Submitted:
03 November 2025
Posted:
04 November 2025
You are already at the latest version
Abstract
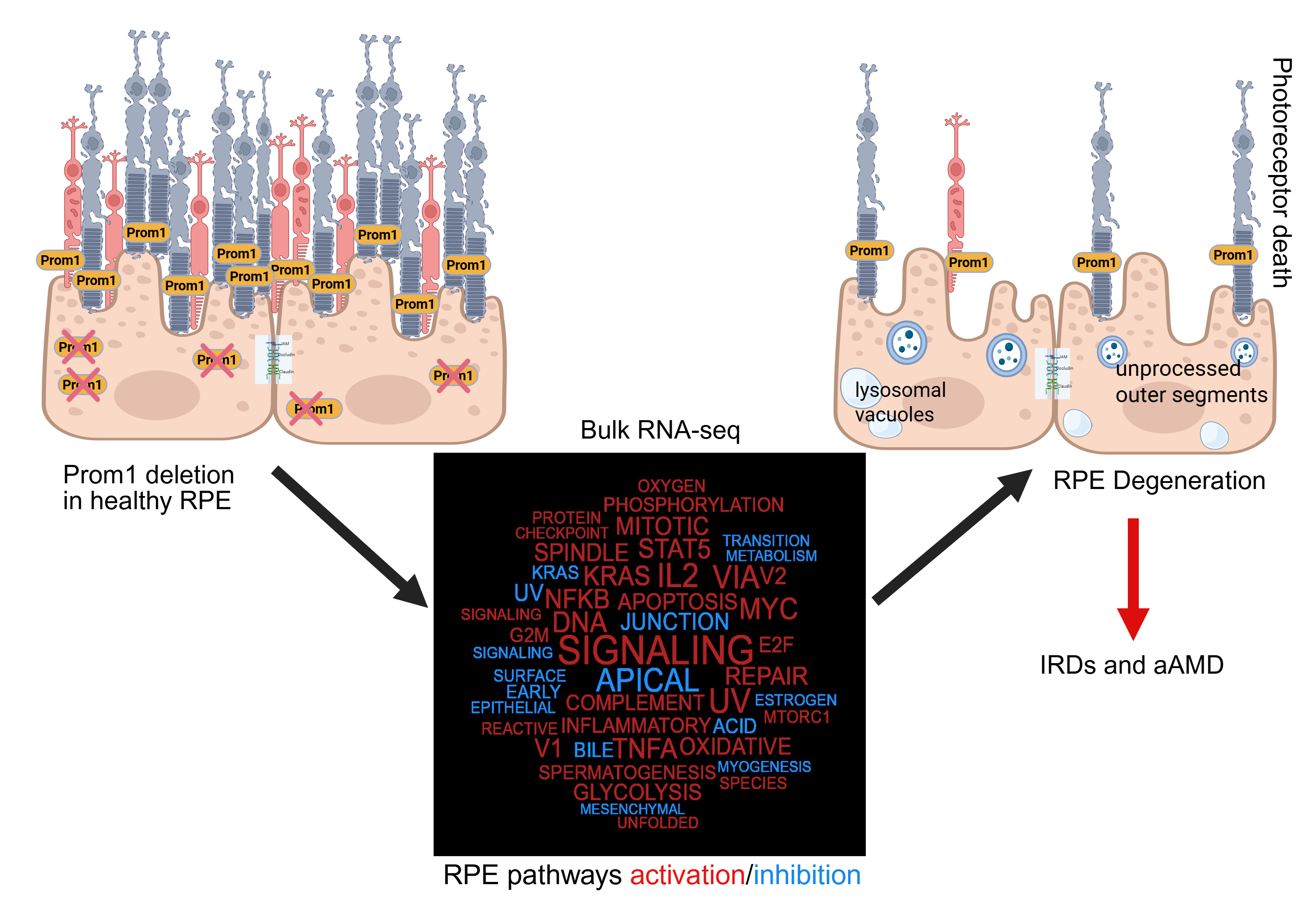
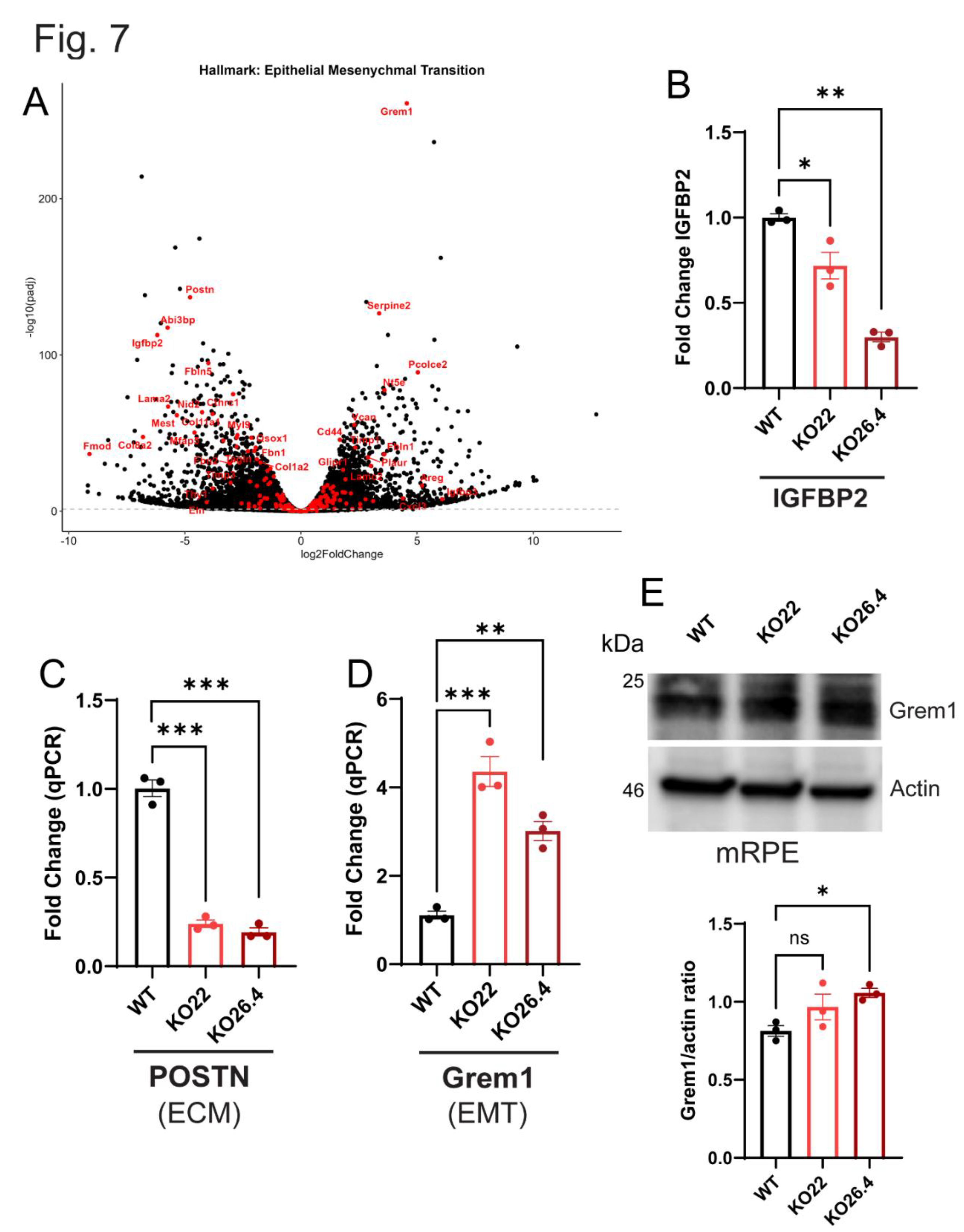
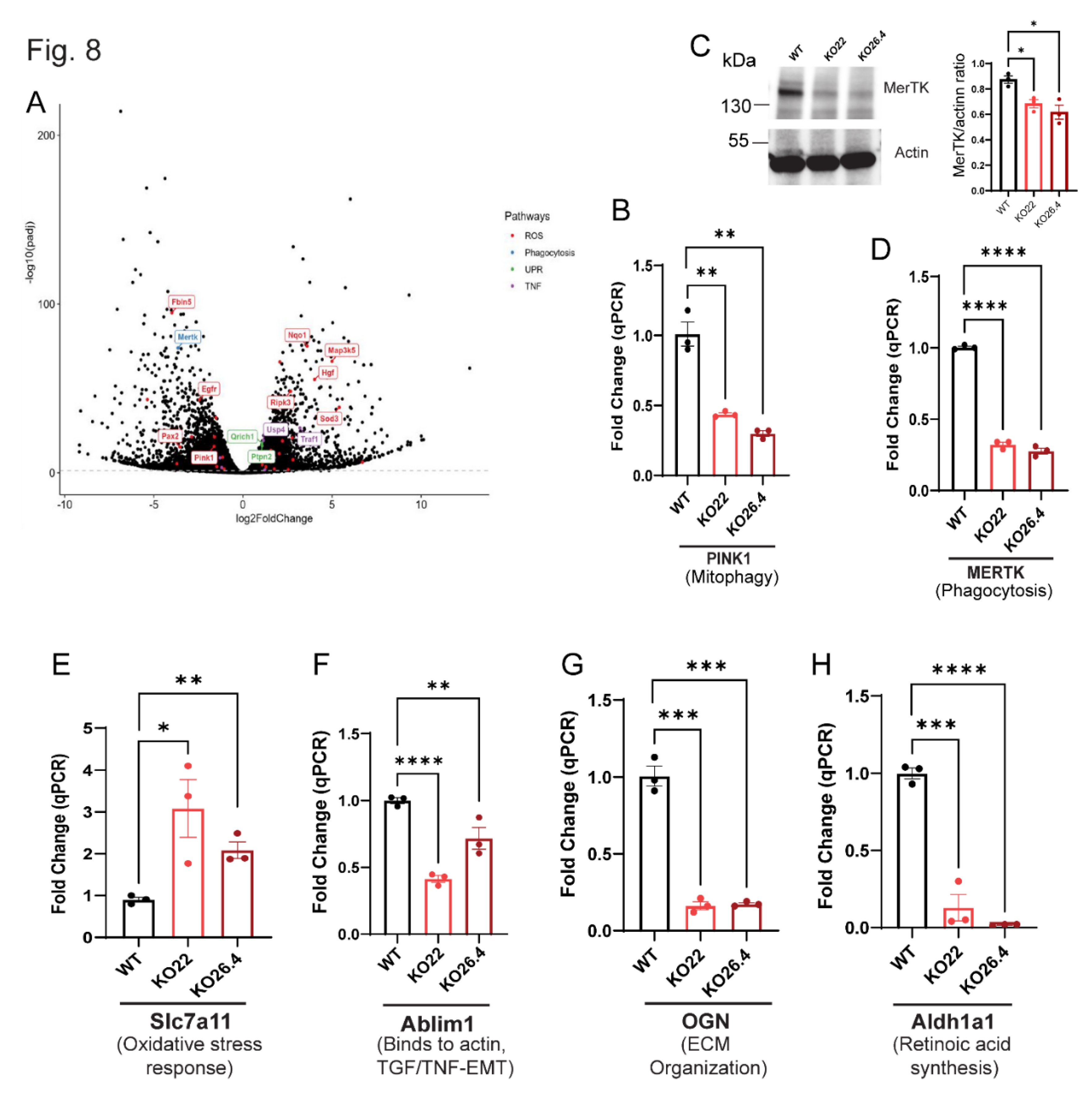
Inherited retinal degenerations (IRDs), driven by pathogenic mutations, often involve primary dysfunction of the retinal pigment epithelium (RPE) — a pathogenic feature shared with atrophic age-related macular degeneration (aAMD), despite aAMD’s multifactorial etiology. Prominin-1 (Prom1), traditionally linked to photoreceptor pathology, has an unclear role in RPE homeostasis. We assessed Prom1 expression in C57BL/6J mouse retina sections and RPE flat mounts using immunohistochemistry and generated Prom1-knockout (KO) mouse RPE cells via CRISPR/Cas9. Bulk RNA sequencing with DESeq2 and gene set enrichment analysis (GSEA) revealed Prom1-regulated pathways. Prom1-KO cells exhibited upregulation of Grem1, Slc7a11, Serpine2, Il1r1, and IL33, and downregulation of Ablim1, Cldn2, IGFBP-2, BMP3, and OGN. Hallmark pathway interrogation identified reduced expression of PINK1 (mitophagy) and MerTK (phagocytosis), implicating defects in mitochondrial quality control and outer segment clearance. Enrichment analysis indicated activation of E2F/MYC targets, mTORC1 signaling, oxidative phosphorylation, and TNFα/NF-κB signaling, alongside suppression of apical junction, bile acid metabolism, and EMT pathways. These findings suggest Prom1 safeguards RPE integrity by modulating stress responses, mitochondrial turnover, phagocytosis, metabolism, and junctional stability. Our study uncovers Prom1-dependent signaling networks, providing mechanistic insights into RPE degeneration relevant to both IRD and aAMD, and highlights potential therapeutic targets for preserving retinal health.
Keywords:
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Mice and Colony Management
2.3. Cell Culture
2.4. Generation of Prom1-Deficient mRPE Cells via CRISPR/Cas9
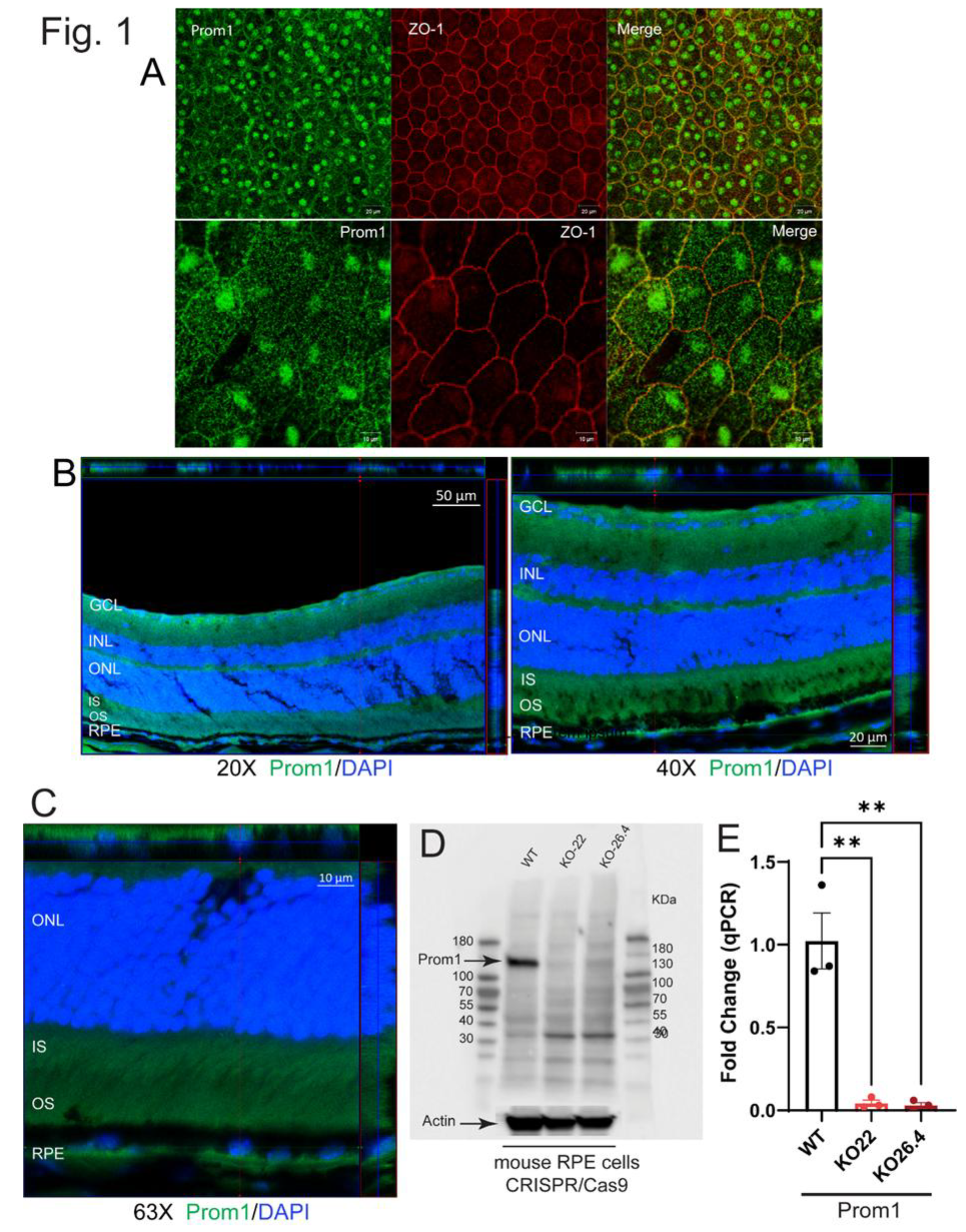
2.5. Mouse RPE Flat Mount Preparation and Immunohistochemistry
2.6. Mouse Retina Sections and Confocal Imaging
2.7. Western Blotting
2.8. Real-Time Quantitative PCR
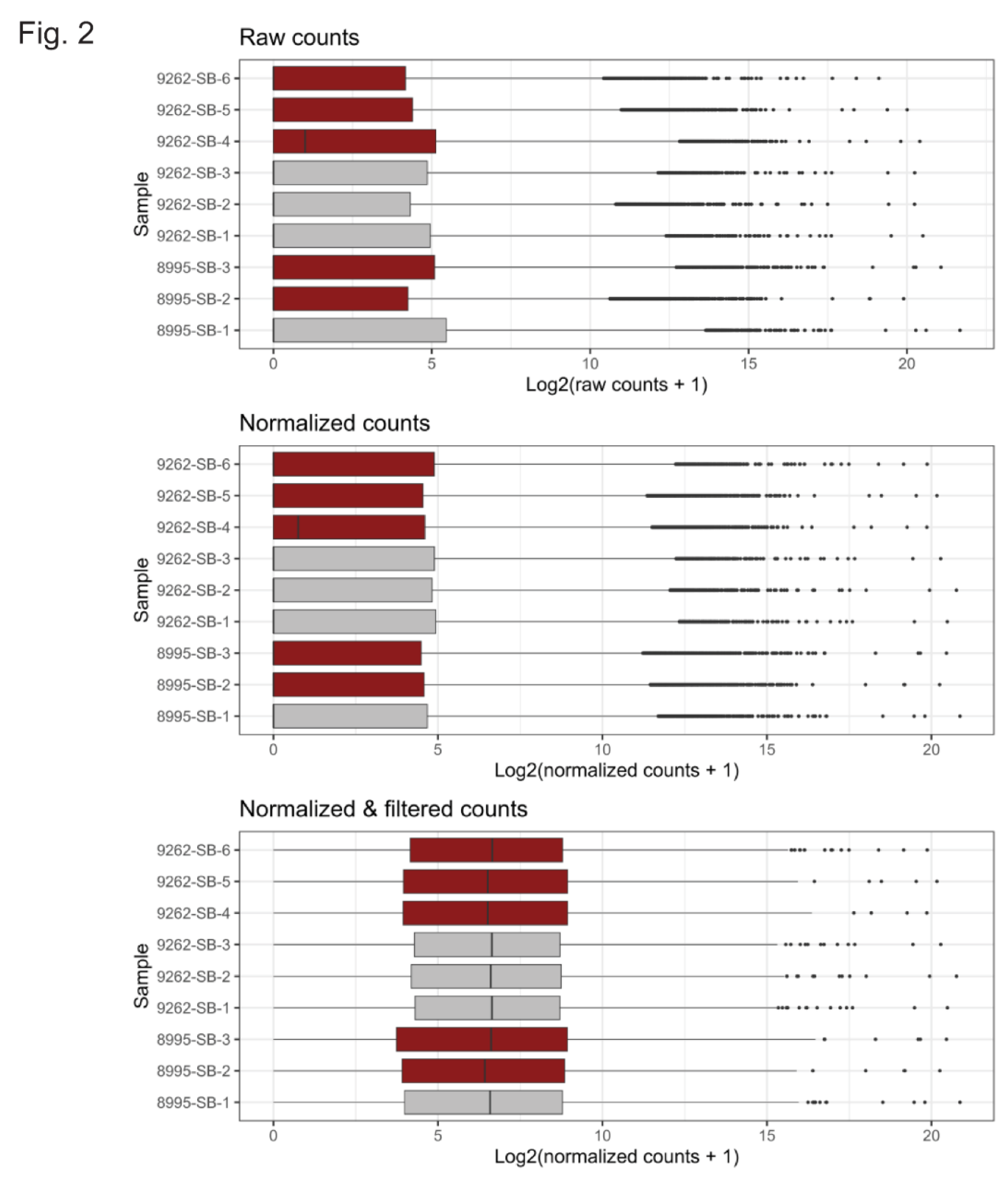
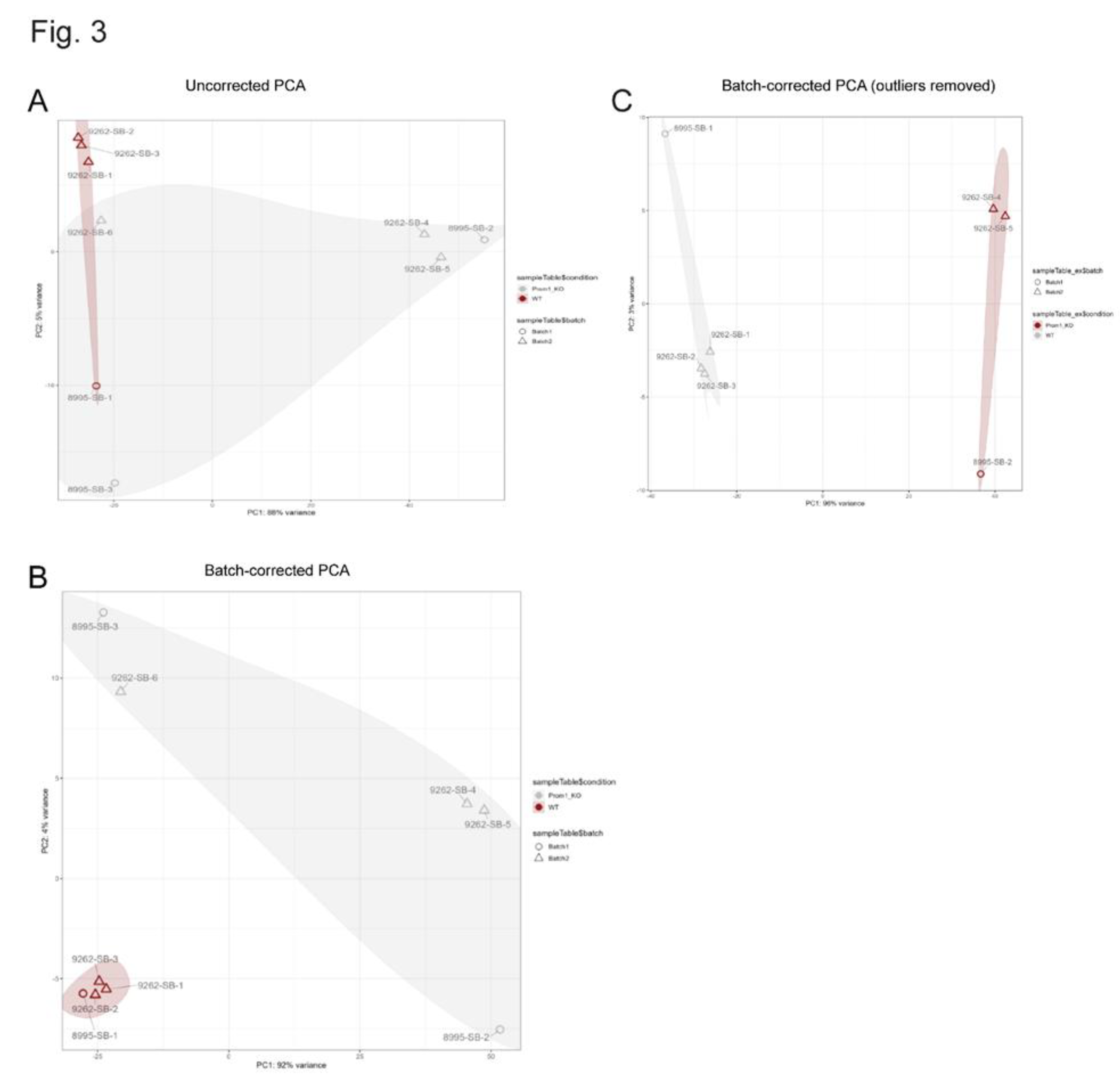
2.9. Bulk RNA Sequencing and Data Analysis
2.10. Statistical Analysis
3. Results
3.1. Prom1 is Expressed in Mouse RPE In Situ
3.2. Bulk RNA-Sequencing of WT and Prom1-KO mRPE Cells
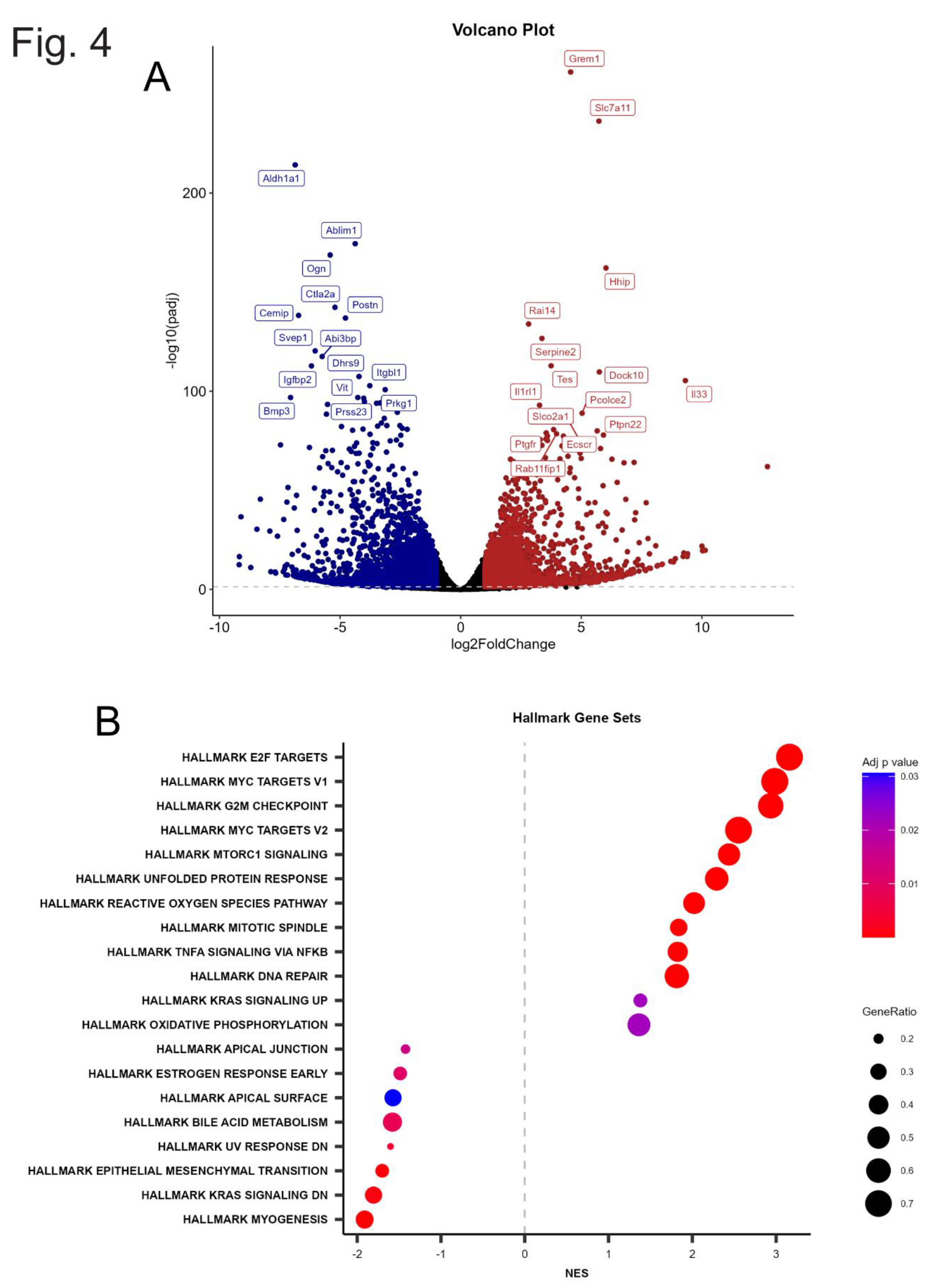
3.3. Differential Expression and Gene Set Enrichment Analyses
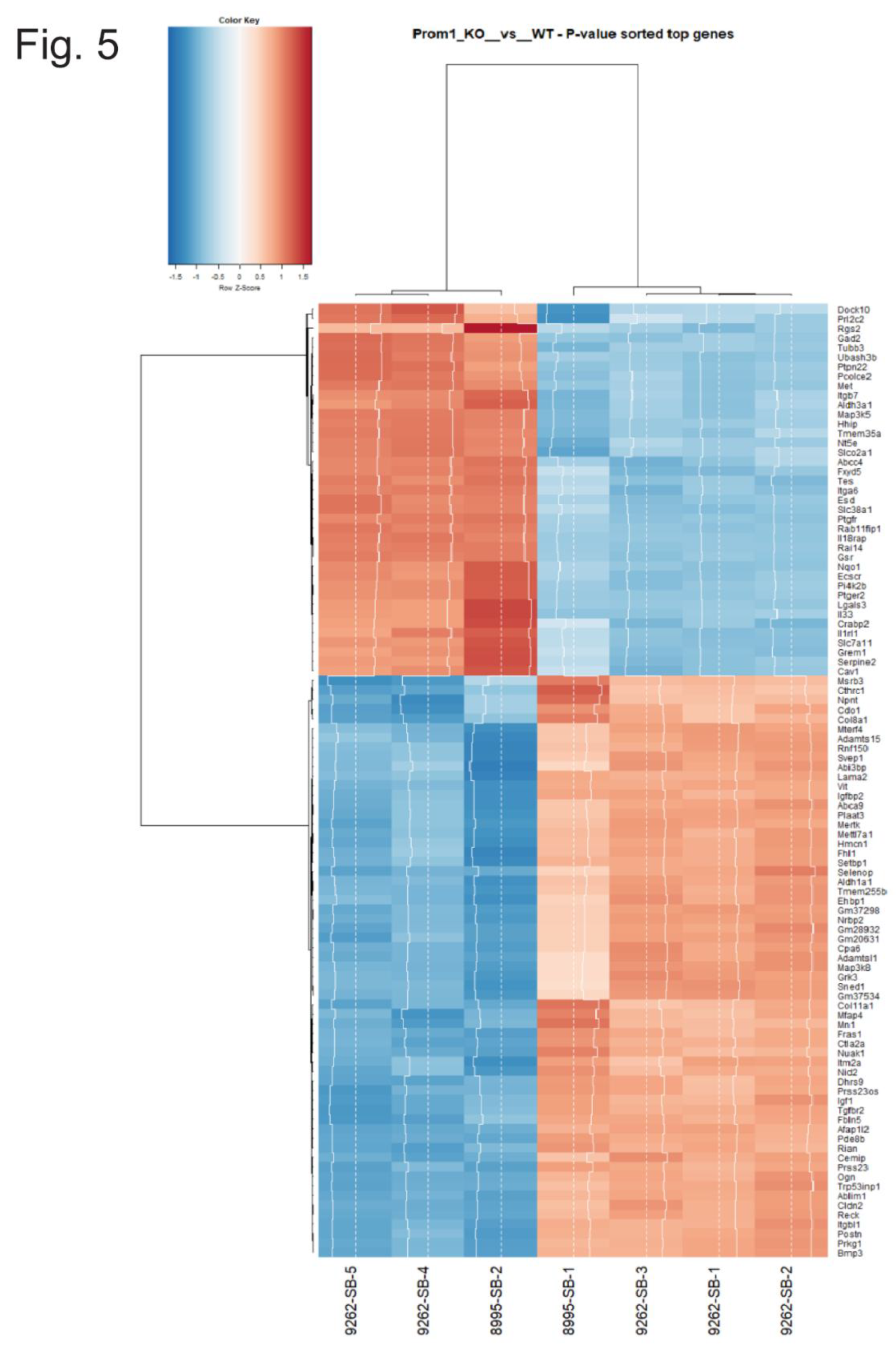
3.4. Heatmap Analysis and Biological Context
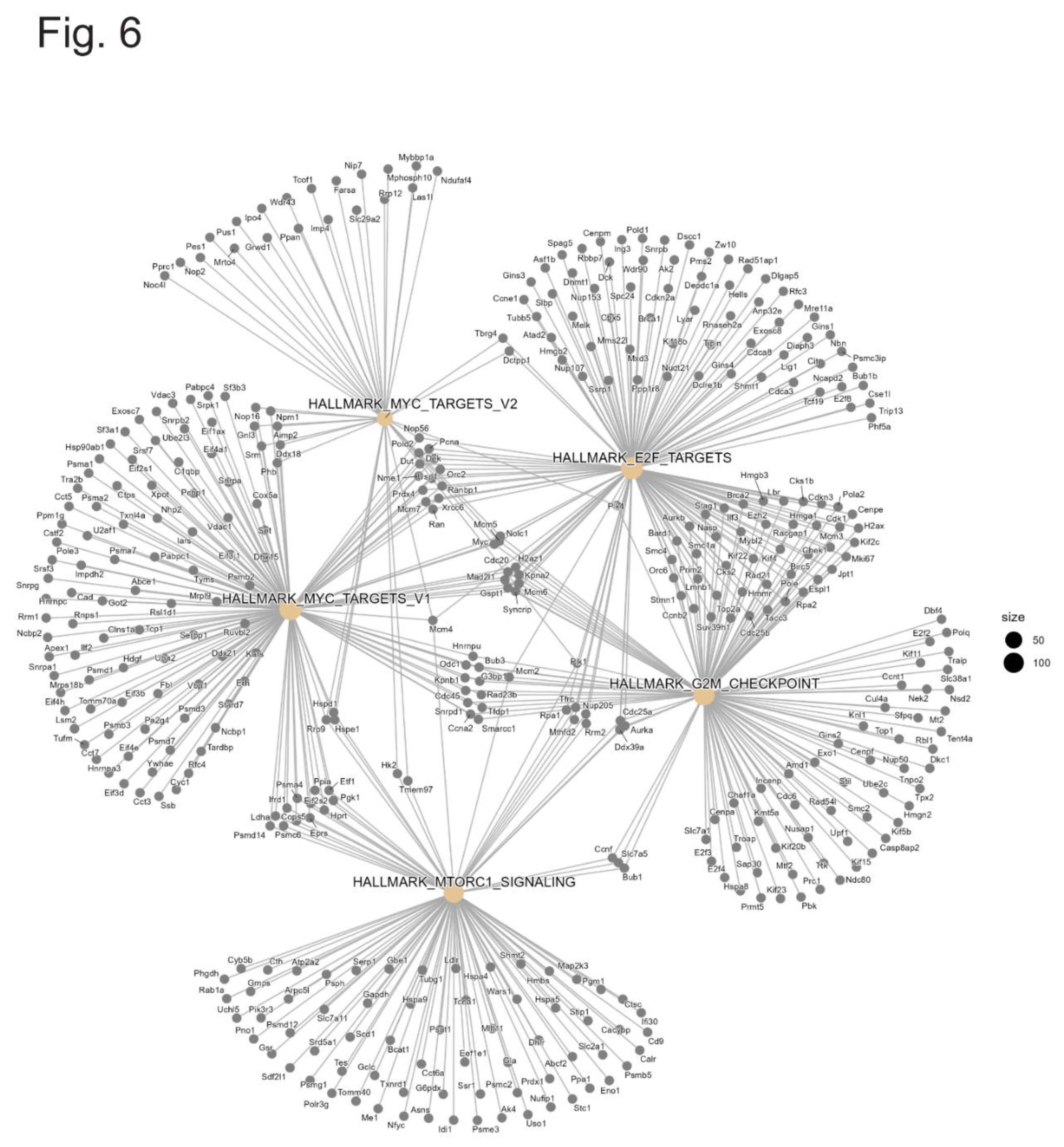
3.5. Gene Network Map in Prom1-KO versus WT mRPE
3.6. Validation of Transcriptomic Data
4. Discussion
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| IRDs | Inherited Retinal Dystrophies |
| aAMD | Atrophic Age-related Macular Degeneration |
| RPE | Retinal Pigment Epithelium |
| mTORC1 | Mammalian Target of Rapamycin Complex 1 |
| Prom1 | Prominin-1 (CD133) |
| STGD4 | Stargardt disease 4 |
| EMT | Epithelial-Mesenchymal Transitio |
| GSEA | Gene Set Enrichment Analysis |
| DEG | Differential Gene Expression |
| qPCR | Quantitative Polymerase Chain Reaction |
| PCA | Principal Component Analysis |
| ECM | Extracellular Matrix |
| UPR | Unfolded Protein Response |
References
- Corradetti, G.; Verma, A.; Tojjar, J.; Almidani, L.; Oncel, D.; Emamverdi, M.; Bradley, A.; Lindenberg, S.; Nittala, M.G.; Sadda, S.R. Retinal Imaging Findings in Inherited Retinal Diseases. J Clin Med 2024, 13. [Google Scholar] [CrossRef]
- Samelska, K.; Szaflik, J.P.; Guszkowska, M.; Kurowska, A.K.; Zaleska-Zmijewska, A. Characteristics of Rare Inherited Retinal Dystrophies in Adaptive Optics-A Study on 53 Eyes. Diagnostics (Basel) 2023, 13. [Google Scholar] [CrossRef]
- Sanie-Jahromi, F.; Nowroozzadeh, M.H. RPE based gene and cell therapy for inherited retinal diseases: A review. Exp Eye Res 2022, 217, 108961. [Google Scholar] [CrossRef]
- Tebbe, L.; Sakthivel, H.; Makia, M.S.; Kakakhel, M.; Conley, S.M.; Al-Ubaidi, M.R.; Naash, M.I. Prph2 disease mutations lead to structural and functional defects in the RPE. FASEB J 2022, 36, e22284. [Google Scholar] [CrossRef]
- Hwang, S.; Kang, S.W.; Jang, J.H.; Kim, S.J. Genetic and clinical characteristics of PROM1-related retinal degeneration in Korean. Sci Rep 2023, 13, 21877. [Google Scholar] [CrossRef] [PubMed]
- Puertas-Neyra, K.; Coco-Martin, R.M.; Hernandez-Rodriguez, L.A.; Gobelli, D.; Garcia-Ferrer, Y.; Palma-Vecino, R.; Telleria, J.J.; Simarro, M.; de la Fuente, M.A.; Fernandez-Bueno, I. Clinical exome analysis and targeted gene repair of the c.1354dupT variant in iPSC lines from patients with PROM1-related retinopathies exhibiting diverse phenotypes. Stem Cell Res Ther 2024, 15, 192. [Google Scholar] [CrossRef] [PubMed]
- Miraglia, S.; Godfrey, W.; Yin, A.H.; Atkins, K.; Warnke, R.; Holden, J.T.; Bray, R.A.; Waller, E.K.; Buck, D.W. A novel five-transmembrane hematopoietic stem cell antigen: isolation, characterization, and molecular cloning. Blood 1997, 90, 5013–5021. [Google Scholar] [CrossRef]
- Weigmann, A.; Corbeil, D.; Hellwig, A.; Huttner, W.B. Prominin, a novel microvilli-specific polytopic membrane protein of the apical surface of epithelial cells, is targeted to plasmalemmal protrusions of non-epithelial cells. Proc Natl Acad Sci U S A 1997, 94, 12425–12430. [Google Scholar] [CrossRef] [PubMed]
- Pleskac, P.; Fargeas, C.A.; Veselska, R.; Corbeil, D.; Skoda, J. Emerging roles of prominin-1 (CD133) in the dynamics of plasma membrane architecture and cell signaling pathways in health and disease. Cell Mol Biol Lett 2024, 29, 41. [Google Scholar] [CrossRef]
- Ragi, S.D.; Lima de Carvalho, J.R., Jr.; Tanaka, A.J.; Park, K.S.; Mahajan, V.B.; Maumenee, I.H.; Tsang, S.H. Compound heterozygous novel frameshift variants in the PROM1 gene result in Leber congenital amaurosis. Cold Spring Harb Mol Case Stud 2019, 5. [Google Scholar] [CrossRef]
- Maw, M.A.; Corbeil, D.; Koch, J.; Hellwig, A.; Wilson-Wheeler, J.C.; Bridges, R.J.; Kumaramanickavel, G.; John, S.; Nancarrow, D.; Roper, K.; et al. A frameshift mutation in prominin (mouse)-like 1 causes human retinal degeneration. Hum Mol Genet 2000, 9, 27–34. [Google Scholar] [CrossRef]
- Yang, Z.; Chen, Y.; Lillo, C.; Chien, J.; Yu, Z.; Michaelides, M.; Klein, M.; Howes, K.A.; Li, Y.; Kaminoh, Y.; et al. Mutant prominin 1 found in patients with macular degeneration disrupts photoreceptor disk morphogenesis in mice. J Clin Invest 2008, 118, 2908–2916. [Google Scholar] [CrossRef]
- Zhang, Q.; Zulfiqar, F.; Xiao, X.; Riazuddin, S.A.; Ahmad, Z.; Caruso, R.; MacDonald, I.; Sieving, P.; Riazuddin, S.; Hejtmancik, J.F. Severe retinitis pigmentosa mapped to 4p15 and associated with a novel mutation in the PROM1 gene. Hum Genet 2007, 122, 293–299. [Google Scholar] [CrossRef]
- Pras, E.; Abu, A.; Rotenstreich, Y.; Avni, I.; Reish, O.; Morad, Y.; Reznik-Wolf, H.; Pras, E. Cone-rod dystrophy and a frameshift mutation in the PROM1 gene. Mol Vis 2009, 15, 1709–1716. [Google Scholar]
- Eidinger, O.; Leibu, R.; Newman, H.; Rizel, L.; Perlman, I.; Ben-Yosef, T. An intronic deletion in the PROM1 gene leads to autosomal recessive cone-rod dystrophy. Mol Vis 2015, 21, 1295–1306. [Google Scholar]
- Khan, A.O.; Bolz, H.J. Pediatric Cone-Rod Dystrophy with High Myopia and Nystagmus Suggests Recessive PROM1 Mutations. Ophthalmic Genet 2015, 36, 349–352. [Google Scholar] [CrossRef] [PubMed]
- Imani, S.; Cheng, J.; Shasaltaneh, M.D.; Wei, C.; Yang, L.; Fu, S.; Zou, H.; Khan, M.A.; Zhang, X.; Chen, H.; et al. Genetic identification and molecular modeling characterization reveal a novel PROM1 mutation in Stargardt4-like macular dystrophy. Oncotarget 2018, 9, 122–141. [Google Scholar] [CrossRef]
- Carr, B.J.; Skitsko, D.; Kriese, L.M.; Song, J.; Li, Z.; Ju, M.J.; Moritz, O.L. Prominin-1 null Xenopus laevis develop subretinal drusenoid-like deposits, cone-rod dystrophy, and RPE atrophy. J Cell Sci 2024. [Google Scholar] [CrossRef] [PubMed]
- Strauss, R.W.; Munoz, B.; Ahmed, M.I.; Bittencourt, M.; Schonbach, E.M.; Michaelides, M.; Birch, D.; Zrenner, E.; Ervin, A.M.; Charbel Issa, P.; et al. The Progression of the Stargardt Disease Type 4 (ProgStar-4) Study: Design and Baseline Characteristics (ProgStar-4 Report No. 1). Ophthalmic Res 2018, 60, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Kniazeva, M.; Chiang, M.F.; Morgan, B.; Anduze, A.L.; Zack, D.J.; Han, M.; Zhang, K. A new locus for autosomal dominant stargardt-like disease maps to chromosome 4. Am J Hum Genet 1999, 64, 1394–1399. [Google Scholar] [CrossRef]
- Abalem, M.F.; Omari, A.A.; Schlegel, D.; Khan, N.W.; Jayasundera, T. Macular hyperpigmentary changes in ABCA4-Stargardt disease. Int J Retina Vitreous 2019, 5, 9. [Google Scholar] [CrossRef]
- Lee, W.; Paavo, M.; Zernant, J.; Stong, N.; Laurente, Z.; Bearelly, S.; Nagasaki, T.; Tsang, S.H.; Goldstein, D.B.; Allikmets, R. Modification of the PROM1 disease phenotype by a mutation in ABCA4. Ophthalmic Genet 2019, 40, 369–375. [Google Scholar] [CrossRef]
- Permanyer, J.; Navarro, R.; Friedman, J.; Pomares, E.; Castro-Navarro, J.; Marfany, G.; Swaroop, A.; Gonzalez-Duarte, R. Autosomal recessive retinitis pigmentosa with early macular affectation caused by premature truncation in PROM1. Invest Ophthalmol Vis Sci 2010, 51, 2656–2663. [Google Scholar] [CrossRef]
- Paavo, M.; Lee, W.; Parmann, R.; Lima de Carvalho, J.R., Jr.; Zernant, J.; Tsang, S.H.; Allikmets, R.; Sparrow, J.R. Insights Into PROM1-Macular Disease Using Multimodal Imaging. Invest Ophthalmol Vis Sci 2023, 64, 27. [Google Scholar] [CrossRef]
- Ricca, A.M.; Han, I.C.; Hoffmann, J.; Stone, E.M.; Sohn, E.H. Macular Atrophy and Phenotypic Variability in Autosomal Dominant Stargardt-Like Macular Dystrophy Due to Prom1 Mutation. Retina 2023, 43, 1165–1173. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Yin, J.; Winborn, C.S.; Zhang, Q.; Yue, J.; Chaum, E. Prominin-1 Is a Novel Regulator of Autophagy in the Human Retinal Pigment Epithelium. Invest Ophthalmol Vis Sci 2017, 58, 2366–2387. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Yang, T.S.; Nabit, B.P.; Krystofiak, E.S.; Rex, T.S.; Chaum, E. Prominin-1 Knockdown Causes RPE Degeneration in a Mouse Model. Cells 2024, 13. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Yin, J.; Huo, W.; Chaum, E. Loss of Prom1 impairs autophagy and promotes epithelial-mesenchymal transition in mouse retinal pigment epithelial cells. J Cell Physiol 2023. [Google Scholar] [CrossRef]
- Fernandez-Godino, R.; Garland, D.L.; Pierce, E.A. Isolation, culture and characterization of primary mouse RPE cells. Nat Protoc 2016, 11, 1206–1218. [Google Scholar] [CrossRef]
- Boatright, J.H.; Dalal, N.; Chrenek, M.A.; Gardner, C.; Ziesel, A.; Jiang, Y.; Grossniklaus, H.E.; Nickerson, J.M. Methodologies for analysis of patterning in the mouse RPE sheet. Mol Vis 2015, 21, 40–60. [Google Scholar]
- Bhattacharya, S.; Chaum, E.; Johnson, D.A.; Johnson, L.R. Age-related susceptibility to apoptosis in human retinal pigment epithelial cells is triggered by disruption of p53-Mdm2 association. Invest Ophthalmol Vis Sci 2012, 53, 8350–8366. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Yuan, D.; Shen, H.; Mao, X.; Yuan, S.; Liu, Q. Gremlin-1: An endogenous BMP antagonist induces epithelial-mesenchymal transition and interferes with redifferentiation in fetal RPE cells with repeated wounds. Mol Vis 2019, 25, 625–635. [Google Scholar]
- Winokur, P.N.; Subramanian, P.; Bullock, J.L.; Arocas, V.; Becerra, S.P. Comparison of two neurotrophic serpins reveals a small fragment with cell survival activity. Mol Vis 2017, 23, 372–384. [Google Scholar]
- Cheng, X.; He, D.; Liao, C.; Lin, S.; Tang, L.; Wang, Y.L.; Hu, J.; Li, W.; Liu, Z.; Wu, Y.; et al. IL-1/IL-1R signaling induced by all-trans-retinal contributes to complement alternative pathway activation in retinal pigment epithelium. J Cell Physiol 2021, 236, 3660–3674. [Google Scholar] [CrossRef] [PubMed]
- Xi, H.; Katschke, K.J., Jr.; Li, Y.; Truong, T.; Lee, W.P.; Diehl, L.; Rangell, L.; Tao, J.; Arceo, R.; Eastham-Anderson, J.; et al. IL-33 amplifies an innate immune response in the degenerating retina. J Exp Med 2016, 213, 189–207. [Google Scholar] [CrossRef]
- Kutty, R.K.; Samuel, W.; Chen, S.; Vijayasarathy, C.; Dun, Y.; Mysona, B.; Wiggert, B.; Smith, S.B. Immunofluorescence analysis of the expression of Norpeg (Rai14) in retinal Muller and ganglion cells. Neurosci Lett 2006, 404, 294–298. [Google Scholar] [CrossRef]
- Shen, X.; Zhang, J.; Zhang, X.; Wang, Y.; Hu, Y.; Guo, J. Retinoic Acid-Induced Protein 14 (RAI14) Promotes mTOR-Mediated Inflammation Under Inflammatory Stress and Chemical Hypoxia in a U87 Glioblastoma Cell Line. Cell Mol Neurobiol 2019, 39, 241–254. [Google Scholar] [CrossRef]
- Barrientos, T.; Frank, D.; Kuwahara, K.; Bezprozvannaya, S.; Pipes, G.C.; Bassel-Duby, R.; Richardson, J.A.; Katus, H.A.; Olson, E.N.; Frey, N. Two novel members of the ABLIM protein family, ABLIM-2 and -3, associate with STARS and directly bind F-actin. J Biol Chem 2007, 282, 8393–8403. [Google Scholar] [CrossRef]
- Li, G.; Huang, S.; Yang, S.; Wang, J.; Cao, J.; Czajkowsky, D.M.; Shao, Z.; Zhu, X. abLIM1 constructs non-erythroid cortical actin networks to prevent mechanical tension-induced blebbing. Cell Discov 2018, 4, 42. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Chaum, E. A reassessment of insulin-like growth factor binding protein gene expression in the human retinal pigment epithelium. J Cell Biochem 2003, 89, 933–943. [Google Scholar] [CrossRef]
- Mukherjee, S.; King, J.L.; Guidry, C. Phenotype-associated changes in retinal pigment epithelial cell expression of insulin-like growth factor binding proteins. Invest Ophthalmol Vis Sci 2009, 50, 5449–5455. [Google Scholar] [CrossRef]
- Low, S.W.Y.; Connor, T.B.; Kassem, I.S.; Costakos, D.M.; Chaurasia, S.S. Small Leucine-Rich Proteoglycans (SLRPs) in the Retina. Int J Mol Sci 2021, 22. [Google Scholar] [CrossRef]
- Mercau, M.E.; Akalu, Y.T.; Mazzoni, F.; Gyimesi, G.; Alberto, E.J.; Kong, Y.; Hafler, B.P.; Finnemann, S.C.; Rothlin, C.V.; Ghosh, S. Inflammation of the retinal pigment epithelium drives early-onset photoreceptor degeneration in Mertk-associated retinitis pigmentosa. Sci Adv 2023, 9, eade9459. [Google Scholar] [CrossRef] [PubMed]
- Louer, E.M.M.; Gunzel, D.; Rosenthal, R.; Carmone, C.; Yi, G.; Stunnenberg, H.G.; den Hollander, A.I.; Deen, P.M.T. Differential day-night expression of tight junction components in murine retinal pigment epithelium. Exp Eye Res 2020, 193, 107985. [Google Scholar] [CrossRef] [PubMed]
- Schlecht, A.; Leimbeck, S.V.; Jagle, H.; Feuchtinger, A.; Tamm, E.R.; Braunger, B.M. Deletion of Endothelial Transforming Growth Factor-beta Signaling Leads to Choroidal Neovascularization. Am J Pathol 2017, 187, 2570–2589. [Google Scholar] [CrossRef]
- Shimizu, H.; Yamada, K.; Suzumura, A.; Kataoka, K.; Takayama, K.; Sugimoto, M.; Terasaki, H.; Kaneko, H. Caveolin-1 Promotes Cellular Senescence in Exchange for Blocking Subretinal Fibrosis in Age-Related Macular Degeneration. Invest Ophthalmol Vis Sci 2020, 61, 21. [Google Scholar] [CrossRef]
- Matsuzawa, A.; Saegusa, K.; Noguchi, T.; Sadamitsu, C.; Nishitoh, H.; Nagai, S.; Koyasu, S.; Matsumoto, K.; Takeda, K.; Ichijo, H. ROS-dependent activation of the TRAF6-ASK1-p38 pathway is selectively required for TLR4-mediated innate immunity. Nat Immunol 2005, 6, 587–592. [Google Scholar] [CrossRef]
- Sun, Y.; Hu, Y.; Luo, S. Complement C5a promotes human retinal pigment epithelial cell viability and migration through SLC38A1-mediated glutamine metabolism. Med Microbiol Immunol 2025, 214, 22. [Google Scholar] [CrossRef]
- Chen, X.; Tzekov, R.; Su, M.; Zhu, Y.; Han, A.; Li, W. Hydrogen peroxide-induced oxidative damage and protective role of peroxiredoxin 6 protein via EGFR/ERK signaling pathway in RPE cells. Front Aging Neurosci 2023, 15, 1169211. [Google Scholar] [CrossRef]
- Chen, X.D.; Su, M.Y.; Chen, T.T.; Hong, H.Y.; Han, A.D.; Li, W.S. Oxidative stress affects retinal pigment epithelial cell survival through epidermal growth factor receptor/AKT signaling pathway. Int J Ophthalmol 2017, 10, 507–514. [Google Scholar] [CrossRef]
- Wang, M.; Topalovski, M.; Toombs, J.E.; Wright, C.M.; Moore, Z.R.; Boothman, D.A.; Yanagisawa, H.; Wang, H.; Witkiewicz, A.; Castrillon, D.H.; et al. Fibulin-5 Blocks Microenvironmental ROS in Pancreatic Cancer. Cancer Res 2015, 75, 5058–5069. [Google Scholar] [CrossRef]
- Bosze, B.; Suarez-Navarro, J.; Soofi, A.; Lauderdale, J.D.; Dressler, G.R.; Brown, N.L. Multiple roles for Pax2 in the embryonic mouse eye. Dev Biol 2021, 472, 18–29. [Google Scholar] [CrossRef]
- Zhang, S.M.; Fan, B.; Li, Y.L.; Zuo, Z.Y.; Li, G.Y. Oxidative Stress-Involved Mitophagy of Retinal Pigment Epithelium and Retinal Degenerative Diseases. Cell Mol Neurobiol 2023, 43, 3265–3276. [Google Scholar] [CrossRef]
- Ross, D.; Siegel, D. Functions of NQO1 in Cellular Protection and CoQ(10) Metabolism and its Potential Role as a Redox Sensitive Molecular Switch. Front Physiol 2017, 8, 595. [Google Scholar] [CrossRef] [PubMed]
- Kojima, K.; Ichijo, H.; Naguro, I. Molecular functions of ASK family in diseases caused by stress-induced inflammation and apoptosis. J Biochem 2021, 169, 395–407. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Barron, E.; He, S.; Ryan, S.J.; Hinton, D.R. Regulation of RPE intercellular junction integrity and function by hepatocyte growth factor. Invest Ophthalmol Vis Sci 2002, 43, 2782–2790. [Google Scholar] [PubMed]
- Zhou, S.R.; Zhu, Y.S.; Yuan, W.T.; Pan, X.Y.; Wang, T.; Chen, X.D. Hepatocyte growth factor promotes retinal pigment epithelium cell activity through MET/AKT signaling pathway. Int J Ophthalmol 2024, 17, 806–814. [Google Scholar] [CrossRef]
- Huang, X.; Li, T.; Zhang, G.; Chen, J.; Li, T.; Yang, S.; Bo, Q.; Zhao, X.; Wan, X.; Zhu, X.; et al. AIM2 activation mediated by RIPK1/3-dependent mitochondrial DNA release drives Abeta(1-40)-Induced retinal pigment epithelium injury. Cell Commun Signal 2025, 23, 301. [Google Scholar] [CrossRef]
- Wert, K.J.; Velez, G.; Cross, M.R.; Wagner, B.A.; Teoh-Fitzgerald, M.L.; Buettner, G.R.; McAnany, J.J.; Olivier, A.; Tsang, S.H.; Harper, M.M.; et al. Extracellular superoxide dismutase (SOD3) regulates oxidative stress at the vitreoretinal interface. Free Radic Biol Med 2018, 124, 408–419. [Google Scholar] [CrossRef]
- You, K.; Wang, L.; Chou, C.H.; Liu, K.; Nakata, T.; Jaiswal, A.; Yao, J.; Lefkovith, A.; Omar, A.; Perrigoue, J.G.; et al. QRICH1 dictates the outcome of ER stress through transcriptional control of proteostasis. Science 2021, 371. [Google Scholar] [CrossRef]
- Elvira, B.; Vandenbempt, V.; Bauza-Martinez, J.; Crutzen, R.; Negueruela, J.; Ibrahim, H.; Winder, M.L.; Brahma, M.K.; Vekeriotaite, B.; Martens, P.J.; et al. PTPN2 Regulates the Interferon Signaling and Endoplasmic Reticulum Stress Response in Pancreatic beta-Cells in Autoimmune Diabetes. Diabetes 2022, 71, 653–668. [Google Scholar] [CrossRef] [PubMed]
- Edilova, M.I.; Abdul-Sater, A.A.; Watts, T.H. TRAF1 Signaling in Human Health and Disease. Front Immunol 2018, 9, 2969. [Google Scholar] [CrossRef]
- Strick, D.J.; Vollrath, D. Focus on molecules: MERTK. Exp Eye Res 2010, 91, 786–787. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.C.; Tsai, S.H.; Yeh, S.I.; Sun, M.H.; Tsao, Y.P. A PEDF-Derived Short Peptide Prevents Sodium Iodate-Induced Retinal Degeneration in Rats by Activating the SLC7A11/GSH/GPX4 Pathway in the RPE Cells. J Cell Mol Med 2025, 29, e70693. [Google Scholar] [CrossRef] [PubMed]
- Goto, S.; Onishi, A.; Misaki, K.; Yonemura, S.; Sugita, S.; Ito, H.; Ohigashi, Y.; Ema, M.; Sakaguchi, H.; Nishida, K.; et al. Neural retina-specific Aldh1a1 controls dorsal choroidal vascular development via Sox9 expression in retinal pigment epithelial cells. Elife 2018, 7. [Google Scholar] [CrossRef]
- Voigt, A.P.; Mulfaul, K.; Mullin, N.K.; Flamme-Wiese, M.J.; Giacalone, J.C.; Stone, E.M.; Tucker, B.A.; Scheetz, T.E.; Mullins, R.F. Single-cell transcriptomics of the human retinal pigment epithelium and choroid in health and macular degeneration. Proc Natl Acad Sci U S A 2019, 116, 24100–24107. [Google Scholar] [CrossRef]
- Dhirachaikulpanich, D.; Li, X.; Porter, L.F.; Paraoan, L. Integrated Microarray and RNAseq Transcriptomic Analysis of Retinal Pigment Epithelium/Choroid in Age-Related Macular Degeneration. Front Cell Dev Biol 2020, 8, 808. [Google Scholar] [CrossRef]
- Chowdhury, O.; Bammidi, S.; Gautam, P.; Babu, V.S.; Liu, H.; Shang, P.; Xin, Y.; Mahally, E.; Nemani, M.; Koontz, V.; et al. Activated mTOR Signaling in the RPE Drives EMT, Autophagy, and Metabolic Disruption, Resulting in AMD-Like Pathology in Mice. Aging Cell 2025, e70018. [Google Scholar] [CrossRef]
- Datta, S.; Cano, M.; Satyanarayana, G.; Liu, T.; Wang, L.; Wang, J.; Cheng, J.; Itoh, K.; Sharma, A.; Bhutto, I.; et al. Mitophagy initiates retrograde mitochondrial-nuclear signaling to guide retinal pigment cell heterogeneity. Autophagy 2023, 19, 966–983. [Google Scholar] [CrossRef]
- Zhao, X.; Gao, M.; Liang, J.; Chen, Y.; Wang, Y.; Wang, Y.; Xiao, Y.; Zhao, Z.; Wan, X.; Jiang, M.; et al. SLC7A11 Reduces Laser-Induced Choroidal Neovascularization by Inhibiting RPE Ferroptosis and VEGF Production. Front Cell Dev Biol 2021, 9, 639851. [Google Scholar] [CrossRef]
- Perino, A.; Demagny, H.; Velazquez-Villegas, L.; Schoonjans, K. Molecular Physiology of Bile Acid Signaling in Health, Disease, and Aging. Physiol Rev 2021, 101, 683–731. [Google Scholar] [CrossRef] [PubMed]
- Zubieta, D.; Warden, C.; Bhattacharya, S.; Brantley, M.A., Jr. Tauroursodeoxycholic Acid Confers Protection Against Oxidative Stress via Autophagy Induction in Retinal Pigment Epithelial Cells. Curr Issues Mol Biol 2025, 47. [Google Scholar] [CrossRef] [PubMed]
- Win, A.; Delgado, A.; Jadeja, R.N.; Martin, P.M.; Bartoli, M.; Thounaojam, M.C. Pharmacological and Metabolic Significance of Bile Acids in Retinal Diseases. Biomolecules 2021, 11. [Google Scholar] [CrossRef]
- Alvarez-Barrios, A.; Alvarez, L.; Saenz de Santa Maria, P.; Garcia, M.; Alvarez-Buylla, J.R.; Pereiro, R.; Gonzalez-Iglesias, H. Dysregulated lipid metabolism in a retinal pigment epithelial cell model and serum of patients with age-related macular degeneration. BMC Biol 2025, 23, 96. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.D.; Mondon, I.; Ellis, C.; Muir, A.M.; Turner, S.; Keeling, E.; Wai, H.A.; Chatelet, D.S.; Johnson, D.A.; Tumbarello, D.A.; et al. Whole RNA-Seq Analysis Reveals Longitudinal Proteostasis Network Responses to Photoreceptor Outer Segment Trafficking and Degradation in RPE Cells. Cells 2025, 14. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Geathers, J.S.; Grillo, S.L.; Weber, S.R.; Wang, W.; Zhao, Y.; Sundstrom, J.M. Role of Epithelial-Mesenchymal Transition in Retinal Pigment Epithelium Dysfunction. Front Cell Dev Biol 2020, 8, 501. [Google Scholar] [CrossRef]
| Gene (Mouse) | Forward Primer | Reverse Primer |
| Beta-actin | CCTGGATAGCAACGTAGATGC | ACCTTCTACAATGACCTGGC |
| Prom1 | AACATATGCGCGGGAGAG | CAGTTTCTGGGTCCCTTTGA |
| Pink1 | CTGATCGAGGAGAAGCAGGC | GCCAATGGCTTGCCCTATGA |
| Ogn | CGCAGCTGGACTCACATGTT | TCTTTCTTGGTTGGTAATGATGCT |
| Mertk | TGGATACGTGCATCTGTCCG | GAGGAGCAGAGAATGGGCTG |
| Grem1 | CTTCGCAGACCTGGAGACG | CAGGTTGTGGTGGGGACTG |
| Slc7a11 | CAGGCATCTTCATCTCCCCC | GAGCAGTTCCACCCAGACTC |
| Ablim1 | GAGGCCATCGGTCTGCTTC | GAAATGCTTGGTCTGCACCC |
| Igfbp2 | CACAGGTGACACTGCAGACG | GAACACAGCCAGCTCCTTCA |
| Aldh1a1 | TGAGCCTGTCACCTGTGTTC | CCTTCTTCCACGTGGCAGAT |
| Postn | ATGACAAGGTCCTGGCTCAC | CCCGCAGATAGCACCTTGAT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





