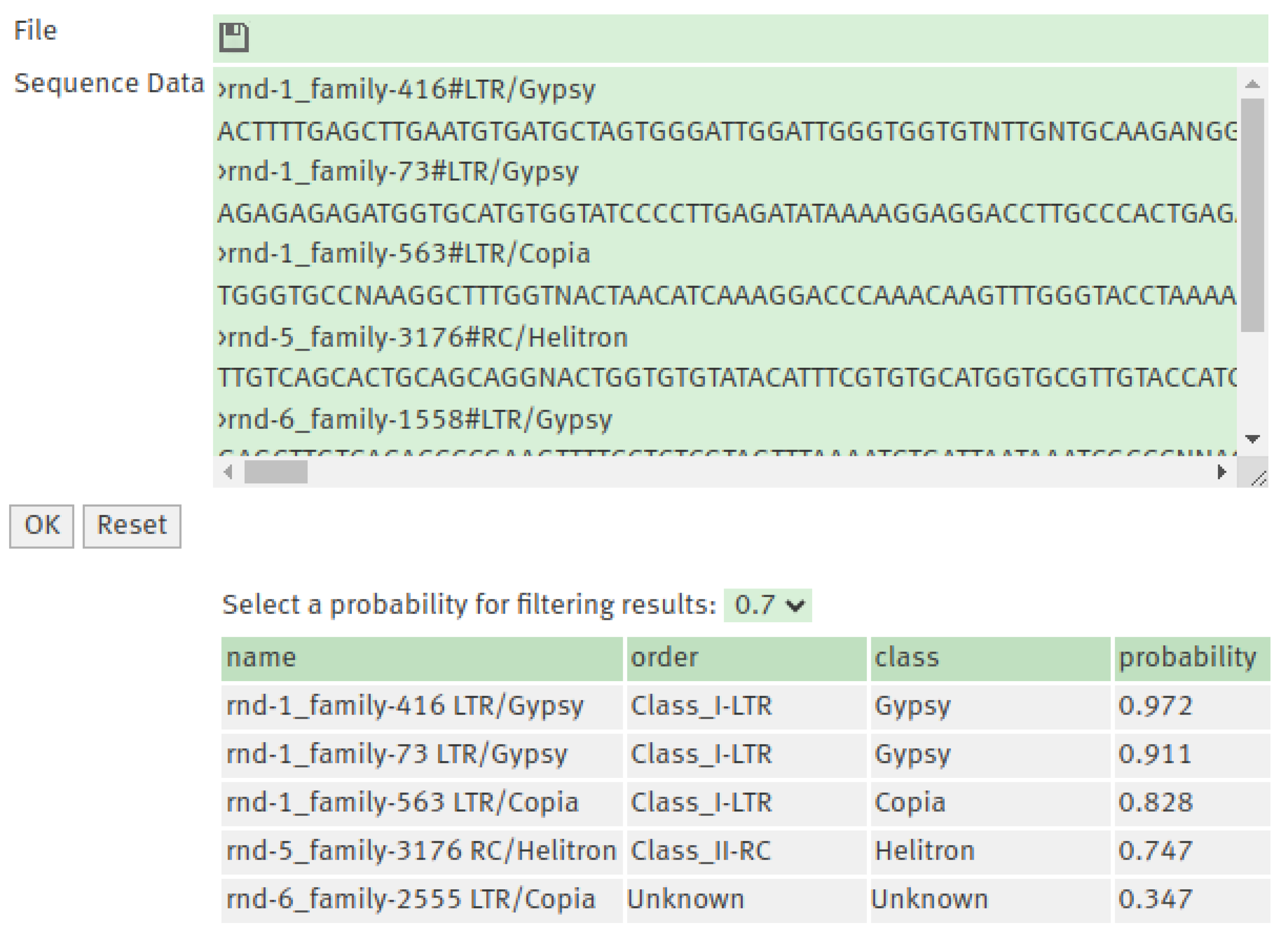
3.4. TEclass2 Web Interface
3.5. Known Limitations
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hoyt, S.J.; et al. From telomere to telomere: The transcriptional and epigenetic state of human repeat elements. Science 2022, 376, eabk3112. [Google Scholar] [CrossRef]
- Schnable, P.S.; et al. The B73 Maize Genome: Complexity, Diversity, and Dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef] [PubMed]
- Wicker, T.; et al. Impact of transposable elements on genome structure and evolution in bread wheat. Genome Biol 2018, 19, 103. [Google Scholar] [CrossRef] [PubMed]
- Moran, J.V.; DeBerardinis, R.J.; Kazazian, H.H., Jr. Exon shuffling by L1 retrotransposition. Science 1999, 283, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Piegu, B.; et al. Doubling genome size without polyploidization:: Dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Research 2006, 16, 1262–1269. [Google Scholar] [CrossRef]
- Ungerer, M.C.; Strakosh, S.C.; Zhen, Y. Genome expansion in three hybrid sunflower species is associated with retrotransposon proliferation. Current Biology 2006, 16, R872–R873. [Google Scholar] [CrossRef]
- Makalowski, W. Genomic scrap yard: how genomes utilize all that junk. Gene 2000, 259, 61–67. [Google Scholar] [CrossRef]
- Rodriguez, M.; Makalowski, W. Software evaluation for de novo detection of transposons. Mobile DNA 2022, 13. [Google Scholar] [CrossRef]
- Makalowski, W.; et al. Transposable Elements: Classification, Identification, and Their Use As a Tool For Comparative Genomics. Evolutionary Genomics, 2 Edition 2019, 1910, 177–207. [Google Scholar]
- Piégu, B.; et al. A survey of transposable element classification systems—A call for a fundamental update to meet the challenge of their diversity and complexity. Molecular Phylogenetics and Evolution 2015, 86, 90–109. [Google Scholar] [CrossRef]
- Finnegan, D.J. Eukaryotic transposable elements and genome evolution. Trends Genet 1989, 5, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Jurka, J.; et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 2005, 110, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Kapitonov, V.V.; Jurka, J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat Rev Genet 2008, 9, 411–412. [Google Scholar] [CrossRef] [PubMed]
- Wicker, T.; et al. A unified classification system for eukaryotic transposable elements. Nature Reviews Genetics 2007, 8, 973–982. [Google Scholar] [CrossRef]
- Bao, W.; Kojima, K.K.; Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA 2015, 6, 11. [Google Scholar] [CrossRef]
- Hoede, C.; et al. PASTEC: an automatic transposable element classification tool. PLoS One 2014, 9, e91929. [Google Scholar] [CrossRef]
- Wu, J.; Zhao, Y.Q. Machine learning technology in the application of genome analysis: A systematic review. Gene 2019, 705, 149–156. [Google Scholar] [CrossRef]
- Lan, K.; et al. A Survey of Data Mining and Deep Learning in Bioinformatics. J Med Syst 2018, 42, 139. [Google Scholar] [CrossRef]
- Li, R.; et al. Machine learning meets omics: applications and perspectives. Brief Bioinform 2022, 23. [Google Scholar] [CrossRef]
- Abrusan, G.; et al. TEclass--a tool for automated classification of unknown eukaryotic transposable elements. Bioinformatics 2009, 25, 1329–1330. [Google Scholar] [CrossRef]
- Yan, H.; Bombarely, A.; Li, S. DeepTE: a computational method for de novo classification of transposons with convolutional neural network. Bioinformatics 2020, 36, 4269–4275. [Google Scholar] [CrossRef]
- da Cruz, M.H.P.; et al. TERL: classification of transposable elements by convolutional neural networks. Briefings in bioinformatics 2021, 22, bbaa185. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, R.; et al. Terrier: a deep learning repeat classifier. Brief Bioinform 2025, 26. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Gu, X.; Peterson, T. TIR-Learner, a new ensemble method for TIR transposable element annotation, provides evidence for abundant new transposable elements in the maize genome. Molecular plant 2019, 12, 447–460. [Google Scholar] [CrossRef] [PubMed]
- Schietgat, L.; et al. A machine learning based framework to identify and classify long terminal repeat retrotransposons. PLoS computational biology 2018, 14, e1006097. [Google Scholar] [CrossRef]
- Orozco-Arias, S.; et al. Inpactor2: a software based on deep learning to identify and classify LTR-retrotransposons in plant genomes. Brief Bioinform 2023, 24. [Google Scholar] [CrossRef]
- Vaswani, A.; et al. Attention is all you need. Advances in neural information processing systems 2017, 30. [Google Scholar]
- Wolf, T.; et al. Transformers: State-of-the-art natural language processing. In Proceedings of the 2020 conference on empirical methods in natural language processing: system demonstrations. 2020.
- Ji, Y.; et al. DNABERT: pre-trained Bidirectional Encoder Representations from Transformers model for DNA-language in genome. Bioinformatics 2021, 37, 2112–2120. [Google Scholar] [CrossRef]
- Devlin, J.; et al. Bert: Pre-training of deep bidirectional transformers for language understanding. In Proceedings of the 2019 conference of the North American chapter of the association for computational linguistics: human language technologies, volume 1 (long and short papers), volume 1 (long and short papers). 2019, 2019. [Google Scholar]
- He, S.; et al. Nucleic transformer: Deep learning on nucleic acids with self-attention and convolutions. bioRxiv 2021. [Google Scholar] [CrossRef]
- Dalla-Torre, H.; et al. Nucleotide Transformer: building and evaluating robust foundation models for human genomics. Nat Methods 2025, 22, 287–297. [Google Scholar] [CrossRef]
- Wang, S.; et al. Linformer: Self-attention with linear complexity (2020). arXiv 2006, arXiv:2006.04768. [Google Scholar]
- Beltagy, I.; Peters, M.E.; Cohan, A. Longformer: The long-document transformer. arXiv 2020, arXiv:2004.05150. [Google Scholar] [CrossRef]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning Representations by Back-Propagating Errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Sutskever, I.; Vinyals, O.; Le, Q.V. Sequence to sequence learning with neural networks. Advances in neural information processing systems 2014, 27. [Google Scholar]
- Luong, M.-T.; Pham, H.; Manning, C.D. Effective approaches to attention-based neural machine translation. arXiv 2015, arXiv:1508.04025. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. Journal of Big Data 2019, 6. [Google Scholar] [CrossRef]
- Shaw, P.; Uszkoreit, J.; Vaswani, A. Self-attention with relative position representations. arXiv arXiv:1803.02155, 2018. [CrossRef]
- Suzuki, S.; et al. Genomic language models with k-mer tokenization strategies for plant genome annotation and regulatory element strength prediction. Plant Mol Biol 2025, 115, 100. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep learning. Adaptive computation and machine learning; The MIT Press: Cambridge, MA, USA, 2016; Volume xxii, 775p. [Google Scholar]
- Harris, C.R.; et al. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; et al. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research 2011, 12, 2825–2830. [Google Scholar]
- Paszke, A.; et al. Pytorch: An imperative style, high-performance deep learning library. Advances in neural information processing systems 2019, 32. [Google Scholar]
- Abadi, M.; et al. Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv arXiv:1603.04467, 2016.
- Flynn, J.M.; et al. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci U S A 2020, 117, 9451–9457. [Google Scholar] [CrossRef]
- Dong, Y.; Cordonnier, J.-B.; Loukas, A. Attention is not all you need: Pure attention loses rank doubly exponentially with depth. International conference on machine learning. 2021. [Google Scholar]
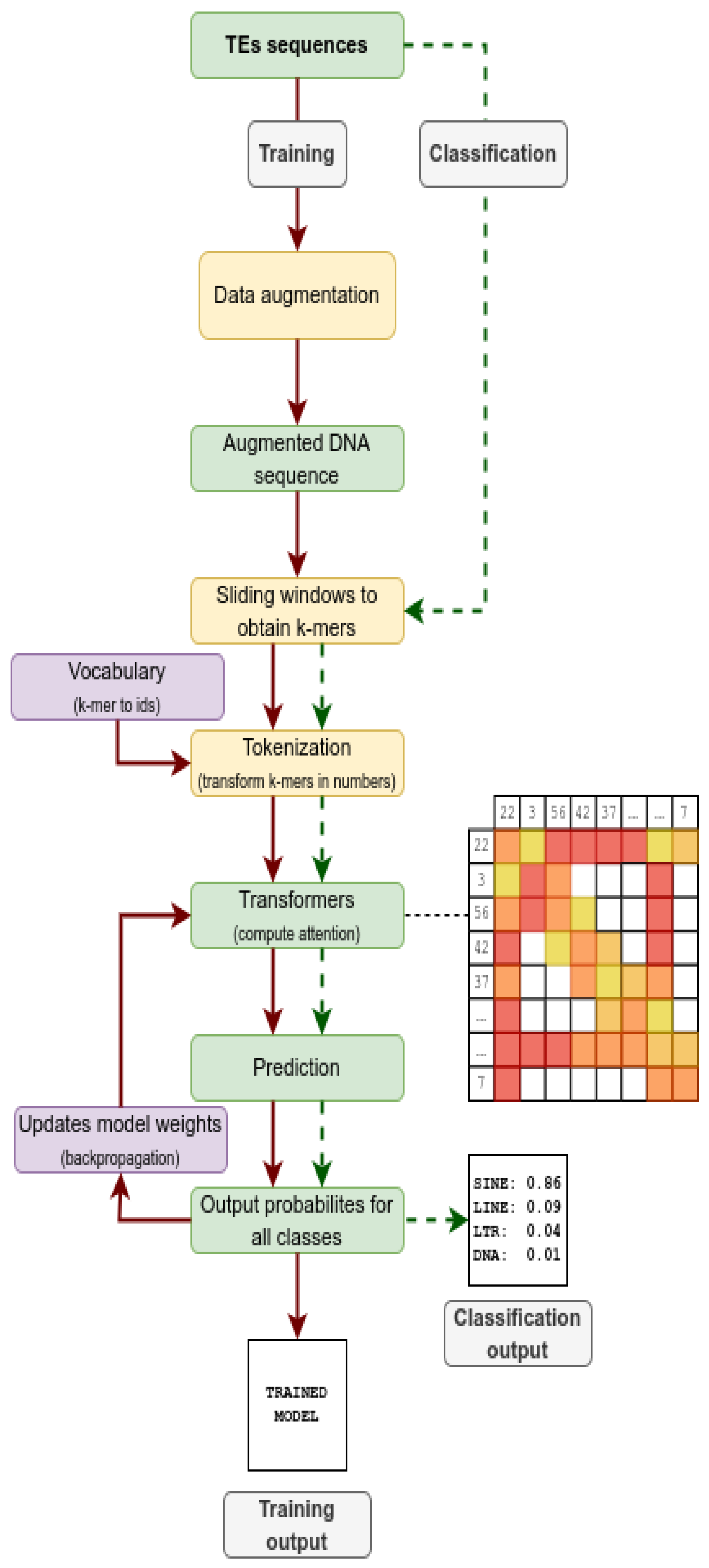
| Augmentation | Description |
|---|---|
| SNP | Replace randomly a single nucleotide. |
| Masking | Replace a base with an N. |
| Insertion | Insert random nucleotides in a sequence. |
| Deletion | Delete a random part of a sequence. |
| Repeat | Repeats a random part of the sequence. |
| Reverse | Reverses the sequence. |
| Complement | Computes the complement sequence. |
| Reverse complement | Computes the opposite strand. |
| Add tail | Adds poly-A tail to the sequence. |
| Remove tail | Removes poly-A tail if present. |
| Group | Training | Validation | Test | Total |
|---|---|---|---|---|
| Copia | 18584 | 3717 | 2478 | 24779 |
| Crypton | 5453 | 1091 | 727 | 7270 |
| ERV | 61539 | 12319 | 8212 | 82124 |
| Gypsy | 79674 | 15935 | 10623 | 106232 |
| hAT | 114707 | 22941 | 15294 | 152943 |
| Helitron | 48980 | 9796 | 6531 | 65307 |
| Jockey | 14201 | 2840 | 1894 | 18935 |
| L1/L2 | 113708 | 22742 | 15161 | 151610 |
| Maverick | 7218 | 1444 | 962 | 9624 |
| Merlin | 2971 | 594 | 396 | 3961 |
| P | 4692 | 938 | 626 | 6256 |
| Pao | 22192 | 4438 | 2959 | 29589 |
| RTE | 43500 | 8700 | 5800 | 58000 |
| SINE | 31960 | 6392 | 4261 | 42613 |
| TcMar | 86925 | 17385 | 11590 | 115900 |
| Transib | 4279 | 856 | 571 | 5705 |
| Total: | 660636 | 132127 | 88085 | 880848 |
| Parameters | Values | Description |
|---|---|---|
| num_epochs | 100 | Times the model is trained on the same data. |
| max_embeddings | 2048 | Maximum length of the input sequence, but can be overcome with the Longformer model. |
| num_hidden_layers | 8 | Dimensions of the encoder hidden state for processing the input. |
| num_attention_heads | 8 | Number of heads (neurons) used to process input data and make predictions. |
| global_att_tokens | [0, 256, 512] | Positions of the input where all the sequence is considered |
| intermediate_size | 3078 | Dimensionality of the feed-forward layer |
| kmer_size | 5 | Length of the words for dividing the input sequence. |
| Group | Precision | Recall | F1-Score | Support |
|---|---|---|---|---|
| Copia | 0.69 | 0.60 | 0.64 | 3716 |
| Crypton | 0.62 | 0.69 | 0.65 | 1090 |
| ERV | 0.86 | 0.91 | 0.88 | 12318 |
| Gypsy | 0.81 | 0.71 | 0.76 | 15934 |
| hAT | 0.84 | 0.82 | 0.83 | 22941 |
| Helitron | 0.72 | 0.81 | 0.76 | 9796 |
| Jockey | 0.61 | 0.71 | 0.66 | 2840 |
| L1/L2 | 0.89 | 0.80 | 0.84 | 22741 |
| Maverick | 0.56 | 0.64 | 0.60 | 1443 |
| Merlin | 0.64 | 0.70 | 0.67 | 594 |
| P | 0.35 | 0.60 | 0.44 | 938 |
| Pao | 0.71 | 0.70 | 0.70 | 4438 |
| RTE | 0.76 | 0.78 | 0.77 | 8700 |
| SINE | 0.76 | 0.80 | 0.78 | 6391 |
| TcMar | 0.76 | 0.77 | 0.76 | 17385 |
| Transib | 0.67 | 0.75 | 0.71 | 855 |
| Accuracy: | 0.79 | 132120 | ||
| Macro avg: | 0.70 | 0.74 | 0.72 | 132120 |
| Weighted avg: | 0.79 | 0.79 | 0.79 | 132120 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





