Introduction
Chest radiography (CXR) is one of the most widely performed imaging investigations in clinical medicine due to its speed, affordability, and accessibility. (1), (2) It is central to diagnosing a wide range of thoracic diseases including tuberculosis, pneumonia, pulmonary edema, pneumothorax, lung cancer, interstitial lung disease, and cardiomegaly. (3), (4), (5), (6), (7) Over two billion chest X-rays are obtained globally each year, yet timely and accurate interpretation remains a major bottleneck. (8) Accurate interpretation requires trained radiologists, who are scarce in many LMICs, for example, Malaysia has only 3.9 radiologists per 100,000 population, while Tanzania, with 58 million people, has just about 60 radiologists. (9), (10) In Nepal, the demand for chest radiographs in emergency, ICU, and outpatient settings exceeds the capacity of available specialists. Even in high-income countries, radiologists face mounting workloads, leading to delays, reporting burnouts, and missed findings. (11)
Manual CXR interpretation suffers from (i) human error in subtle or overlapping findings, (ii) fatigue-induced inconsistency with high daily caseloads, and (iii) limited specialist availability in remote regions. (12), (13), (14) Second opinions are known to change diagnoses in up to 21% of cases, underscoring the need for assistive tools that can enhance diagnostic consistency and reduce human error. (15)
Deep learning-based CNNs have demonstrated potential for automated interpretation of medical imaging. (16) Unlike costly high-performance systems, efficient CNN architectures can be optimized for low-cost hardware and offline operation, making them suitable for deployment in LMIC contexts. (17), (18) Beyond diagnostic accuracy, AI integration may reduce reporting time, energy use, and costs associated with centralized image processing. (19)
AI-based CNNs show promise for automating chest radiograph interpretation. (18) Yet most prior models rely exclusively on clean public datasets, focus on PA views, and do not address co-occurring abnormalities or real-world imaging artifacts. (20) Moreover, few are adapted to LMIC workflows requiring offline functionality and energy efficiency.
This study addresses these gaps by evaluating a CNN pretrained on large public datasets and fine-tuned on a large, adult-only local dataset from Nepal. Unique contributions include inclusion of ER, ICU, and OPD cases; inclusion of AP and lateral views; resilience to poor-quality imaging; and interpretability features including 14-color bounding boxes and co-occurrence detection of 14 pathologies: aortic enlargement, atelectasis, calcification, ILD, infiltration, lung mass/nodule, other lesion (bronchiectasis, hilar lymphadenopathy) mass/nodule, pleural effusion, pleural thickening, consolidation, pneumothorax, lung opacity, fibrosis, and cardiomegaly on frontal and lateral chest radiographs.
This study evaluates the diagnostic performance of a CNN trained on large public datasets (N1 publicly available images from the VinBig and NIH ChestX-ray14 and fine-tuned on local CXRs from adults in Nepal, tested on 522 adult (frontal and lateral view Xrays) assessing not only sensitivity and specificity but also its impact on radiologist workflow, and real-world deployment feasibility. The system is deployed through a lightweight, web based React/NodeJS interface compatible with edge GPUs.
Methods
Study Design and Datasets
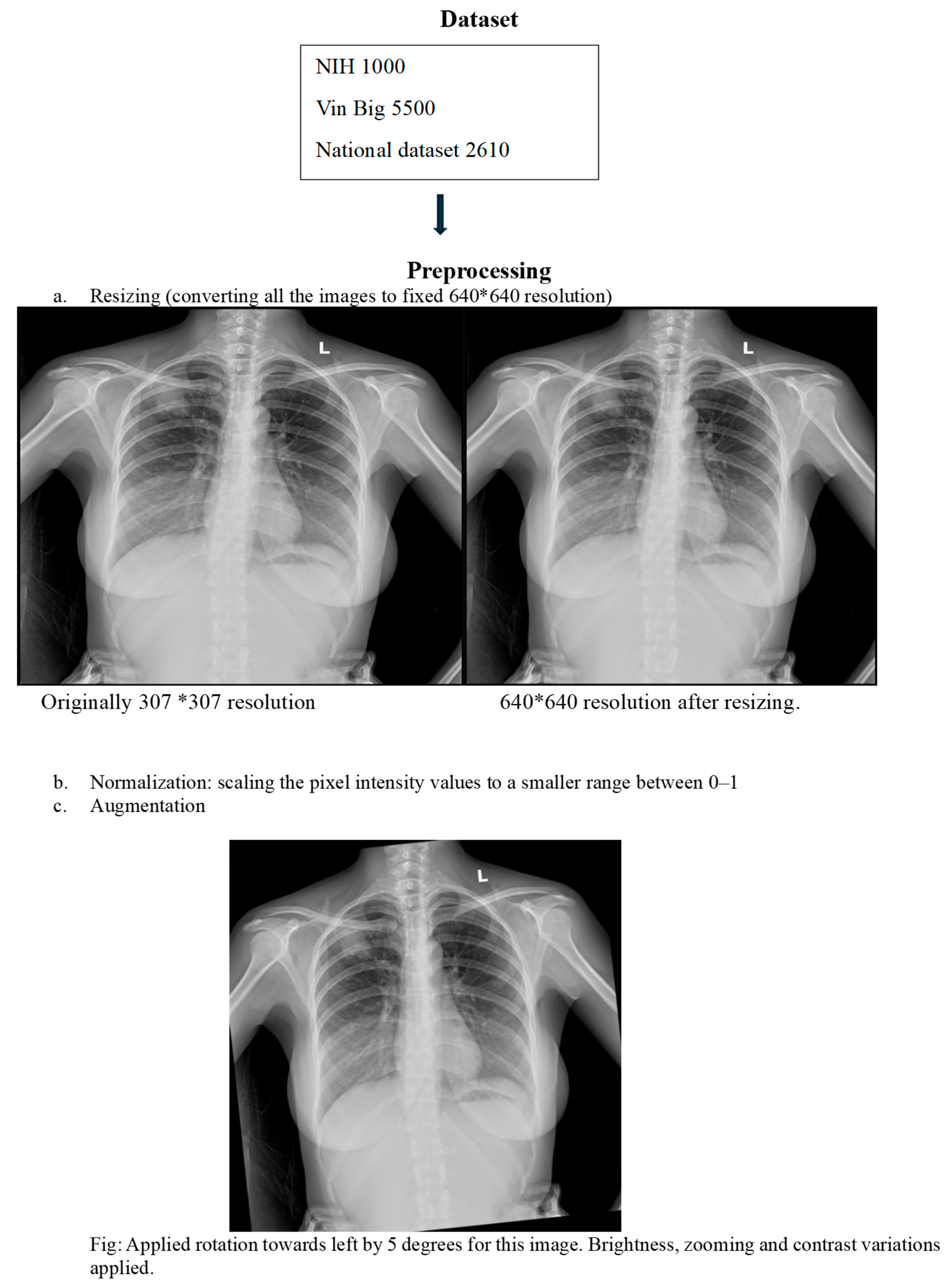
This retrospective study developed an AI model using a hybrid dataset approach. Initial pretraining was performed on two public datasets: the Vin Bigdata Chest X-ray collection (5500 images) NIH Chest X-ray dataset (1000 images). (21), (22) For domain-specific fine-tuning, we curated a local dataset of adult chest X-rays from Tribhuvan University Teaching Hospital, sourced from Emergency Room, Intensive Care Unit, and Outpatient Department settings. Pediatric cases were excluded to avoid anatomical confounding. The local dataset included PA, AP, and lateral views, annotated for 14 thoracic pathologies by at least two qualified radiologists. The dataset was partitioned with strict patient-wise splitting into 70% training, 10% validation, and 20% testing sets to prevent data leakage (
Figure 1).
Preprocessing and Augmentation
We implemented comprehensive preprocessing and augmentation strategies to enhance model generalization. All images underwent standardized normalization. To simulate real-world clinical challenges, we applied transformations including random rotation, noise injection, low exposure simulation, and Gaussian blur. The model was specifically trained to maintain performance despite common clinical artifacts such as text labels, wires, and ICU bed structures, ensuring robustness in diverse hospital environments.
Model Architecture
A computationally efficient lightweight Convolutional Neural Network architecture was selected to facilitate deployment in resource-constrained settings. The model was designed to generate precise bounding box predictions for localization of all 14 target pathologies. We implemented the Weighted Boxes Fusion technique to refine these predictions and enhance localization accuracy by intelligently combining overlapping detections. The final output presents refined bounding boxes overlaid on original X-ray images, with each pathology class assigned a distinctive color for immediate visual identification, accompanied by confidence scores for each detection.
Evaluation Metrics
Model performance was rigorously assessed using comprehensive metrics including Area Under the Receiver Operating Characteristic Curve, sensitivity, specificity, and mean Average Precision. We conducted a reader study to evaluate clinical utility, measuring the AI system’s integration ease with radiologists’ workflow.
Deployment Testing
The system was successfully integrated into the hospital’s Picture Archiving and Communication System (PACS), operating seamlessly within standard diagnostic workflows. We also developed a fully functional standalone offline mode capable of operating on standard laptops without internet connectivity. All AI-generated annotations, including color-coded bounding boxes and confidence scores, were made exportable as overlays within standard DICOM files, ensuring full interoperability with existing medical imaging infrastructure.
Results
Co-Occurrence Detection and Visualization
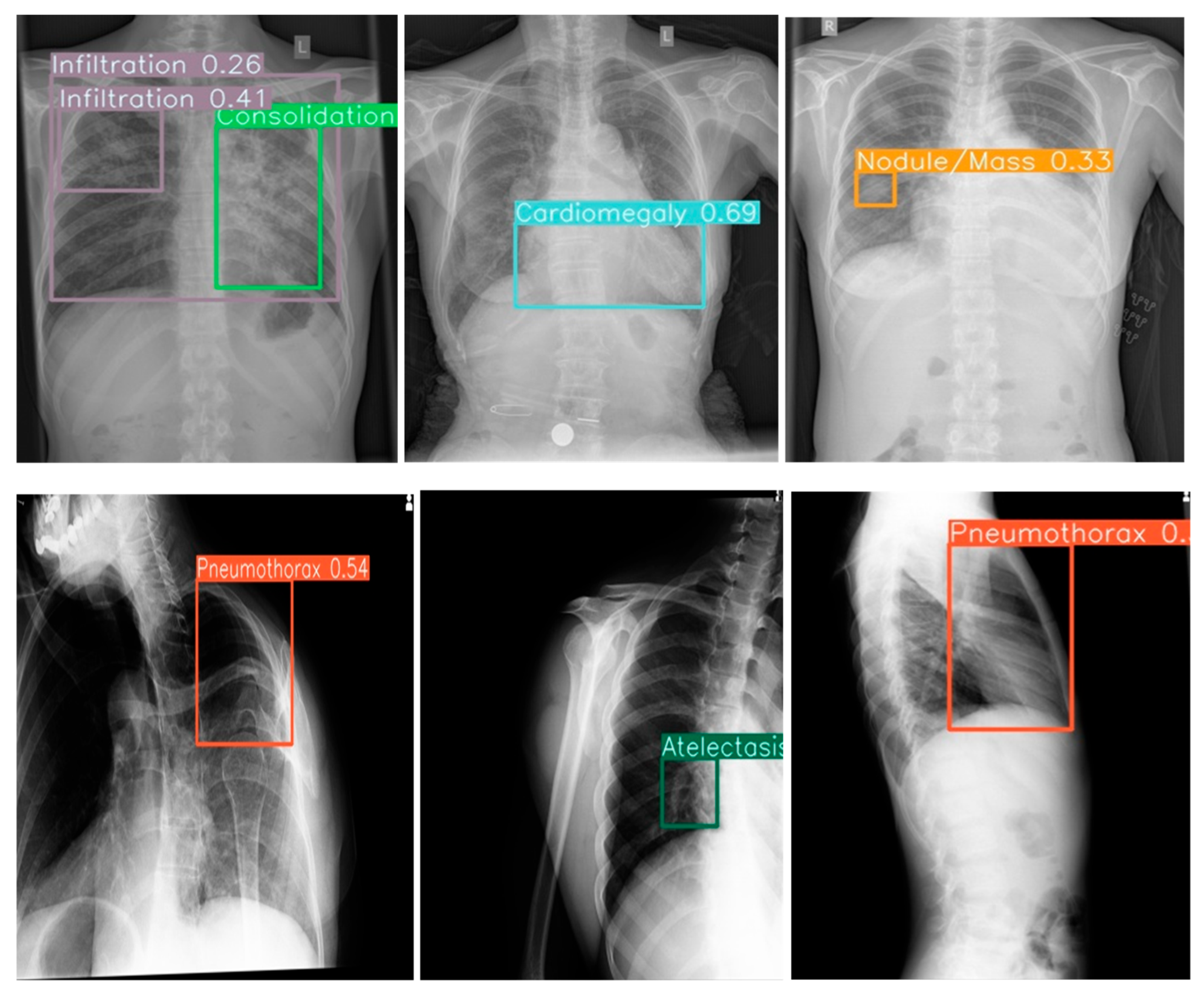
The model effectively identified singular and multiple co-occurrences of pathologies in one Xray in clinical setting. The unique 14-color bounding box system provided immediate visual differentiation, enabling rapid interpretation of different pathologies in a single Xray. Confidence score overlays further enhanced clinical utility by allowing prioritization of high-certainty detections during time-constrained readings.
Robustness and Generalization
The model demonstrated potential for consistent performance across challenging imaging conditions, including poor-quality ICU anteroposterior films and lateral views with artifacts. This suggests possible robustness to technical variations that could support deployment in diverse clinical environments where ideal imaging conditions are not always achievable.
Figure 2.
Example outputs with 14-color bounding boxes on multi-pathology cases in Row 1(Frontal view) and Row 2 (Lateral view).
Figure 2.
Example outputs with 14-color bounding boxes on multi-pathology cases in Row 1(Frontal view) and Row 2 (Lateral view).
Deployment and Integration
The system was successfully integrated into the hospital’s PACS, operating seamlessly within existing clinical workflows. Offline functionality was confirmed on standard laptops without internet connectivity, ensuring reliable operation in resource-constrained settings. The exportable DICOM overlay capability-maintained compatibility with existing medical imaging infrastructure, facilitating smooth adoption into routine diagnostic processes.
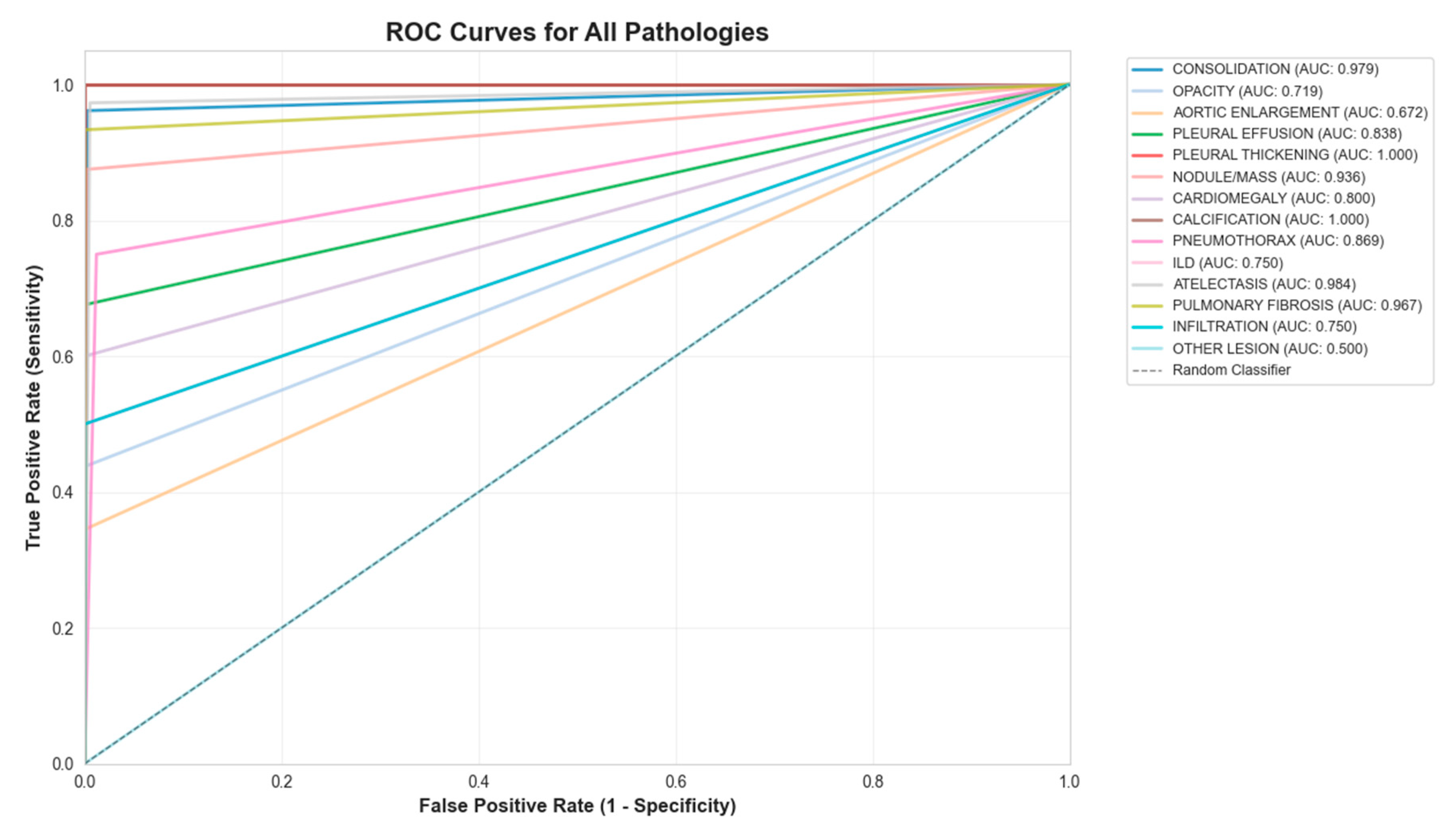
Figure 3.
ROC curve for the 14-class model.
Figure 3.
ROC curve for the 14-class model.
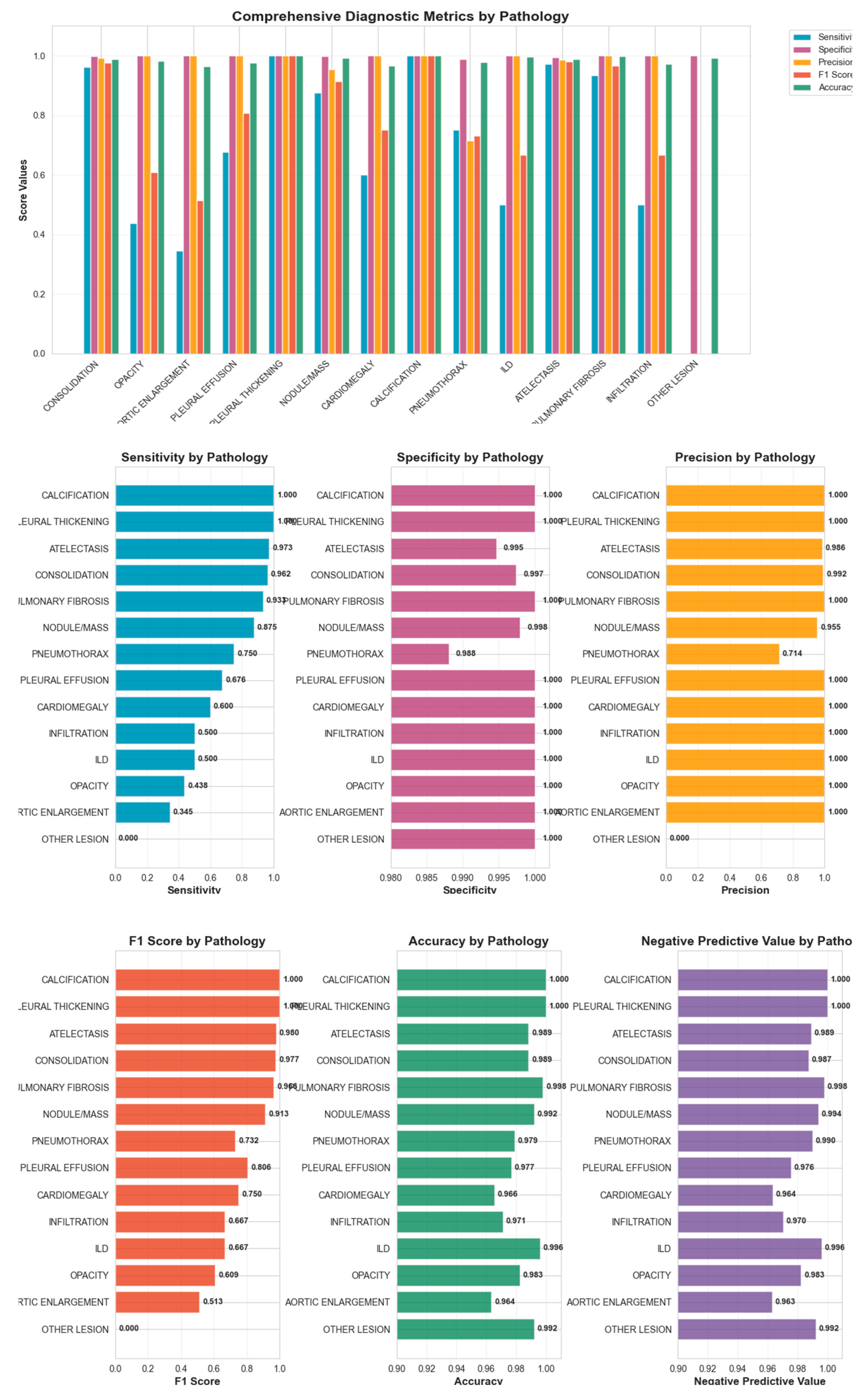
Figure 4.
Comprehensive diagnostic metrics for 14 classes of pathologies.
Figure 4.
Comprehensive diagnostic metrics for 14 classes of pathologies.
Discussion
AI tools have rapidly advanced chest radiograph interpretation, with CNNs demonstrating high diagnostic accuracy across large datasets such as NIH ChestX-ray14 (Wang et al., 2017) and MIMIC-CXR (Johnson et al., 2019). (22), (23) Studies by Rajpurkar et al. (2017) and Hofmeister et al. (2024) found that deep learning models achieved near-radiologist performance on curated datasets. (24), (25) However, their generalizability to low- and middle-income countries (LMICs) remains limited. Most of these frameworks were developed using standardized posteroanterior (PA) films obtained in tertiary hospitals with consistent imaging quality and equipment calibration. (20) In contrast, LMIC hospitals frequently face variability in patient positioning, exposure, and machine type, which substantially affects image quality and limits the applicability of these models in real-world clinical practice. (26)
Unlike prior CNN models limited to single-view datasets, our system incorporated multiview (anteroposterior, lateral, and portable) radiographs from diverse clinical environments, replicating real-world imaging diversity and challenges not included in traditional public datasets. Certain pathologies such as retrocardiac consolidation, mediastinal widening, or small pleural effusions may only be apparent on the lateral projection, clarifying equivocal frontal findings. (27) Hashir et al. (2020) similarly found that training on lateral images reduced false negatives. (28)
Our results demonstrated high average specificity (99%) and precision (90%), indicating reliable identification of normal studies and low false-positive rates. Sensitivity was variable across pathologies (average 68%), with the model performing best on common and well-defined findings such as consolidation, atelectasis, and pulmonary fibrosis, all of which showed F1-scores above 0.90. These pathologies likely benefited from clearer radiographic features and higher representation in the training set. Conversely, rarer conditions such as ILD, aortic enlargement, and other lesions exhibited low sensitivity, reflecting limited training examples and the inherent difficulty of recognizing subtle or heterogeneous imaging patterns, consistent with prior reports (Majkowska 2020; Seyyed-Kalantari et al., 2021). (29), (30) Co-occurrence analysis revealed clinically plausible associations, such as consolidation with atelectasis or pleural effusion, supporting the model’s capacity to recognize physiologic relationships rather than isolated features.
Studies by Liong-Rung et al (2022) and Kaewwilai et al. (2025) reported that clinicians often have to await formal radiology reports which can delay the detection of urgent findings such as pneumothorax, pleural effusion and pulmonary tuberculosis. (31), (32) Several images may not be reviewed by a radiologist at all in LMICs, in high volume setting. Integrating AI systems for rapid, automated screening ensures that these studies are at least evaluated, with high-confidence cases flagged for radiologist review and immediate clinical attention. The use of color-coded bounding boxes and confidence-weighted outputs in our model facilitates transparent, interpretable triage, helping prioritize critical cases and support timely patient care. Similar confidence-driven approaches have proven effective in other screening contexts, such as detection of intracranial hemorrhage. (33)
Although Transformer-based models (Chen et al., 2021, Touvron et al., 2020) offer high representational power, they require advanced hardware and stable internet access, conditions often lacking in LMIC settings. (34), (35), (36) In contrast, our lightweight CNN functions efficiently offline on standard hospital workstations, reducing infrastructure and energy demands and improving scalability.
Limitations
This study was conducted retrospectively at a single tertiary hospital, which may restrict generalizability across regions and imaging devices. Class imbalance, particularly for rare conditions such as ILD and calcification, likely contributed to reduced sensitivity. Pediatric and neonatal populations were not included, despite their high disease burden in LMICs. Additionally, prospective workflow evaluation is needed to determine optimal confidence thresholds and their impact on real-world triage efficiency.
Future Directions
Future studies should focus on multicenter validation across varied LMIC healthcare settings, inclusion of pediatric cohorts, and temporal performance assessment on prospective data. Expanding datasets for low-prevalence conditions and exploring class reweighting or synthetic augmentation may improve detection balance. Incorporating advanced explainability approaches such as attention-based or information-theoretic visualization could further enhance clinician trust and integration into routine diagnostic pathways.
Conclusion
This study presents a CNN-based system for detecting 14 thoracic pathologies, designed to address the technical challenges of diverse clinical environments seen in everyday practice. Trained on a hybrid, multi-view dataset (including AP, lateral, and portable films) and enhanced through data augmentation, the system demonstrated robust performance on poor-quality radiographs, achieving a mean average precision (mAP@0.5) of 93%. Its interpretability features of color-coded bounding boxes and visible confidence scores enable efficient multi-pathology detection and case prioritization. Deployable via both PACS-integrated and standalone interfaces, the system demonstrates potential as a scalable tool for triage and workflow efficiency, with a foundation for future expansion to rarer conditions.
Author Contributions
LG, PRR, SG, SC, SA, and MU contributed to the conceptualization of the study. SG, SC, and SA performed software development, implemented and tested the AI tool, LG, GG, GG3 and PRR contributed to validation and data curation. MU additionally provided guidance and feedback for improvement. LG, PRR, SM, GG, SA, GG3 and MU provided clinical expertise through accuracy testing, formal analysis, and development of the dataset for training and testing the AI tool. Data entry and organization were performed by LG, GG3, SA, SM, SG, SC, and SA. LG, SA, GG3 and SM drafted the original manuscript, while PRR, GG and MU contributed to critical review, editing, and revision of the manuscript. GG, MU, and PRR provided supervision throughout the study, and project administration was managed by LG and PRR. All authors contributed to interpretation of the results and approved the final version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
This retrospective study was approved by the Institutional Review Board. (IRB approval number: 25 (6-11)E2 082/083 ).
Informed Consent Statement
Patient consent was waived due to the retrospective nature of the study, in accordance with IRB guidelines.
Data Availability Statement
The data supporting the findings of this study are available from the corresponding authors upon reasonable request.
Acknowledgments
The authors gratefully acknowledge Mr. Binod Bhattarai, Radio technician at TUTH, Nepal, for his assistance with dataset availability, PACS feasibility testing, offline testing and deployment of the AI tool.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| LMIC – Low- and Middle-Income Countries |
| AI – Artificial Intelligence |
| CNN – Convolutional Neural Network |
| AP – Average Precision |
| PA – Posteroanterior (commonly used in radiology for chest X-rays) |
| ER – Emergency Room |
| OPD – Outpatient Department |
| mAP – Mean Average Precision |
| PACS – Picture Archiving and Communication System |
| ILD – Interstitial Lung Disease |
References
- Speets AM, van der Graaf Y, Hoes AW, Kalmijn S, Sachs AP, Rutten MJ, et al. Chest radiography in general practice: indications, diagnostic yield and consequences for patient management. Br J Gen Pract J R Coll Gen Pract. 2006 Aug;56(529):574–8.
- Imaging the Chest. In: Diagnostic Imaging for the Emergency Physician [Internet]. Elsevier; 2011 [cited 2025 Jul 9]. p. 185–296. Available from: https://linkinghub.elsevier.com/retrieve/pii/B9781416061137100055. [CrossRef]
- Macdonald C, Jayathissa S, Leadbetter M. Is post-pneumonia chest X-ray for lung malignancy useful? Results of an audit of current practice. Intern Med J. 2015 Mar;45(3):329–34. [CrossRef]
- Lindow T, Quadrelli S, Ugander M. Noninvasive Imaging Methods for Quantification of Pulmonary Edema and Congestion: A Systematic Review. JACC Cardiovasc Imaging. 2023 Nov;16(11):1469–84. [CrossRef]
- Tran J, Haussner W, Shah K. Traumatic Pneumothorax: A Review of Current Diagnostic Practices And Evolving Management. J Emerg Med. 2021 Nov;61(5):517–28. [CrossRef]
- Vizioli L, Ciccarese F, Forti P, Chiesa AM, Giovagnoli M, Mughetti M, et al. Integrated Use of Lung Ultrasound and Chest X-Ray in the Detection of Interstitial Lung Disease. Respir Int Rev Thorac Dis. 2017;93(1):15–22. [CrossRef]
- Del Ciello A, Franchi P, Contegiacomo A, Cicchetti G, Bonomo L, Larici AR. Missed lung cancer: when, where, and why? Diagn Interv Radiol Ank Turk. 2017;23(2):118–26. [CrossRef]
- Akhter Y, Singh R, Vatsa M. AI-based radiodiagnosis using chest X-rays: A review. Front Big Data. 2023;6:1120989. [CrossRef]
- Ramli NM, Mohd Zain NR. The Growing Problem of Radiologist Shortage: Malaysia’s Perspective. Korean J Radiol. 2023 Oct;24(10):936–7. [CrossRef]
- Marey A, Mehrtabar S, Afify A, Pal B, Trvalik A, Adeleke S, et al. From Echocardiography to CT/MRI: Lessons for AI Implementation in Cardiovascular Imaging in LMICs—A Systematic Review and Narrative Synthesis. Bioengineering. 2025 Sep 27;12(10):1038. [CrossRef]
- Nakajima Y, Yamada K, Imamura K, Kobayashi K. Radiologist supply and workload: international comparison: Working Group of Japanese College of Radiology. Radiat Med. 2008 Oct;26(8):455–65. [CrossRef]
- Gefter WB, Hatabu H. Reducing Errors Resulting From Commonly Missed Chest Radiography Findings. Chest. 2023 Mar;163(3):634–49. [CrossRef]
- Rimmer A. Radiologist shortage leaves patient care at risk, warns royal college. BMJ. 2017 Oct 11;359:j4683. [CrossRef]
- Fawzy NA, Tahir MJ, Saeed A, Ghosheh MJ, Alsheikh T, Ahmed A, et al. Incidence and factors associated with burnout in radiologists: A systematic review. Eur J Radiol Open. 2023 Dec;11:100530. [CrossRef]
- Eakins C, Ellis WD, Pruthi S, Johnson DP, Hernanz-Schulman M, Yu C, et al. Second opinion interpretations by specialty radiologists at a pediatric hospital: rate of disagreement and clinical implications. AJR Am J Roentgenol. 2012 Oct;199(4):916–20. [CrossRef]
- Sufian MA, Hamzi W, Sharifi T, Zaman S, Alsadder L, Lee E, et al. AI-Driven Thoracic X-ray Diagnostics: Transformative Transfer Learning for Clinical Validation in Pulmonary Radiography. J Pers Med. 2024 Aug 12;14(8):856. [CrossRef]
- Anari PY, Obiezu F, Lay N, Firouzabadi FD, Chaurasia A, Golagha M, et al. Using YOLO v7 to Detect Kidney in Magnetic Resonance Imaging. ArXiv. 2024 Feb 12;arXiv:2402.05817v2. [CrossRef]
- Chen Z, Pawar K, Ekanayake M, Pain C, Zhong S, Egan GF. Deep Learning for Image Enhancement and Correction in Magnetic Resonance Imaging—State-of-the-Art and Challenges. J Digit Imaging. 2022 Nov 2;36(1):204–30. [CrossRef]
- Zhong B, Yi J, Jin Z. AC-Faster R-CNN: an improved detection architecture with high precision and sensitivity for abnormality in spine x-ray images. Phys Med Biol. 2023 Sep 26;68(19). [CrossRef]
- Rajaraman S, Zamzmi G, Folio LR, Antani S. Detecting Tuberculosis-Consistent Findings in Lateral Chest X-Rays Using an Ensemble of CNNs and Vision Transformers. Front Genet. 2022;13:864724. [CrossRef]
- Nguyen HQ, Lam K, Le LT, Pham HH, Tran DQ, Nguyen DB, et al. VinDr-CXR: An open dataset of chest X-rays with radiologist’s annotations. Sci Data. 2022 Jul 20;9(1):429. [CrossRef]
- Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM. ChestX-Ray8: Hospital-Scale Chest X-Ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) [Internet]. Honolulu, HI: IEEE; 2017 [cited 2025 Oct 25]. p. 3462–71. Available from: http://ieeexplore.ieee.org/document/8099852/.
- Johnson AEW, Pollard TJ, Berkowitz SJ, Greenbaum NR, Lungren MP, Deng CY, et al. MIMIC-CXR, a de-identified publicly available database of chest radiographs with free-text reports. Sci Data. 2019 Dec 12;6(1):317. [CrossRef]
- Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T, et al. CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning [Internet]. arXiv; 2017 [cited 2025 Jun 25]. Available from: http://arxiv.org/abs/1711.05225. [CrossRef]
- Hofmeister J, Garin N, Montet X, Scheffler M, Platon A, Poletti PA, et al. Validating the accuracy of deep learning for the diagnosis of pneumonia on chest x-ray against a robust multimodal reference diagnosis: a post hoc analysis of two prospective studies. Eur Radiol Exp. 2024 Feb 2;8(1):20. [CrossRef]
- Obermeyer Z, Emanuel EJ. Predicting the Future — Big Data, Machine Learning, and Clinical Medicine. N Engl J Med. 2016 Sep 29;375(13):1216–9. [CrossRef]
- Gaber KA, McGavin CR, Wells IP. Lateral chest X-ray for physicians. J R Soc Med. 2005 Jul;98(7):310–2.
- Hashir M, Bertrand H, Cohen JP. Quantifying the Value of Lateral Views in Deep Learning for Chest X-rays [Internet]. arXiv; 2020 [cited 2025 Oct 25]. Available from: https://arxiv.org/abs/2002.02582. [CrossRef]
- Majkowska A, Mittal S, Steiner DF, Reicher JJ, McKinney SM, Duggan GE, et al. Chest Radiograph Interpretation with Deep Learning Models: Assessment with Radiologist-adjudicated Reference Standards and Population-adjusted Evaluation. Radiology. 2020 Feb;294(2):421–31. [CrossRef]
- Seyyed-Kalantari L, Zhang H, McDermott MBA, Chen IY, Ghassemi M. Underdiagnosis bias of artificial intelligence algorithms applied to chest radiographs in under-served patient populations. Nat Med. 2021 Dec;27(12):2176–82. [CrossRef]
- Liong-Rung L, Hung-Wen C, Ming-Yuan H, Shu-Tien H, Ming-Feng T, Chia-Yu C, et al. Using Artificial Intelligence to Establish Chest X-Ray Image Recognition Model to Assist Crucial Diagnosis in Elder Patients With Dyspnea. Front Med. 2022;9:893208. [CrossRef]
- Kaewwilai L, Yoshioka H, Choppin A, Prueksaritanond T, Ayuthaya TPN, Brukesawan C, et al. Development and evaluation of an artificial intelligence (AI) -assisted chest x-ray diagnostic system for detecting, diagnosing, and monitoring tuberculosis. Glob Transit. 2025;7:87–93. [CrossRef]
- Gibson E, Georgescu B, Ceccaldi P, Trigan PH, Yoo Y, Das J, et al. Artificial Intelligence with Statistical Confidence Scores for Detection of Acute or Subacute Hemorrhage on Noncontrast CT Head Scans. Radiol Artif Intell. 2022 May;4(3):e210115. [CrossRef]
- Chen CF, Fan Q, Panda R. CrossViT: Cross-Attention Multi-Scale Vision Transformer for Image Classification [Internet]. arXiv; 2021 [cited 2025 Oct 25]. Available from: https://arxiv.org/abs/2103.14899. [CrossRef]
- Touvron H, Cord M, Douze M, Massa F, Sablayrolles A, Jégou H. Training data-efficient image transformers & distillation through attention [Internet]. arXiv; 2020 [cited 2025 Oct 25]. Available from: https://arxiv.org/abs/2012.12877. [CrossRef]
- Kong Z, Xu D, Li Z, Dong P, Tang H, Wang Y, et al. AutoViT: Achieving Real-Time Vision Transformers on Mobile via Latency-aware Coarse-to-Fine Search. Int J Comput Vis. 2025 Sep;133(9):6170–86. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).