Submitted:
12 November 2025
Posted:
13 November 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Origin of the Intratumoral Microbiota and Relationships with Other Human Microbiomes
3. The Role of the Intratumoral Microbiome in Cancer Progression
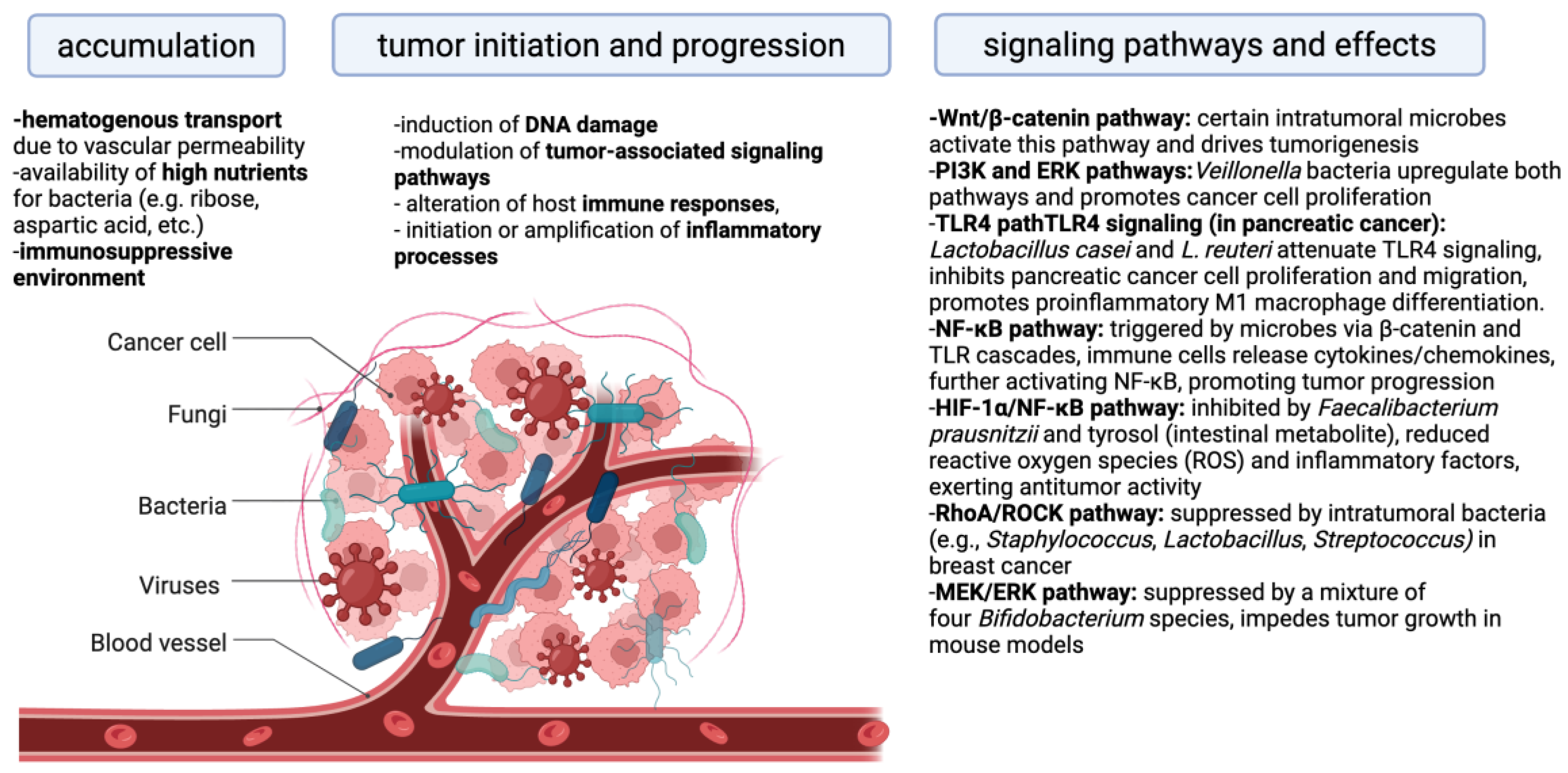
4. The Relationship Between Immune Cells and Intratumoral Microbiota in the Context of Treatment Outcomes
| Year | Disease/Model | Population | Brief Results | References |
|---|---|---|---|---|
| 2024 | Colorectal cancer | Data from The Cancer Genome Atlas (TCGA) database | Network analysis revealed significant interactions between microbial abundance and genes involved in CTL evasion. Among these, suppressor of cytokine signaling 1 (SOCS1) exhibited the highest number of negative correlations, particularly with the genera Phascolarctobacterium, Sneathia, and Intestinimonas. Additionally, the genus Oscillibacter was negatively associated with exon skipping in the CD74 gene, indicating that the tumor-associated microbiota may influence the regulation of antigen presentation and thereby modulate the antitumor immune response. | Liu et al. [100] |
| 2024 | Breast cancer | Female MMTV-PyMT transgenic mice | Both three-dimensional imaging and X-Y optical sections revealed spatial segregation between the intratumor microbiome and CD4⁺ and/or CD8⁺ T cell clusters, indicating the exclusion of activated T cells from bacterially colonized tumor regions. These data are consistent with the observed isolation of TLS and NK cells from microbe-enriched areas, highlighting the spatial compartmentalization of immune and microbial niches within tumor tissue. | Wang et al. [96] |
| 2022 | Lung cancer | 12 patients with early-stage lung cancer |
The bacterial load was significantly higher in tumor cells compared to T cells, macrophages, other immune cells, and stromal components, forming a gradient that increased from normal lung tissue and tertiary lymphoid structures to tumor cells and the airways. This pattern suggests potential penetration of intratumoral bacteria through the respiratory tract. Moreover, bacterial load levels showed a positive correlation with the expression of oncogenic β-catenin, tumor histological type, and environmental exposures. | Wong-Rolle et al. [101] |
| 2024 | Colorectal cancer | C57BL/6 (B6) and Balb/c mice | The immunogenic chemotherapeutic agent oxaliplatin synergizes with E. coli, activating the innate and adaptive immune response in the colorectal tumor microenvironment, leading to complete remission and the formation of stable antitumor immunological memory in mice. The combined action of oxaliplatin and bacteria significantly enhances the expression of costimulatory and antigen-presenting molecules on antigen-presenting cells, facilitating the effective activation of cytotoxic T lymphocytes against tumor cells. | Lim et al. [102] |
| 2022 | Colorectal cancer | C57BL/6 (B6) Thy 1.1 mice | The live attenuated Brucella melitensis strain (BmΔvjbR) was found to selectively colonize tumor tissue and remodel the tumor microenvironment by inducing proinflammatory polarization of M1 macrophages and enhancing both the number and activity of CD8⁺ cytotoxic T cells. In a colorectal adenocarcinoma model, treatment combining BmΔvjbR with adoptive transfer of tumor-specific CD8⁺ T cells almost completely suppressed tumor growth and achieved 100% animal survival. These findings highlight the potential of live attenuated bacteria to overcome tumor resistance to CAR-T therapy by remodeling the tumor microenvironment and activating macrophage-T-cell antitumor immunity. | Guo et al. [103] |
| 2023 | Leukemia | Non-obese diabetic scid gamma mice | A probiotic-targeted CAR-T cell (ProCAR) platform was developed in which tumor-colonizing probiotics secrete synthetic targets that mark tumor tissue for local lysis by CAR-T cells. Using the Escherichia coli Nissle 1917 strain with a synchronized lysis system (SLIC) enabled the release of synthetic targets directly into the tumor microenvironment, inducing safe and effective CAR-T cell activation in various cancer models. Additionally, an engineered strain co-expressing the chemokine mutant CXCL16K42A enhanced ProCAR-T cell recruitment and antitumor activity, resulting in increased hCD45⁺CD3⁺ T cell infiltration and significant tumor growth inhibition without toxic effects. | Vincent et al. [104][ |
| 2021 | Melanoma | C57BL/6NTac germ-free, BALB/cAnNCrl, B6-Ly5.1/Cr, B6-Ifnar1 (Ifnar1 KO) and C57BL/6J-Tmem173/J (STING KO) mice, 6 patients with melanoma | The microbiota regulates the immune compartment of the tumor microenvironment, reprogramming mononuclear phagocytes into immune-stimulatory monocytes and dendritic cells. The absence of microbiota shifts the balance of the tumor microbiome toward pro-tumorigenic macrophages, while microbial STING agonists induce type I interferon production, regulating macrophage polarization and NK cell–dendritic cell interactions. Modulation of the microbiota with a high-fiber diet activated the IFN-I–NK–DC axis and enhanced the efficacy of immune checkpoint blockade therapy, as confirmed in both experimental models and patients with melanoma. | Lam et al. [105] |
| 2022 | Pancreatic cancer | C57BL/6 mice | Oncogenic KrasG12D was shown to induce IL-33 expression in pancreatic ductal adenocarcinoma cells, which promotes the recruitment and activation of TH2 and ILC2 cells, which stimulate tumor growth. Ablation of IL-33 in tumor cells or antifungal therapy reduced TH2 and ILC2 cell infiltration, induced tumor regression, and increased survival. Thus, the intratumor mycobiome regulates IL-33 secretion and promotes the formation of a protumorigenic environment, opening up opportunities for targeted therapy for PDAC. | Alam et al. [97] |
| 2025 | Breast cancer | 89 female patients | In patients with early-stage TNBC treated with neoadjuvant chemo-immunotherapy, the pCR group exhibited higher intratumoral microbiota diversity and load compared to the non-pCR group. Single-cell RNA sequencing revealed enhanced T cell infiltration and reduced tumor-associated macrophages in the pCR group. Microbiota load positively correlated with CD4⁺CXCL13⁺ T cells and negatively with CD68⁺SPP1⁺ macrophages. Combined 16S and scRNA-seq analyses confirmed bacterial presence in both cancer and immune cells. A predictive model integrating microbial and clinicopathological data accurately forecasted pCR outcomes. | Chen et al. [98] |
| 2022 | Cutaneous melanoma | Data from The Cancer Genome Atlas (TCGA) database |
Low CD8⁺ T cell counts were associated with worse patient survival (OR = 1.57; 95% CI: 1.17–2.10; p = 0.002). The Lachnoclostridium genus showed the highest positive correlation with CD8⁺ T cell infiltration and expression of chemokines CXCL9, CXCL10, and CCL5, and its high level was associated with a reduced risk of mortality (p = 0.0003). | Zhu et al. [106] |
| 2021 | Prostate cancer | 137 men | Macrophages stimulated with Cutibacterium acnes in vitro were shown to increase the expression of PD-L1, CCL17, and CCL18 (p < 0.05), and the presence of C. acnes in prostate cancer patients was positively correlated with Tregs infiltration in the tumor stroma and epithelium (p = 0.0004 and p = 0.046). These data suggest that C. acnes contribute to the formation of an immunosuppressive tumor microenvironment that promotes prostate cancer progression. | Davidsson et al. [99] |
| 2018 | Pancreatic cancer | KC, C57BL/6 (H-2Kb) mice, | Pancreatic tumor tissue in mice and humans contains a significantly more abundant microbiome compared to normal tissue, with certain bacterial taxa selectively enriched compared to the intestine. Microbiome ablation prevented the development of pancreatic ductal adenocarcinoma and induced immune reprogramming of the microenvironment with increased M1 macrophage polarization and activation of CD4⁺ Th1 and CD8⁺ T cells. Furthermore, microbiome ablation enhanced the efficacy of immunotherapy by upregulating PD-1 expression, while the PDA microbiome induced a tolerogenic phenotype through activation of specific Toll-like receptors. | Pushalkar et al. [53] |
| 2019 | Pancreatic cancer | 68 patients | Long-term survivor patients with pancreatic ductal adenocarcinoma exhibited higher alpha diversity of the tumor microbiome and a characteristic microbial signature (Pseudoxanthomonas–Streptomyces–Saccharopolyspora–Bacillus clausii) predicting a favorable prognosis. Transplantation of microbiota from long-term survivor donors into mice slowed tumor growth and enhanced immune infiltration. Immunohistochemistry analysis revealed that long-term survivor patients had significantly higher densities of CD3⁺, CD8⁺, and GzmB⁺ T cells (p = 0.0273; p < 0.0001; p = 0.04), which positively correlated with both overall survival and microbiome diversity, suggesting a link between microbial diversity, CD8⁺ T cell activation, and the antitumor immune response. | Riquelme et al. [13] |
5. Current limitation and Future Direction
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CAR-T | Chimeric Antigen Receptor-Positive T-cell |
| NK | Natural Killer (cells) |
| NF-κB | NF-kappa B (Signaling Pathway) |
| PI3K | PI3K (Signaling Pathway) |
| TAMs | Tumor-Associated Macrophages |
| ACT | Adoptive Cell Transfer |
| OCI | Oncology Cellular Immunotherapy |
| DCs | Dendritic Cells |
| NKCs | Natural Killer Cells |
| CIKs | Cytokine-Induced Killer Cells |
| TILs | Tumor-Infiltrating Lymphocytes |
| LAKs | Lymphocyte-Activated Killer Cells |
| MAKs | Killer-Induced Macrophages |
| TCR-T | T-cell Receptor-Transfer |
| FDA | U.S. Food and Drug Administration |
| NGS | Next-Generation Sequencing |
| rRNA | Ribosomal RNA |
| WGS | Whole-Genome Shotgun sequencing |
| FISH | Fluorescence In Situ Hybridization |
| SCFAs | Short-Chain Fatty Acids |
| ROS | Reactive Oxygen Species |
| TLR | Toll-like receptors |
| ERK | ERK (Signaling Pathway) |
| RhoA/ROCK | RhoA/ROCK signaling pathway |
| TLR4 | Toll-like Receptor 4 |
| HIF-1α | Hypoxia-Inducible Factor 1 Alpha |
| HLA | Human Leukocyte Antigen |
| PAMPs | Pathogen-Associated Molecular Patterns |
| LPS | Lipopolysaccharides |
| PRRs | Pattern Recognition Receptors |
| TLRs | Toll-like Receptors |
| NLRs | Nod-like Receptors |
| VacA | Vacuolating cytotoxin A |
| TIGIT | T cell Immunoreceptor with Ig and ITIM domains |
| CEACAM1 | Carcinoembryonic antigen-related cell adhesion molecule 1 |
| OSCC | Oral Squamous Cell Carcinoma |
| GLUT1 | Glucose Transporter 1 |
| PDAC | Pancreatic Ductal Adenocarcinoma |
| ILC2 | Innate Lymphoid Cell 2 |
| PD-L1 | Programmed Death-Ligand 1 |
| TCGA | The Cancer Genome Atlas |
| CTL | Cytotoxic T Lymphocyte |
| TLS | Tertiary Lymphoid Structures |
| SLIC | Synchronized Lysis System |
| IFN-I | Type I Interferon |
| STING | Stimulator of Interferon Genes |
References
- Mukherjee, A.G.; Wanjari, U.R.; Namachivayam, A.; Murali, R.; Prabakaran, D.S.; Ganesan, R.; Renu, K.; Dey, A.; Vellingiri, B.; Ramanathan, G.; et al. Role of Immune Cells and Receptors in Cancer Treatment: An Immunotherapeutic Approach. Vaccines 2022, 10, 1493, doi:10.3390/vaccines10091493. [CrossRef]
- Zhang, Y.; Zhang, Z. The History and Advances in Cancer Immunotherapy: Understanding the Characteristics of Tumor-Infiltrating Immune Cells and Their Therapeutic Implications. Cell. Mol. Immunol. 2020, 17, 807–821, doi:10.1038/s41423-020-0488-6. [CrossRef]
- Hayes, C. Cellular Immunotherapies for Cancer. Ir. J. Med. Sci. 2021, 190, 41–57, doi:10.1007/s11845-020-02264-w. [CrossRef]
- Zhao, Y.; Deng, J.; Rao, S.; Guo, S.; Shen, J.; Du, F.; Wu, X.; Chen, Y.; Li, M.; Chen, M.; et al. Tumor Infiltrating Lymphocyte (TIL) Therapy for Solid Tumor Treatment: Progressions and Challenges. Cancers 2022, 14, 4160, doi:10.3390/cancers14174160. [CrossRef]
- Sharma, A.; Ren, X.; Rosato, A.; Sangiolo, D.; Wang, Z.; Tettamanti, S.; Zhang, Y.; Rettinger, E.; Fenix, K.A.; Sommaggio, R.; et al. Cytokine-Induced Killer (CIK) Cells, Successes and Challenges: Report on the First International Conference Dedicated to the Clinical Translation of This Unique Adoptive Cell Immunotherapy. Cancer Immunol. Immunother. CII 2024, 73, 21, doi:10.1007/s00262-023-03605-1. [CrossRef]
- Chu, J.; Gao, F.; Yan, M.; Zhao, S.; Yan, Z.; Shi, B.; Liu, Y. Natural Killer Cells: A Promising Immunotherapy for Cancer. J. Transl. Med. 2022, 20, 240, doi:10.1186/s12967-022-03437-0. [CrossRef]
- Wculek, S.K.; Cueto, F.J.; Mujal, A.M.; Melero, I.; Krummel, M.F.; Sancho, D. Dendritic Cells in Cancer Immunology and Immunotherapy. Nat. Rev. Immunol. 2020, 20, 7–24, doi:10.1038/s41577-019-0210-z. [CrossRef]
- Chen, P.; Chen, Y.; Wang, Y.; Sharma, A.; Veronika, L.-K.; Weiher, H.; Maria, A.G.-C.; Schmidt-Wolf, I.G.H. Macrophage-Derived pro-Inflammatory Cytokines Augment the Cytotoxicity of Cytokine-Induced Killer Cells by Strengthening the NKG2D Pathway in Multiple Myeloma. Sci. Rep. 2025, 15, 16739, doi:10.1038/s41598-025-99289-x. [CrossRef]
- Tan, S.; Li, D.; Zhu, X. Cancer Immunotherapy: Pros, Cons and Beyond. Biomed. Pharmacother. Biomedecine Pharmacother. 2020, 124, 109821, doi:10.1016/j.biopha.2020.109821. [CrossRef]
- Jogalekar, M.P.; Rajendran, R.L.; Khan, F.; Dmello, C.; Gangadaran, P.; Ahn, B.-C. CAR T-Cell-Based Gene Therapy for Cancers: New Perspectives, Challenges, and Clinical Developments. Front. Immunol. 2022, 13, 925985, doi:10.3389/fimmu.2022.925985. [CrossRef]
- Khan, A.N.; Asija, S.; Pendhari, J.; Purwar, R. CAR-T Cell Therapy in Hematological Malignancies: Where Are We Now and Where Are We Heading For? Eur. J. Haematol. 2024, 112, 6–18, doi:10.1111/ejh.14076. [CrossRef]
- Hu, L.; Fan, C.; Bross, P.; Das, A.; Cho, E.S.; Knudson, K.M.; Tegenge, M.; Gao, Q.; Brewer, J.R.; Theoret, M.R.; et al. FDA Approval Summary: Lifileucel for Unresectable or Metastatic Melanoma Previously Treated with an Anti–PD-1–Based Immunotherapy. Clin. Cancer Res. 2025, 31, 4004–4009, doi:10.1158/1078-0432.CCR-25-0880. [CrossRef]
- Zhao, T.; You, J.; Wang, C.; Li, B.; Liu, Y.; Shao, M.; Zhao, W.; Zhou, C. Cell-Based Immunotherapies for Solid Tumors: Advances, Challenges, and Future Directions. Front. Oncol. 2025, 15, 1551583, doi:10.3389/fonc.2025.1551583. [CrossRef]
- Bates, S.M.; Evans, K.V.; Delsing, L.; Wong, R.; Cornish, G.; Bahjat, M. Immune Safety Challenges Facing the Preclinical Assessment and Clinical Progression of Cell Therapies. Drug Discov. Today 2024, 29, 104239, doi:10.1016/j.drudis.2024.104239. [CrossRef]
- Leonov, G.E.; Grinchevskaya, L.R.; Makhnach, O.V.; Samburova, M.V.; Goldshtein, D.V.; Salikhova, D.I. Safety Assessment of Stem Cell-Based Therapies: Current Standards and Advancing Frameworks. Cells 2025, 14, 1660, doi:10.3390/cells14211660. [CrossRef]
- Jayathilaka, B.; Mian, F.; Franchini, F.; Au-Yeung, G.; IJzerman, M. Cancer and Treatment Specific Incidence Rates of Immune-Related Adverse Events Induced by Immune Checkpoint Inhibitors: A Systematic Review. Br. J. Cancer 2025, 132, 51–57, doi:10.1038/s41416-024-02887-1. [CrossRef]
- Mw, H.; Sy, J.; Ch, S.; H, P.; Jp, G.; Ry, H.; Kw, K.; Dh, Y. Incidence of Immune Effector Cell-Associated Neurotoxicity among Patients Treated with CAR T-Cell Therapy for Hematologic Malignancies: Systematic Review and Meta-Analysis. Front. Neurol. 2024, 15, doi:10.3389/fneur.2024.1392831. [CrossRef]
- Garg, P.; Pareek, S.; Kulkarni, P.; Horne, D.; Salgia, R.; Singhal, S.S. Next-Generation Immunotherapy: Advancing Clinical Applications in Cancer Treatment. J. Clin. Med. 2024, 13, 6537, doi:10.3390/jcm13216537. [CrossRef]
- Ahmad, A.; Mahmood, N.; Raza, M.A.; Mushtaq, Z.; Saeed, F.; Afzaal, M.; Hussain, M.; Amjad, H.W.; Al-Awadi, H.M. Gut Microbiota and Their Derivatives in the Progression of Colorectal Cancer: Mechanisms of Action, Genome and Epigenome Contributions. Heliyon 2024, 10, e29495, doi:10.1016/j.heliyon.2024.e29495. [CrossRef]
- Rahman, M.M.; Islam, M.R.; Shohag, S.; Ahasan, M.T.; Sarkar, N.; Khan, H.; Hasan, A.M.; Cavalu, S.; Rauf, A. Microbiome in Cancer: Role in Carcinogenesis and Impact in Therapeutic Strategies. Biomed. Pharmacother. Biomedecine Pharmacother. 2022, 149, 112898, doi:10.1016/j.biopha.2022.112898. [CrossRef]
- Hou, K.; Wu, Z.-X.; Chen, X.-Y.; Wang, J.-Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in Health and Diseases. Signal Transduct. Target. Ther. 2022, 7, 135, doi:10.1038/s41392-022-00974-4. [CrossRef]
- Zhang, M.; Liu, J.; Xia, Q. Role of Gut Microbiome in Cancer Immunotherapy: From Predictive Biomarker to Therapeutic Target. Exp. Hematol. Oncol. 2023, 12, 84, doi:10.1186/s40164-023-00442-x. [CrossRef]
- Clavijo-Salomon, M.A.; Trinchieri, G. Unlocking the Power of the Microbiome for Successful Cancer Immunotherapy. J. Immunother. Cancer 2025, 13, doi:10.1136/jitc-2024-011281. [CrossRef]
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The Human Tumor Microbiome Is Composed of Tumor Type–Specific Intracellular Bacteria. Science 2020, doi:10.1126/science.aay9189. [CrossRef]
- Xue, C.; Chu, Q.; Zheng, Q.; Yuan, X.; Su, Y.; Bao, Z.; Lu, J.; Li, L. Current Understanding of the Intratumoral Microbiome in Various Tumors. Cell Rep. Med. 2023, 4, 100884, doi:10.1016/j.xcrm.2022.100884. [CrossRef]
- Eisenhofer, R.; Minich, J.J.; Marotz, C.; Cooper, A.; Knight, R.; Weyrich, L.S. Contamination in Low Microbial Biomass Microbiome Studies: Issues and Recommendations. Trends Microbiol. 2019, 27, 105–117, doi:10.1016/j.tim.2018.11.003. [CrossRef]
- Gihawi, A.; Ge, Y.; Lu, J.; Puiu, D.; Xu, A.; Cooper, C.S.; Brewer, D.S.; Pertea, M.; Salzberg, S.L. Major Data Analysis Errors Invalidate Cancer Microbiome Findings. mBio 2023, 14, e01607-23, doi:10.1128/mbio.01607-23. [CrossRef]
- Triner, D.; Devenport, S.N.; Ramakrishnan, S.K.; Ma, X.; Frieler, R.A.; Greenson, J.K.; Inohara, N.; Nunez, G.; Colacino, J.A.; Mortensen, R.M.; et al. Neutrophils Restrict Tumor-Associated Microbiota to Reduce Growth and Invasion of Colon Tumors in Mice. Gastroenterology 2019, 156, 1467–1482, doi:10.1053/j.gastro.2018.12.003. [CrossRef]
- Singh, R.P.; Kumari, N.; Gupta, S.; Jaiswal, R.; Mehrotra, D.; Singh, S.; Mukherjee, S.; Kumar, R. Intratumoral Microbiota Changes with Tumor Stage and Influences the Immune Signature of Oral Squamous Cell Carcinoma. Microbiol. Spectr. 2023, doi:10.1128/spectrum.04596-22. [CrossRef]
- Reynoso-García, J.; Miranda-Santiago, A.E.; Meléndez-Vázquez, N.M.; Acosta-Pagán, K.; Sánchez-Rosado, M.; Díaz-Rivera, J.; Rosado-Quiñones, A.M.; Acevedo-Márquez, L.; Cruz-Roldán, L.; Tosado-Rodríguez, E.L.; et al. A Complete Guide to Human Microbiomes: Body Niches, Transmission, Development, Dysbiosis, and Restoration. Front. Syst. Biol. 2022, 2, doi:10.3389/fsysb.2022.951403. [CrossRef]
- Kitamoto, S.; Nagao-Kitamoto, H.; Hein, R.; Schmidt, T.M.; Kamada, N. The Bacterial Connection between the Oral Cavity and the Gut Diseases. J. Dent. Res. 2020, 99, 1021–1029, doi:10.1177/0022034520924633. [CrossRef]
- Minalyan, A.; Gabrielyan, L.; Scott, D.; Jacobs, J.; Pisegna, J.R. The Gastric and Intestinal Microbiome: Role of Proton Pump Inhibitors. Curr. Gastroenterol. Rep. 2017, 19, 42, doi:10.1007/s11894-017-0577-6. [CrossRef]
- Li, X.; Zhang, S.; Sheng, H.; Zhen, Y.; Wu, B.; Li, Z.; Chen, D.; Zhou, H. Oral Fusobacterium Nucleatum Resists the Acidic pH of the Stomach Due to Membrane Erucic Acid Synthesized via Enoyl-CoA Hydratase-Related Protein FnFabM. J. Oral Microbiol. 2025, 17, 2453964, doi:10.1080/20002297.2025.2453964. [CrossRef]
- Woelfel, S.; Silva, M.S.; Stecher, B. Intestinal Colonization Resistance in the Context of Environmental, Host, and Microbial Determinants. Cell Host Microbe 2024, 32, 820–836, doi:10.1016/j.chom.2024.05.002. [CrossRef]
- Rashidi, A.; Ebadi, M.; Weisdorf, D.J.; Costalonga, M.; Staley, C. No Evidence for Colonization of Oral Bacteria in the Distal Gut in Healthy Adults. Proc. Natl. Acad. Sci. 2021, 118, e2114152118, doi:10.1073/pnas.2114152118. [CrossRef]
- Rashidi, A.; Koyama, M.; Dey, N.; McLean, J.S.; Hill, G.R. Colonization Resistance Is Dispensable for Segregation of Oral and Gut Microbiota. BMC Med. Genomics 2023, 16, 31, doi:10.1186/s12920-023-01449-3. [CrossRef]
- Kunath, B.J.; De Rudder, C.; Laczny, C.C.; Letellier, E.; Wilmes, P. The Oral–Gut Microbiome Axis in Health and Disease. Nat. Rev. Microbiol. 2024, 22, 791–805, doi:10.1038/s41579-024-01075-5. [CrossRef]
- Tortora, S.C.; Agurto, M.G.; Martello, L.A. The Oral-Gut-Circulatory Axis: From Homeostasis to Colon Cancer. Front. Cell. Infect. Microbiol. 2023, 13, doi:10.3389/fcimb.2023.1289452. [CrossRef]
- De Pessemier, B.; Grine, L.; Debaere, M.; Maes, A.; Paetzold, B.; Callewaert, C. Gut–Skin Axis: Current Knowledge of the Interrelationship between Microbial Dysbiosis and Skin Conditions. Microorganisms 2021, 9, 353, doi:10.3390/microorganisms9020353. [CrossRef]
- Elkafas, H.; Walls, M.; Al-Hendy, A.; Ismail, N. Gut and Genital Tract Microbiomes: Dysbiosis and Link to Gynecological Disorders. Front. Cell. Infect. Microbiol. 2022, 12, 1059825, doi:10.3389/fcimb.2022.1059825. [CrossRef]
- Mahmud, Md.R.; Akter, S.; Tamanna, S.K.; Mazumder, L.; Esti, I.Z.; Banerjee, S.; Akter, S.; Hasan, Md.R.; Acharjee, M.; Hossain, Md.S.; et al. Impact of Gut Microbiome on Skin Health: Gut-Skin Axis Observed through the Lenses of Therapeutics and Skin Diseases. Gut Microbes 14, 2096995, doi:10.1080/19490976.2022.2096995. [CrossRef]
- Somodi, C.; Dora, D.; Horváth, M.; Szegvari, G.; Lohinai, Z. Gut Microbiome Changes and Cancer Immunotherapy Outcomes Associated with Dietary Interventions: A Systematic Review of Preclinical and Clinical Evidence. J. Transl. Med. 2025, 23, 756, doi:10.1186/s12967-025-06586-0. [CrossRef]
- Dokoshi, T.; Chen, Y.; Cavagnero, K.J.; Rahman, G.; Hakim, D.; Brinton, S.; Schwarz, H.; Brown, E.A.; O’Neill, A.; Nakamura, Y.; et al. Dermal Injury Drives a Skin to Gut Axis That Disrupts the Intestinal Microbiome and Intestinal Immune Homeostasis in Mice. Nat. Commun. 2024, 15, 3009, doi:10.1038/s41467-024-47072-3. [CrossRef]
- Dang, A.T.; Marsland, B.J. Microbes, Metabolites, and the Gut–Lung Axis. Mucosal Immunol. 2019, 12, 843–850, doi:10.1038/s41385-019-0160-6. [CrossRef]
- Ivleva, E.A.; Grivennikov, S.I. Microbiota-Driven Mechanisms at Different Stages of Cancer Development. Neoplasia N. Y. N 2022, 32, 100829, doi:10.1016/j.neo.2022.100829. [CrossRef]
- Dapito, D.H.; Mencin, A.; Gwak, G.-Y.; Pradere, J.-P.; Jang, M.-K.; Mederacke, I.; Caviglia, J.M.; Khiabanian, H.; Adeyemi, A.; Bataller, R.; et al. Promotion of Hepatocellular Carcinoma by the Intestinal Microbiota and TLR4. Cancer Cell 2012, 21, 504–516, doi:10.1016/j.ccr.2012.02.007. [CrossRef]
- Bertocchi, A.; Carloni, S.; Ravenda, P.S.; Bertalot, G.; Spadoni, I.; Lo Cascio, A.; Gandini, S.; Lizier, M.; Braga, D.; Asnicar, F.; et al. Gut Vascular Barrier Impairment Leads to Intestinal Bacteria Dissemination and Colorectal Cancer Metastasis to Liver. Cancer Cell 2021, 39, 708-724.e11, doi:10.1016/j.ccell.2021.03.004. [CrossRef]
- Bullman, S.; Pedamallu, C.S.; Sicinska, E.; Clancy, T.E.; Zhang, X.; Cai, D.; Neuberg, D.; Huang, K.; Guevara, F.; Nelson, T.; et al. Analysis of Fusobacterium Persistence and Antibiotic Response in Colorectal Cancer. Science 2017, 358, 1443–1448, doi:10.1126/science.aal5240. [CrossRef]
- Senthakumaran, T.; Moen, A.E.F.; Tannæs, T.M.; Endres, A.; Brackmann, S.A.; Rounge, T.B.; Bemanian, V.; Tunsjø, H.S. Microbial Dynamics with CRC Progression: A Study of the Mucosal Microbiota at Multiple Sites in Cancers, Adenomatous Polyps, and Healthy Controls. Eur. J. Clin. Microbiol. Infect. Dis. 2023, 42, 305–322, doi:10.1007/s10096-023-04551-7. [CrossRef]
- Menéndez, G.G.; Sichel, L.; López, M. del C.; Hernández, Y.; Arteaga, E.; Rodríguez, M.; Fleites, V.; Fernández, L.T.; Cano, R.D.J. From Colon Wall to Tumor Niche: Unraveling the Microbiome’s Role in Colorectal Cancer Progression. PLOS ONE 2024, 19, e0311233, doi:10.1371/journal.pone.0311233. [CrossRef]
- Zhu, Y.; Liu, W.; Wang, M.; Wang, X.; Wang, S. Causal Roles of Skin Microbiota in Skin Cancers Suggested by Genetic Study. Front. Microbiol. 2024, 15, doi:10.3389/fmicb.2024.1426807. [CrossRef]
- Tsay, J.-C.J.; Wu, B.G.; Sulaiman, I.; Gershner, K.; Schluger, R.; Li, Y.; Yie, T.-A.; Meyn, P.; Olsen, E.; Perez, L.; et al. Lower Airway Dysbiosis Affects Lung Cancer Progression. Cancer Discov. 2021, 11, 293–307, doi:10.1158/2159-8290.CD-20-0263. [CrossRef]
- Pushalkar, S.; Hundeyin, M.; Daley, D.; Zambirinis, C.P.; Kurz, E.; Mishra, A.; Mohan, N.; Aykut, B.; Usyk, M.; Torres, L.E.; et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2018, 8, 403–416, doi:10.1158/2159-8290.CD-17-1134. [CrossRef]
- Meng, Y.-F.; Fan, Z.-Y.; Zhou, B.; Zhan, H.-X. Role of the Intratumoral Microbiome in Tumor Progression and Therapeutics Implications. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 189014, doi:10.1016/j.bbcan.2023.189014. [CrossRef]
- Xie, Z.; Wu, Z.; Liu, Y.; Gu, Y.; Niu, J.; Lv, K. Intratumoral Microbiota: Implications for Cancer Progression and Treatment. Front. Microbiol. 2025, 16, 1551515, doi:10.3389/fmicb.2025.1551515. [CrossRef]
- Narunsky-Haziza, L.; Sepich-Poore, G.D.; Livyatan, I.; Asraf, O.; Martino, C.; Nejman, D.; Gavert, N.; Stajich, J.E.; Amit, G.; González, A.; et al. Pan-Cancer Analyses Reveal Cancer-Type-Specific Fungal Ecologies and Bacteriome Interactions. Cell 2022, 185, 3789-3806.e17, doi:10.1016/j.cell.2022.09.005. [CrossRef]
- Zhou, Z.; Lv, Y.; Zuo, A.; Zhu, X.; Xu, Y.; Zuo, L.; Xu, H.; Liu, S.; Zhang, Y.; Weng, S.; et al. Interactions between Microbiota and Innate Immunity in Tumor Microenvironment: Novel Insights into Cancer Progression and Immunotherapy. hLife 2025, 3, 462–493, doi:10.1016/j.hlife.2025.05.008. [CrossRef]
- Chen, Y.; Wu, F.-H.; Wu, P.-Q.; Xing, H.-Y.; Ma, T. The Role of The Tumor Microbiome in Tumor Development and Its Treatment. Front. Immunol. 2022, 13, 935846, doi:10.3389/fimmu.2022.935846. [CrossRef]
- Dohlman, A.B.; Mendoza, D.A.; Ding, S.; Gao, M.; Dressman, H.; Iliev, I.D.; Lipkin, S.M.; Shen, X. The Cancer Microbiome Atlas: A Pan-Cancer Comparative Analysis to Distinguish Tissue-Resident Microbiota from Contaminants. Cell Host Microbe 2021, 29, 281-298.e5, doi:10.1016/j.chom.2020.12.001. [CrossRef]
- Li, L.; Chandra, V.; McAllister, F. Tumor-Resident Microbes: The New Kids on the Microenvironment Block. Trends Cancer 2024, 10, 347–355, doi:10.1016/j.trecan.2023.12.002. [CrossRef]
- Wang, Y.; Guo, H.; Gao, X.; Wang, J. The Intratumor Microbiota Signatures Associate With Subtype, Tumor Stage, and Survival Status of Esophageal Carcinoma. Front. Oncol. 2021, 11, doi:10.3389/fonc.2021.754788. [CrossRef]
- Wang, J.; Wang, Y.; Li, Z.; Gao, X.; Huang, D. Global Analysis of Microbiota Signatures in Four Major Types of Gastrointestinal Cancer. Front. Oncol. 2021, 11, 685641, doi:10.3389/fonc.2021.685641. [CrossRef]
- Yu, G.; Gail, M.H.; Consonni, D.; Carugno, M.; Humphrys, M.; Pesatori, A.C.; Caporaso, N.E.; Goedert, J.J.; Ravel, J.; Landi, M.T. Characterizing Human Lung Tissue Microbiota and Its Relationship to Epidemiological and Clinical Features. Genome Biol. 2016, 17, 163, doi:10.1186/s13059-016-1021-1. [CrossRef]
- Xie, Y.; Xie, F.; Zhou, X.; Zhang, L.; Yang, B.; Huang, J.; Wang, F.; Yan, H.; Zeng, L.; Zhang, L.; et al. Microbiota in Tumors: From Understanding to Application. Adv. Sci. 2022, 9, 2200470, doi:10.1002/advs.202200470. [CrossRef]
- Rajasekaran, K.; Carey, R.M.; Lin, X.; Seckar, T.D.; Wei, Z.; Chorath, K.; Newman, J.G.; O’Malley, B.W.; Weinstein, G.S.; Feldman, M.D.; et al. The Microbiome of HPV-Positive Tonsil Squamous Cell Carcinoma and Neck Metastasis. Oral Oncol. 2021, 117, 105305, doi:10.1016/j.oraloncology.2021.105305. [CrossRef]
- Chiba, A.; Bawaneh, A.; Velazquez, C.; Clear, K.Y.J.; Wilson, A.S.; Howard-McNatt, M.; Levine, E.A.; Levi-Polyachenko, N.; Yates-Alston, S.A.; Diggle, S.P.; et al. Neoadjuvant Chemotherapy Shifts Breast Tumor Microbiota Populations to Regulate Drug Responsiveness and the Development of Metastasis. Mol. Cancer Res. MCR 2020, 18, 130–139, doi:10.1158/1541-7786.MCR-19-0451. [CrossRef]
- Zhang, W.; Xiang, Y.; Ren, H.; Liu, Y.; Wang, Q.; Ran, M.; Zhou, W.; Tian, L.; Zheng, X.; Qiao, C.; et al. The Tumor Microbiome in Cancer Progression: Mechanisms and Therapeutic Potential. Mol. Cancer 2025, 24, 195, doi:10.1186/s12943-025-02403-w. [CrossRef]
- Yang, L.; Li, A.; Wang, Y.; Zhang, Y. Intratumoral Microbiota: Roles in Cancer Initiation, Development and Therapeutic Efficacy. Signal Transduct. Target. Ther. 2023, 8, 35, doi:10.1038/s41392-022-01304-4. [CrossRef]
- Goodwin, A.C.; Shields, C.E.D.; Wu, S.; Huso, D.L.; Wu, X.; Murray-Stewart, T.R.; Hacker-Prietz, A.; Rabizadeh, S.; Woster, P.M.; Sears, C.L.; et al. Polyamine Catabolism Contributes to Enterotoxigenic Bacteroides Fragilis-Induced Colon Tumorigenesis. Proc. Natl. Acad. Sci. 2011, 108, 15354–15359, doi:10.1073/pnas.1010203108. [CrossRef]
- Sevcikova, A.; Mladosievicova, B.; Mego, M.; Ciernikova, S. Exploring the Role of the Gut and Intratumoral Microbiomes in Tumor Progression and Metastasis. Int. J. Mol. Sci. 2023, 24, 17199, doi:10.3390/ijms242417199. [CrossRef]
- Ji, Y.; Lv, J.; Sun, D.; Huang, Y. Therapeutic Strategies Targeting Wnt/Β catenin Signaling for Colorectal Cancer (Review). Int. J. Mol. Med. 2022, 49, 1–17, doi:10.3892/ijmm.2021.5056. [CrossRef]
- Yu, R.; Wang, S.; Han, L. Relevance of Harmful Intratumoral Microbiota in Cancer Progression and Its Clinical Application. Biomed. Pharmacother. 2024, 178, 117238, doi:10.1016/j.biopha.2024.117238. [CrossRef]
- Tsay, J.-C.J.; Wu, B.G.; Badri, M.H.; Clemente, J.C.; Shen, N.; Meyn, P.; Li, Y.; Yie, T.-A.; Lhakhang, T.; Olsen, E.; et al. Airway Microbiota Is Associated with Upregulation of the PI3K Pathway in Lung Cancer. Am. J. Respir. Crit. Care Med. 2018, 198, 1188–1198, doi:10.1164/rccm.201710-2118OC. [CrossRef]
- Pandya, G.; Kirtonia, A.; Singh, A.; Goel, A.; Mohan, C.D.; Rangappa, K.S.; Pandey, A.K.; Kapoor, S.; Tandon, S.; Sethi, G.; et al. A Comprehensive Review of the Multifaceted Role of the Microbiota in Human Pancreatic Carcinoma. Semin. Cancer Biol. 2022, 86, 682–692, doi:10.1016/j.semcancer.2021.05.027. [CrossRef]
- Guo, Q.; Jin, Y.; Chen, X.; Ye, X.; Shen, X.; Lin, M.; Zeng, C.; Zhou, T.; Zhang, J. NF-κB in Biology and Targeted Therapy: New Insights and Translational Implications. Signal Transduct. Target. Ther. 2024, 9, 53, doi:10.1038/s41392-024-01757-9. [CrossRef]
- Guo, J.; Meng, F.; Hu, R.; Chen, L.; Chang, J.; Zhao, K.; Ren, H.; Liu, Z.; Hu, P.; Wang, G.; et al. Inhibition of the NF-κB/HIF-1α Signaling Pathway in Colorectal Cancer by Tyrosol: A Gut Microbiota-Derived Metabolite. J. Immunother. Cancer 2024, 12, e008831, doi:10.1136/jitc-2024-008831. [CrossRef]
- Zhang, S.; Huang, J.; Jiang, Z.; Tong, H.; Ma, X.; Liu, Y. Tumor Microbiome: Roles in Tumor Initiation, Progression, and Therapy. Mol. Biomed. 2025, 6, 9, doi:10.1186/s43556-025-00248-9. [CrossRef]
- Fu, A.; Yao, B.; Dong, T.; Chen, Y.; Yao, J.; Liu, Y.; Li, H.; Bai, H.; Liu, X.; Zhang, Y.; et al. Tumor-Resident Intracellular Microbiota Promotes Metastatic Colonization in Breast Cancer. Cell 2022, 185, 1356-1372.e26, doi:10.1016/j.cell.2022.02.027. [CrossRef]
- Naghavian, R.; Faigle, W.; Oldrati, P.; Wang, J.; Toussaint, N.C.; Qiu, Y.; Medici, G.; Wacker, M.; Freudenmann, L.K.; Bonté, P.-E.; et al. Microbial Peptides Activate Tumour-Infiltrating Lymphocytes in Glioblastoma. Nature 2023, 617, 807–817, doi:10.1038/s41586-023-06081-w. [CrossRef]
- Zhang, H.; Fu, L.; Leiliang, X.; Qu, C.; Wu, W.; Wen, R.; Huang, N.; He, Q.; Cheng, Q.; Liu, G.; et al. Beyond the Gut: The Intratumoral Microbiome’s Influence on Tumorigenesis and Treatment Response. Cancer Commun. 2024, 44, 1130–1167, doi:10.1002/cac2.12597. [CrossRef]
- Lin, L.; Zhang, D. Unveiling the Microbial Influence: Bacteria’s Dual Role in Tumor Metastasis. Front. Oncol. 2025, 15, 1524887, doi:10.3389/fonc.2025.1524887. [CrossRef]
- Fan, H.; Wang, Y.; Han, M.; Wang, L.; Li, X.; Kuang, X.; Du, J.; Peng, F. Multi-Omics-Based Investigation of Bifidobacterium’s Inhibitory Effect on Glioma: Regulation of Tumor and Gut Microbiota, and MEK/ERK Cascade. Front. Microbiol. 2024, 15, doi:10.3389/fmicb.2024.1344284. [CrossRef]
- Mogensen, T.H. Pathogen Recognition and Inflammatory Signaling in Innate Immune Defenses. Clin. Microbiol. Rev. 2009, 22, 240–273, doi:10.1128/CMR.00046-08. [CrossRef]
- Zhang, J.; You, Z.; Li, X.; Hu, J.; Li, J.; Jing, Z. Harnessing Intratumoral Microbiota: New Horizons in Immune Microenvironment and Immunotherapy. J. Transl. Med. 2025, 23, 897, doi:10.1186/s12967-025-06916-2. [CrossRef]
- Qiao, H.; Tan, X.-R.; Li, H.; Li, J.-Y.; Chen, X.-Z.; Li, Y.-Q.; Li, W.-F.; Tang, L.-L.; Zhou, G.-Q.; Zhang, Y.; et al. Association of Intratumoral Microbiota With Prognosis in Patients With Nasopharyngeal Carcinoma From 2 Hospitals in China. JAMA Oncol. 2022, 8, 1301–1309, doi:10.1001/jamaoncol.2022.2810. [CrossRef]
- Xu, S.; Wu, X.; Zhang, X.; Chen, C.; Chen, H.; She, F. CagA Orchestrates eEF1A1 and PKCδ to Induce Interleukin-6 Expression in Helicobacter Pylori-Infected Gastric Epithelial Cells. Gut Pathog. 2020, 12, 31, doi:10.1186/s13099-020-00368-3. [CrossRef]
- Gur, C.; Ibrahim, Y.; Isaacson, B.; Yamin, R.; Abed, J.; Gamliel, M.; Enk, J.; Bar-On, Y.; Stanietsky-Kaynan, N.; Coppenhagen-Glazer, S.; et al. Binding of the Fap2 Protein of Fusobacterium Nucleatum to Human Inhibitory Receptor TIGIT Protects Tumors from Immune Cell Attack. Immunity 2015, 42, 344, doi:10.1016/j.immuni.2015.01.010. [CrossRef]
- Sun, J.; Tang, Q.; Yu, S.; Xie, M.; Zheng, W.; Chen, G.; Yin, Y.; Huang, X.; Wo, K.; Lei, H.; et al. F. Nucleatum Facilitates Oral Squamous Cell Carcinoma Progression via GLUT1-Driven Lactate Production. eBioMedicine 2023, 88, 104444, doi:10.1016/j.ebiom.2023.104444. [CrossRef]
- Schöpf, F.; Marongiu, G.L.; Milaj, K.; Sprink, T.; Kikhney, J.; Moter, A.; Roderer, D. Structural Basis of Fusobacterium Nucleatum Adhesin Fap2 Interaction with Receptors on Cancer and Immune Cells. Nat. Commun. 2025, 16, 8104, doi:10.1038/s41467-025-63451-w. [CrossRef]
- Wang, M.; Yu, F.; Li, P. Intratumor Microbiota in Cancer Pathogenesis and Immunity: From Mechanisms of Action to Therapeutic Opportunities. Front. Immunol. 2023, 14, 1269054, doi:10.3389/fimmu.2023.1269054. [CrossRef]
- Nagler, A.; Kalaora, S.; Rosenberg, D.G.; Alon, M.; Barnea, E.; Levy, R.; Vervier, K.; Trabish, S.; Dadosh, T.; Zaidman, S.; et al. 672 Identification of Microbial-Derived HLA-Bound Peptides in Melanoma. J. Immunother. Cancer 2020, 8, doi:10.1136/jitc-2020-SITC2020.0672. [CrossRef]
- Wang, X.; Jia, Y.; Wen, L.; Mu, W.; Wu, X.; Liu, T.; Liu, X.; Fang, J.; Luan, Y.; Chen, P.; et al. Porphyromonas Gingivalis Promotes Colorectal Carcinoma by Activating the Hematopoietic NLRP3 Inflammasome. Cancer Res. 2021, 81, 2745–2759, doi:10.1158/0008-5472.CAN-20-3827. [CrossRef]
- Liu, S.; Zhou, X.; Peng, X.; Li, M.; Ren, B.; Cheng, G.; Cheng, L. Porphyromonas Gingivalis Promotes Immunoevasion of Oral Cancer by Protecting Cancer from Macrophage Attack. J. Immunol. 2020, 205, 282–289, doi:10.4049/jimmunol.1901138. [CrossRef]
- Yijia, Z.; Li, X.; Ma, L.; Wang, S.; Du, H.; Wu, Y.; Yu, J.; Xiang, Y.; Xiong, D.; Shan, H.; et al. Identification of Intratumoral Microbiome-Driven Immune Modulation and Therapeutic Implications in Diffuse Large B-Cell Lymphoma. Cancer Immunol. Immunother. 2025, 74, 131, doi:10.1007/s00262-025-03972-x. [CrossRef]
- Xu, Z.; Lv, Z.; Chen, F.; Zhang, Y.; Xu, Z.; Huo, J.; Liu, W.; Yu, S.; Tuersun, A.; Zhao, J.; et al. Dysbiosis of Human Tumor Microbiome and Aberrant Residence of Actinomyces in Tumor-Associated Fibroblasts in Young-Onset Colorectal Cancer. Front. Immunol. 2022, 13, 1008975, doi:10.3389/fimmu.2022.1008975. [CrossRef]
- Wang, Y.; Jiang, Z.; Zhang, K.; Tang, H.; Wang, G.; Gao, J.; He, G.; Liang, B.; Li, L.; Yang, C.; et al. Whole-Tumor Clearing and Imaging of Intratumor Microbiota in Three Dimensions with miCDaL Strategy. Adv. Sci. 2024, 11, 2400694, doi:10.1002/advs.202400694. [CrossRef]
- Alam, A.; Levanduski, E.; Denz, P.; Villavicencio, H.S.; Bhatta, M.; Alhorebi, L.; Zhang, Y.; Gomez, E.C.; Morreale, B.; Senchanthisai, S.; et al. Fungal Mycobiome Drives IL-33 Secretion and Type 2 Immunity in Pancreatic Cancer. Cancer Cell 2022, 40, 153-167.e11, doi:10.1016/j.ccell.2022.01.003. [CrossRef]
- Chen, Y.; Yang, L.; Huang, Y.; Zhu, T.; Zhang, L.; Cheng, M.; Wu, C.; Li, P.; Liang, M.; Zhang, X.; et al. Intratumoral Microbiota Predicts the Response to Neoadjuvant Chemoimmunotherapy in Triple-Negative Breast Cancer. J. Immunother. Cancer 2025, 13, e010365, doi:10.1136/jitc-2024-010365. [CrossRef]
- Davidsson, S.; Carlsson, J.; Greenberg, L.; Wijkander, J.; Söderquist, B.; Erlandsson, A. Cutibacterium Acnes Induces the Expression of Immunosuppressive Genes in Macrophages and Is Associated with an Increase of Regulatory T-Cells in Prostate Cancer. Microbiol. Spectr. 9, e01497-21, doi:10.1128/spectrum.01497-21. [CrossRef]
- Liu, Z.; Zhang, X.; Zhang, H.; Zhang, H.; Yi, Z.; Zhang, Q.; Liu, Q.; Liu, X. Multi-Omics Analysis Reveals Intratumor Microbes as Immunomodulators in Colorectal Cancer. Microbiol. Spectr. 2023, 11, e05038-22, doi:10.1128/spectrum.05038-22. [CrossRef]
- Wong-Rolle, A.; Dong, Q.; Zhu, Y.; Divakar, P.; Hor, J.L.; Kedei, N.; Wong, M.; Tillo, D.; Conner, E.A.; Rajan, A.; et al. Spatial Meta-Transcriptomics Reveal Associations of Intratumor Bacteria Burden with Lung Cancer Cells Showing a Distinct Oncogenic Signature. J. Immunother. Cancer 2022, 10, e004698, doi:10.1136/jitc-2022-004698. [CrossRef]
- Lim, S.-K.; Lin, W.-C.; Huang, S.-W.; Pan, Y.-C.; Hu, C.-W.; Mou, C.-Y.; Hu, C.-M.J.; Mou, K.Y. Bacteria Colonization in Tumor Microenvironment Creates a Favorable Niche for Immunogenic Chemotherapy. EMBO Mol. Med. 2024, 16, 416–428, doi:10.1038/s44321-023-00022-w. [CrossRef]
- Guo, F.; Das, J.K.; Kobayashi, K.S.; Qin, Q.-M.; A Ficht, T.; Alaniz, R.C.; Song, J.; Figueiredo, P.D. Live Attenuated Bacterium Limits Cancer Resistance to CAR-T Therapy by Remodeling the Tumor Microenvironment. J. Immunother. Cancer 2022, 10, e003760, doi:10.1136/jitc-2021-003760. [CrossRef]
- Vincent, R.L.; Gurbatri, C.R.; Li, F.; Vardoshvili, A.; Coker, C.; Im, J.; Ballister, E.R.; Rouanne, M.; Savage, T.; de los Santos-Alexis, K.; et al. Probiotic-Guided CAR-T Cells for Solid Tumor Targeting. Science 2023, 382, 211–218, doi:10.1126/science.add7034. [CrossRef]
- Lam, K.C.; Araya, R.E.; Huang, A.; Chen, Q.; Modica, M.D.; Rodrigues, R.R.; Lopès, A.; Johnson, S.B.; Schwarz, B.; Bohrnsen, E.; et al. Microbiota Triggers STING-Type I IFN-Dependent Monocyte Reprogramming of the Tumor Microenvironment. Cell 2021, 184, 5338-5356.e21, doi:10.1016/j.cell.2021.09.019. [CrossRef]
- Zhu, G.; Su, H.; Johnson, C.H.; Khan, S.A.; Kluger, H.; Lu, L. Intratumour Microbiome Associated with the Infiltration of Cytotoxic CD8+ T Cells and Patient Survival in Cutaneous Melanoma. Eur. J. Cancer Oxf. Engl. 1990 2021, 151, 25–34, doi:10.1016/j.ejca.2021.03.053. [CrossRef]
- Riquelme, E.; Zhang, Y.; Zhang, L.; Montiel, M.; Zoltan, M.; Dong, W.; Quesada, P.; Sahin, I.; Chandra, V.; Lucas, A.S.; et al. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell 2019, 178, 795-806.e12, doi:10.1016/j.cell.2019.07.008. [CrossRef]
- Sfanos, K.S. Intratumoral Bacteria as Mediators of Cancer Immunotherapy Response. Cancer Res. 2023, 83, 2985–2986, doi:10.1158/0008-5472.CAN-23-1857. [CrossRef]
- Zheng, J.; Chen, H. Effects of Intratumoral Microbiota on Tumorigenesis, Anti-Tumor Immunity, and Microbe-Based Cancer Therapy. Front. Oncol. 2024, 14, 1429722, doi:10.3389/fonc.2024.1429722. [CrossRef]
- Zheng, S.; Su, Y.; Cai, F.; Xu, D.; Xu, Y. From Mechanisms to Precision Medicine: The Role of Organoids in Studying the Gut Microbiota-Tumor Microenvironment Axis. Front. Microbiol. 2025, 16, 1669482, doi:10.3389/fmicb.2025.1669482. [CrossRef]
- Sun, Y.; Zhang, X.; Jin, C.; Yue, K.; Sheng, D.; Zhang, T.; Dou, X.; Liu, J.; Jing, H.; Zhang, L.; et al. Prospective, Longitudinal Analysis of the Gut Microbiome in Patients with Locally Advanced Rectal Cancer Predicts Response to Neoadjuvant Concurrent Chemoradiotherapy. J. Transl. Med. 2023, 21, 221, doi:10.1186/s12967-023-04054-1. [CrossRef]
- Harmak, Z.; Kone, A.-S.; Ghouzlani, A.; Ghazi, B.; Badou, A. Beyond Tumor Borders: Intratumoral Microbiome Effects on Tumor Behavior and Therapeutic Responses. Immune Netw. 2024, 24, e40, doi:10.4110/in.2024.24.e40. [CrossRef]
- Wang, N.; Wu, S.; Huang, L.; Hu, Y.; He, X.; He, J.; Hu, B.; Xu, Y.; Rong, Y.; Yuan, C.; et al. Intratumoral Microbiome: Implications for Immune Modulation and Innovative Therapeutic Strategies in Cancer. J. Biomed. Sci. 2025, 32, 23, doi:10.1186/s12929-025-01117-x. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





