1. Introduction
Chest radiography remains the most frequently performed diagnostic imaging procedure globally, accounting for over 40% of all radiologic examinations [
1,
2]. Its accessibility, relatively low cost, and diagnostic versatility make it indispensable in evaluating cardiopulmonary conditions—particularly in low-resource environments [
2,
3]. A key radiographic parameter frequently derived from posteroanterior (PA) chest X-rays is the cardiothoracic ratio (CTR), a straightforward yet clinically valuable index for assessing potential cardiac enlargement.
CTR is defined as the ratio of the maximal horizontal cardiac diameter to the maximal internal thoracic diameter on a PA chest radiograph. In adults and older children, a CTR greater than 0.5 is commonly interpreted as indicative of cardiomegaly—an early radiographic sign of possible cardiac dysfunction [
3,
4]. Accurate estimation of CTR is essential in the early detection of heart failure and associated conditions such as systemic hypertension, valvular heart disease, and pulmonary hypertension [
4,
5,
6].
Despite its diagnostic value, CTR measurement is still routinely performed manually, introducing observer variability and workflow delays, particularly in healthcare systems with limited radiographic personnel and rising imaging volumes. These challenges are especially pronounced in low- and middle-income countries (LMICs), where radiology services often face staffing shortages and constrained technical infrastructure [
1,
7].
Recent advances in deep learning (DL) offer promising avenues for automating CTR measurement. Several studies have applied convolutional neural networks to this task, yielding high segmentation accuracy and increased consistency compared to manual measurement [
8,
9,
10]. However, most models have been developed using datasets from high-income countries (HICs), such as NIH ChestX-ray14 and MIMIC CXR, which may not generalize well to African settings due to differences in imaging equipment, protocols, and patient demographics [
9,
11].
Furthermore, many DL models assume availability of GPU-based systems—which are often not present in sub-Saharan African healthcare environments. This creates an implementation gap, where AI solutions—despite their potential—remain inaccessible due to hardware limitations [
12].
To bridge this divide, the present study developed and validated a CPU-compatible deep learning model for automated CTR estimation. By training and testing the model on a CPU, the study demonstrates feasibility of deploying functional AI tools within realistic hardware constraints. The model was evaluated on PA chest radiographs collected from two Nigerian institutions: University of Calabar Teaching Hospital (UCTH) and Doren Specialist Hospital, Lagos.
Although automated CTR estimation has been previously explored [
8,
9,
10], few studies have addressed the dual challenge of achieving accurate results and ensuring real-world feasibility in LMIC settings. This study contributes uniquely by validating a CPU-deployable model tailored for clinical use in sub-Saharan Africa, bridging the gap between algorithmic performance and implementation practicality.
The primary aim was to develop a low-cost, accurate, and generalizable AI solution for CTR measurement that functions efficiently in resource-limited environments. Unlike prior models trained on NIH or MIMIC datasets, our model was trained and evaluated using real-world data from Nigerian healthcare institutions, improving context relevance and generalizability in African settings. By incorporating data from a diverse African patient cohort and targeting modest hardware deployment, this study advances contextually relevant, equitable, and scalable AI innovation in radiographic practice.
2. Materials and Methods
2.1. Study Design and Setting
This retrospective cross-sectional study was conducted in Nigeria using chest radiographs collected from two major hospitals: University of Calabar Teaching Hospital (UCTH), Calabar and Doren Specialist Hospital, Lagos. These centres serve patients from diverse ethnic, geographic, and socioeconomic backgrounds across southern and southwestern Nigeria. This diversity strengthens the generalisability of the model to the wider population, as it reflects variations in anatomy, disease patterns, and radiographic acquisition conditions representative of the broader Nigerian and sub-Saharan African context.
2.2. Data Collection and Preprocessing
A total of 5000 adult posteroanterior (PA) chest radiographs were retrospectively retrieved in DICOM format from the radiology archive of University of Calabar Teaching Hospital (UCTH), Calabar and Doren Specialist Hospital, Lagos between January 2020 and December 2023. Inclusion criteria required:
Adequate inspiration, defined by visualization of at least nine posterior ribs;
Proper patient positioning, indicated by symmetrical clavicles and centrally aligned spinous processes;
Absence of significant rotation or artefacts that could affect accurate cardiothoracic ratio (CTR) estimation.
Exclusion criteria included lateral or anteroposterior (AP) projections, pediatric chest X-rays, and cases in which CTR measurement was not feasible (e.g., due to dense opacities obscuring cardiac borders) were excluded from analysis.
All images were anonymised and manually reviewed to ensure compliance with the inclusion criteria. The DICOM images were resized to 512 × 512 pixels, and intensity normalization was applied to standardize brightness and contrast across the dataset.
Ground truth segmentation masks for the lungs and cardiac silhouette were created using the Computer Vision Annotation Tool (CVAT v2.0.0). Three experienced radiographers performed the initial annotations, followed by collaborative review for consensus. A consultant radiologist conducted final validation to ensure anatomical accuracy and minimize inter-observer variability.
Although 5000 radiographs were initially retrieved, a final subset of 3000 high-quality images was selected for model training and evaluation based on the following considerations:
Annotation workload limitations: Due to the labor-intensive nature of manual segmentation and limited expert availability, only 3000 images could be fully annotated and validated within the study timeline.
Quality assurance filtering: Some images were excluded due to subtle artefacts, poor contrast, or suboptimal positioning that could compromise segmentation quality.
Radiologist validation prioritization: The final 3000 images represent the highest-quality cases that passed expert validation, ensuring a reliable dataset for model development.
2.3. Model Development
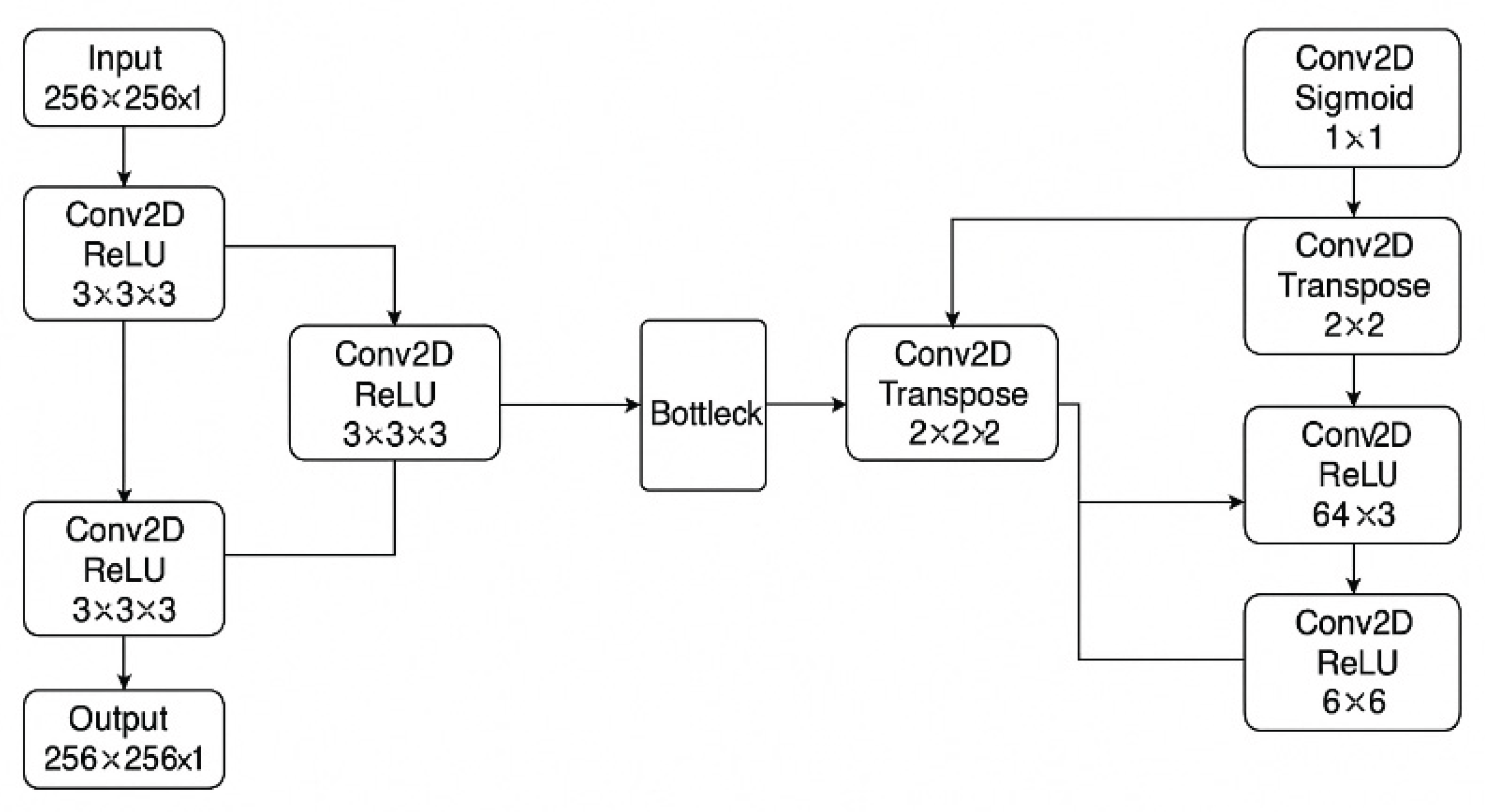
Two separate U-Net convolutional neural networks were implemented in Python (TensorFlow 2.19, Keras): one for heart segmentation and another for lungs. Each network adopted a classic encoder–decoder structure with skip connections to preserve spatial detail [
15] (
Figure 1). The encoder comprised convolutional layers with ReLU activation and max pooling for feature extraction, while the decoder utilized transposed convolutions for spatial upsampling. A sigmoid activation function was applied in the final layer to produce binary segmentation masks.
Out of 3000 curated PA chest X-rays, an 80:20 split was used for training and testing, with 10% of the training set further reserved for validation.
Models were trained on a CPU-only system (Intel Core i7-5500U, 12 GB RAM, Intel HD Graphics 5500) without GPU acceleration, simulating a low-resource deployment environment. Training was conducted using the binary cross-entropy loss function and the Adam optimizer with a learning rate of 0.0001. A batch size of 8 was utilized, and model accuracy was employed as the primary performance metric. To enhance generalization and reduce the risk of overfitting, data augmentation techniques were applied, including horizontal and vertical flipping, rotations within ±15 degrees, and zoom scaling in the range of 0.8× to 1.2×.
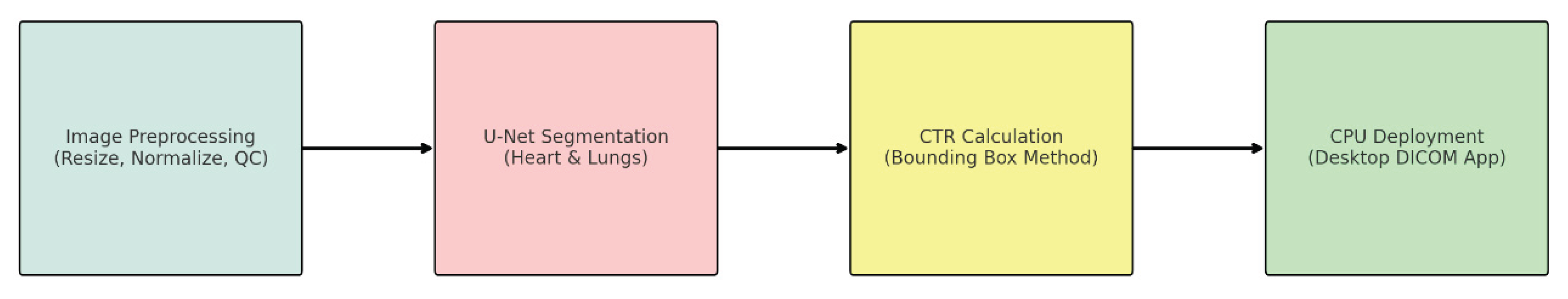
An overview of the complete deep learning pipeline—from image preprocessing through segmentation and CTR calculation to deployment in a lightweight DICOM viewer application—is presented in
Figure 2. This streamlined, CPU-based architecture ensures practical applicability in low-resource clinical settings.
This figure illustrates a simplified U-Net architecture used for heart and lung segmentation from chest radiographs. The network consists of a contracting path (left) with convolutional and ReLU layers for feature extraction, a bottleneck, and an expansive path (right) that performs upsampling via transposed convolutions. The final layer uses a sigmoid activation for pixel-wise classification to generate the binary segmentation mask.
Performance Summary:
Table 1.
Performance Summary of Heart and Lung Segmentation Models.
Table 1.
Performance Summary of Heart and Lung Segmentation Models.
| Model |
Training Phase |
Accuracy (%) |
Recall (%) |
Specificity (%) |
AUC-ROC |
Notes |
| Heart Segmentation |
Initial (15 epochs) |
95.85 |
78.54 |
— |
0.8847 |
Baseline model |
| |
Fine-tuned (2×20 epochs) |
98.54 |
91.42 |
— |
0.9310 |
Selective fine-tuning on challenging cases |
| Lung Segmentation |
Initial |
71.18 |
88.52 |
— |
0.7879 |
Baseline model |
| |
Final (20 epochs) |
97.66 |
97.10 |
98.06 |
0.9758 |
Improved with fine-tuning and data augmentation |
2.4. Cardiothoracic Ratio Calculation
Following segmentation, the maximal horizontal cardiac width and maximal internal thoracic width were computed from the bounding boxes of the predicted masks. The cardiothoracic ratio (CTR) was calculated automatically as:
Manual CTR measurements were also performed using standard PACS tools for 200 randomly selected images by two independent radiologists for comparative analysis.
2.5. Model Evaluation Metrics
Segmentation performance was assessed using the Dice Similarity Coefficient (DSC), Intersection over Union (IoU), and pixel-wise accuracy. Bland–Altman analysis and Pearson correlation were used to compare AI-based and manual CTR measurements. Statistical significance was set at p < 0.05.
2.6. Deployment and Prototype Application
A lightweight prototype DICOM viewer application was developed using Python Dash to enable real-time testing of the model. The application allows side-by-side display of the original image, manual measurement overlay, and AI-predicted CTR, enabling usability assessment in a clinical-like environment.
2.7. Data and Code Availability
The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request. The code used for image processing and analysis (including segmentation and cardiothoracic ratio calculation) is also available for academic and non-commercial purposes, subject to reasonable request and institutional approval if required.
2.8. Generative AI Disclosure
ChatGPT was used for language editing and for the generation of
Figure 1 and
Figure 2 based on the author’s original research and concepts. All data analysis and experimental design were performed independently using standard programming libraries and tools.
2.9. Ethical Approval
This retrospective study was approved by the Institutional Review Boards of the University of Calabar Teaching Hospital and Doren Specialist Hospital. All procedures involving human data were conducted in accordance with the ethical standards of the institutional research committees and the principles outlined in the Declaration of Helsinki and its subsequent amendments.
3. Results
3.1. Dataset Characteristics
A total of 3000 anonymised adult posteroanterior (PA) chest radiographs were included. The images met predefined quality standards and were evenly distributed across the two institutions. Of these, 200 images were randomly selected for manual CTR measurement comparison with AI-predicted values.
3.2. Segmentation Performance
The U-Net models demonstrated strong segmentation performance on the test dataset:
These metrics reflect high spatial overlap between the predicted segmentation masks and ground truth annotations, particularly for the lung regions, which are typically larger and more distinct in X-ray images.
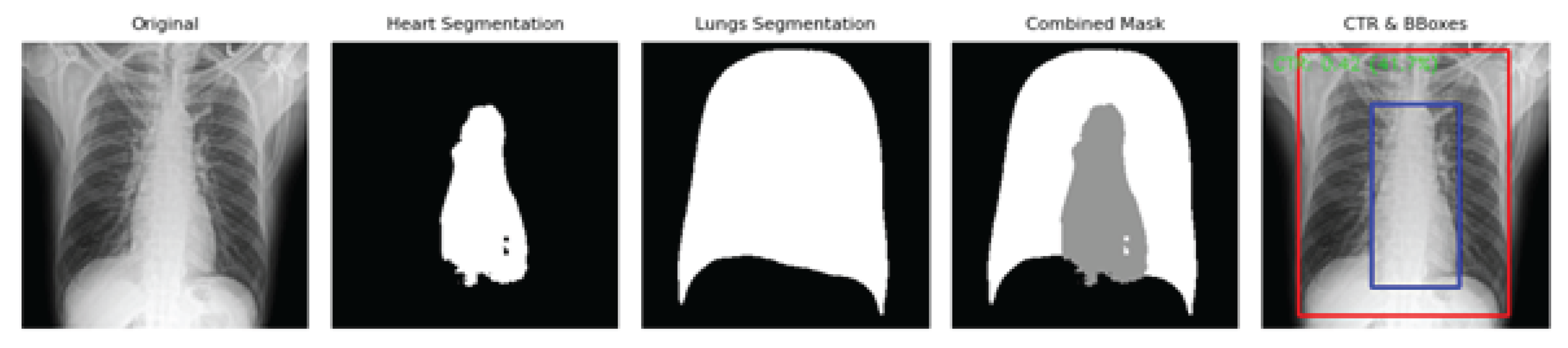
Figure 3 illustrates representative outputs from the AI pipeline, including the original PA chest X-ray, individual binary masks for the heart and lungs, a combined segmentation overlay, and the final CTR measurement visualization with bounding boxes. The clear anatomical delineation supports the accuracy and clinical utility of the automated CTR estimation approach, even on standard CPU systems.
3.3. CTR Prediction Accuracy
The AI-estimated cardiothoracic ratio (CTR) values were compared to manual measurements in a subset of 200 chest radiographs. The mean AI-predicted CTR was 0.50 ± 0.06, while the mean manually measured CTR was 0.51 ± 0.05, indicating close agreement.
Statistical analysis showed:
Pearson correlation coefficient (r): 0.91, p < 0.001 — demonstrating a strong positive linear relationship between AI and manual measurements.
Mean absolute error (MAE): 0.025
-
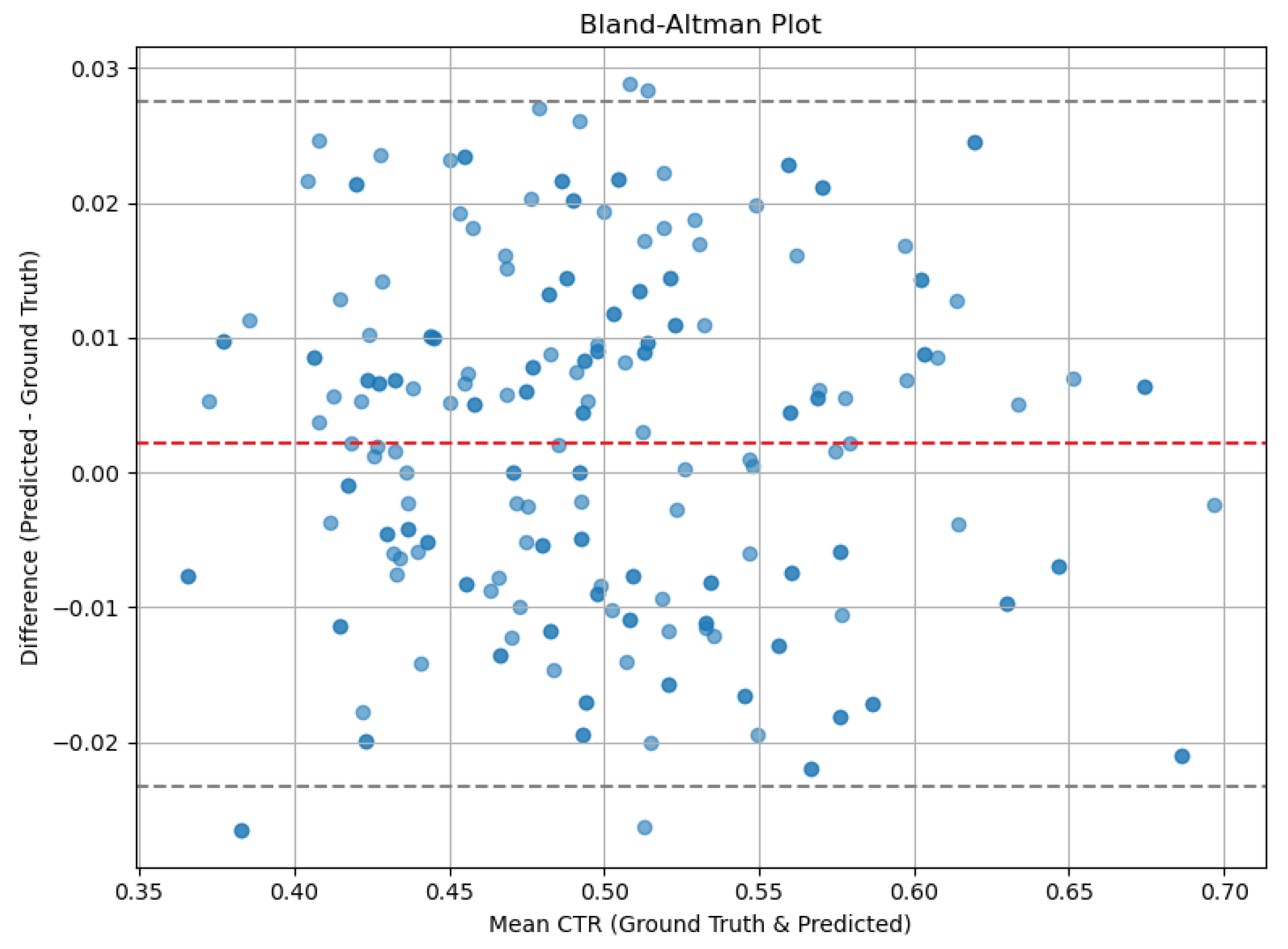
Bland–Altman analysis:
- ○
Mean difference (bias): −0.01
- ○
95% limits of agreement: −0.06 to +0.04
These results suggest that the AI model performs comparably to expert manual measurement, with minimal bias and clinically acceptable error margins.
Figure 4 presents a scatter plot of AI-predicted versus manual CTR values, where the red dashed line represents perfect agreement. The clustering of points around the line reflects high accuracy and strong correlation across the test set.
Figure 5 shows the Bland–Altman plot, highlighting the distribution of differences between manual and AI measurements. Most values fall within the 95% limits of agreement, reinforcing the model’s precision and reliability for clinical use.
3.4. Clinical Threshold Detection
Using the clinical threshold of CTR > 0.50 to indicate cardiomegaly, the AI model showed:
Sensitivity: 90.2%
Specificity: 94.7%
Accuracy: 92.4%
This highlights the model’s clinical utility in screening for cardiomegaly on chest radiographs.
3.5. Prototype Application Demonstration
A lightweight DICOM viewer application integrating the trained model was developed to showcase real-time AI-assisted cardiothoracic ratio (CTR) estimation. The interface enables users to interactively toggle between the original chest X-ray, manual measurement tools, and AI-generated segmentation overlays. It displays both manual and AI-derived CTR values within the same interface for direct comparison. A short demonstration video of the application was shared with radiologists and imaging professionals to obtain expert feedback on its clarity, usability, and clinical relevance.
3.6. Expert Evaluation of the Demonstration Interface
A feedback survey was completed by nine professionals—comprising radiologists and senior radiographers—following a live demonstration of the AI-powered desktop application (
EphraView). The evaluation focused on clarity, usability, accuracy of AI output, and perceived clinical value.
Table 1 summarises the experts’ responses, which were largely positive across all categories, particularly for the application’s user-friendliness and its potential for integration into clinical or educational settings.
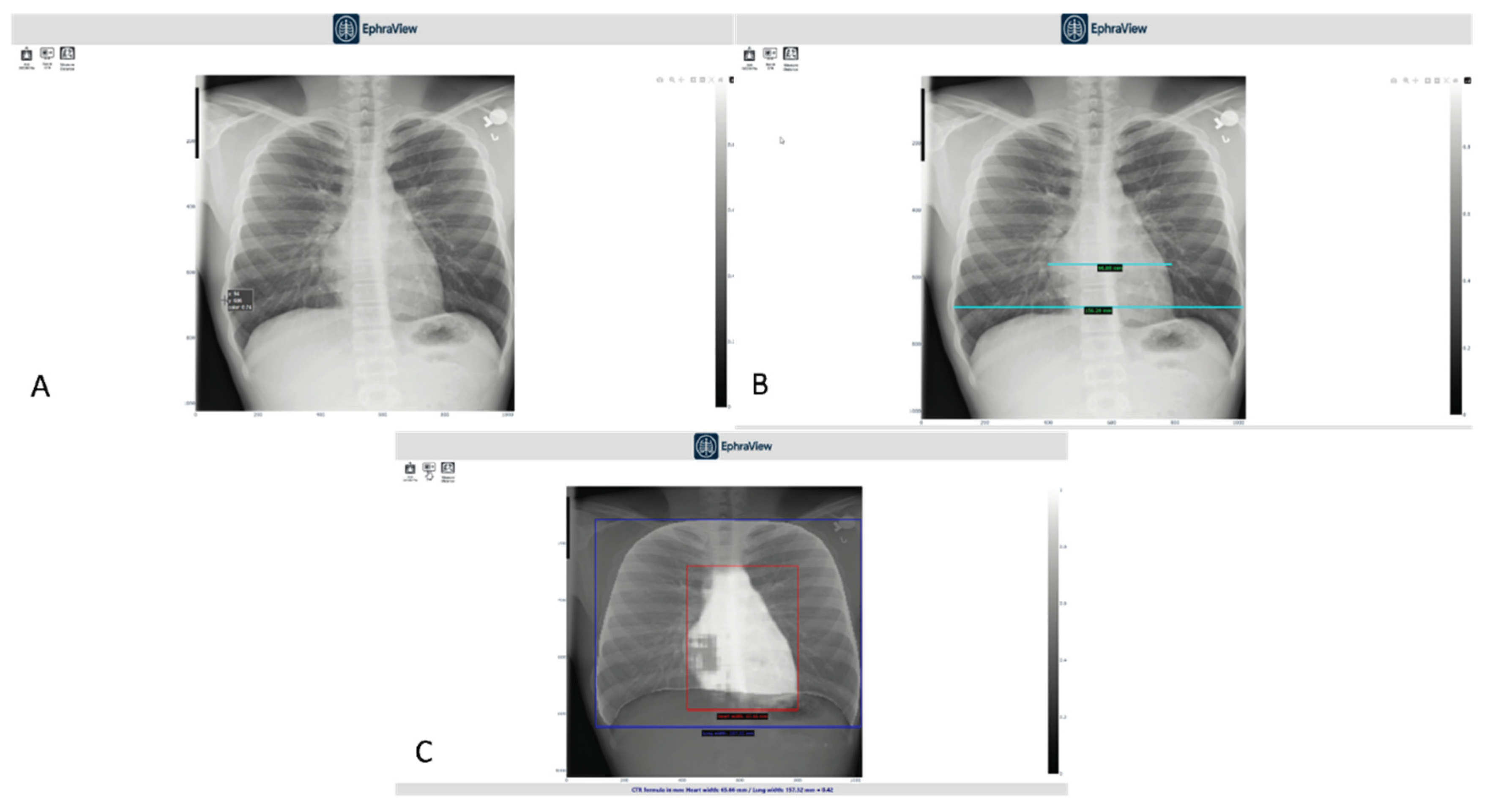
Figure 6 illustrates key aspects of the viewer interface used during the demo.
Panel A shows the manual CTR measurement using traditional PACS tools.
Panel B demonstrates thoracic width measurement with standard digital calipers, used by experts as a reference.
Panel C displays the AI-generated overlay with bounding boxes: red for heart width and blue for thoracic width, alongside an automatically computed CTR.
This real-time visualization enables immediate comparison between manual and AI-generated measurements, reinforcing trust and facilitating potential integration into clinical radiology workflows.
Panels:
A – Manual measurement interface using standard PACS tools for cardiac width estimation.
B – Manual thoracic width measurement using digital calipers, performed by a radiologist for reference CTR computation.
C – AI-generated segmentation overlay with bounding boxes for heart (red) and thorax (blue), including automatically computed CTR displayed at the bottom.
Table 2.
Expert Evaluation of AI Demo (n = 9).
Table 2.
Expert Evaluation of AI Demo (n = 9).
| Evaluation Item |
% Positive (n) |
% Neutral (n) |
Mean Rating (1–5) |
| Demonstration was clear and easy to follow |
89% (8) |
11% (1) |
4.6 |
| Manual measurement reflected good radiographic practice |
78% (7) |
22% (2) |
4.3 |
| Interface was user-friendly and precise |
78% (7) |
22% (2) |
4.2 |
| AI estimation was accurate compared to manual |
67% (6) |
33% (3) |
4.1 |
| AI segmentation of heart and lungs was acceptable |
78% (7) |
22% (2) |
4.3 |
| AI measurement is clinically useful in its current form |
67% (6) |
33% (3) |
4.1 |
| Application is valuable for clinical or educational settings |
89% (8) |
11% (1) |
4.5 |
| Recommendation for further development or pilot testing |
100% (9) |
0% (0) |
4.8 |
This interface enables real-time comparison of manual and AI measurements, facilitating clinical validation and integration into radiology workflows.
Inference: The application was well received, with strong support for its accuracy and clinical relevance. All respondents recommended further development and testing for real-world integration.
4. Discussion
This study developed and evaluated a lightweight deep learning model for automated cardiothoracic ratio (CTR) estimation on chest radiographs using a CPU-based U-Net architecture. The model achieved high segmentation accuracy and showed excellent agreement with manual CTR measurements. Furthermore, the model was successfully deployed in a prototype DICOM viewer application, and expert feedback confirmed its clinical relevance and usability.
The segmentation performance, with Dice coefficients of 0.87 for the heart and 0.93 for the lungs, indicates reliable anatomical delineation. These results are comparable to or better than those reported in similar studies. For instance, Gupte et al. [
9] reported segmentation results with strong CTR accuracy in GPU-based models, and Chamveha et al. [
8] demonstrated acceptable segmentation Dice via U-Net for CTR extraction. Notably, our CPU-based implementation demonstrated comparable performance despite hardware constraints, highlighting its feasibility for low-resource environments like many sub-Saharan African hospitals [
12,
13].
The AI-estimated CTR values closely matched manual measurements (r = 0.91, MAE = 0.025), suggesting that the model can serve as a viable screening tool for cardiomegaly. Bland–Altman analysis confirmed minimal bias, and the classification performance (sensitivity 90.2%, specificity 94.7%) further supports its potential clinical utility. These metrics align with results from an observer-performance study using Attention U-Net by Ajmera et al. [
10], which reported similar sensitivity and specificity ranges.
The use of CTR > 0.50 as a diagnostic threshold is widely accepted in radiology for identifying cardiomegaly. The AI model’s sensitivity and specificity near or above 90% are sufficient for use in triaging or decision support roles, particularly in areas with limited radiology expertise [
3,
6].
One of the novel contributions of this study is the model’s deployment on CPU-only hardware, without reliance on GPUs. Most AI in medical imaging literature assumes access to high-performance computing, limiting applicability in under-resourced settings [
12]. Our success in achieving real-time inference on standard systems offers a paradigm shift for AI accessibility in global health, particularly across Africa where GPU infrastructure is rare [
13,
14].
This practical deployment opens doors for integration into PACS systems, mobile teleradiology solutions, and training centers, addressing the longstanding gap between AI research and clinical implementation in the Global South [
14].
With its low hardware requirement and demonstrated usability, the system could be scaled to regional teleradiology hubs or integrated into radiography training institutions across Africa, enhancing diagnostic reach and educational quality.
The overwhelmingly positive feedback from expert radiologists and senior radiographers validates the clinical usability of the tool. All experts recommended continued development and pilot testing, highlighting its value for routine reporting and training. While some noted that segmentation artifacts occurred in a few suboptimal images, this did not significantly affect measurement accuracy—likely due to the robust preprocessing and model generalisation strategy [
11].
Additionally, the educational value of visual overlays (mask + measurement) was emphasized, supporting its use in academic and training institutions. This is critical in sub-Saharan Africa, where radiography training programs are expanding, but access to digital learning tools is limited (see
Table 1).
While most prior works focused on GPU-based models (e.g., attention U-Net studies by Ajmera et al. [
10], and earlier CNN-based CTR estimations), our CPU-optimised approach shows that lightweight models can still achieve high performance. This study complements previous global initiatives like the NIH ChestX-ray14 dataset and classical deep learning frameworks (e.g., Gupte et al. [
9]; Chamveha et al. [
8]), but goes further by enabling deployment in resource-constrained environments.
Furthermore, our incorporation of a practical DICOM viewer application for side-by-side comparison of manual and AI measurements offers a tangible path toward real-world use, a gap rarely addressed in prior CTR-related research [
14].
4.1. Limitations and Future Work
Despite promising results, some limitations exist. First, the study focused on adult PA chest X-rays, and model generalisation to pediatric, supine, or lateral views remains untested. Second, the number of expert reviewers, while valuable, was small (n=9), limiting generalisability of the feedback.
Future work should include:
Larger, multi-center datasets across varied demographic and technical settings
Expansion to mobile-based deployment
Integration into PACS and teleradiology platforms
Prospective clinical trials to evaluate diagnostic impact
5. Conclusion
This study demonstrates that CPU-based deep learning can be used effectively for cardiothoracic ratio estimation on chest radiographs, offering high accuracy, low computational burden, and practical clinical relevance. It bridges a critical gap between AI research and real-world application in Sub-Saharan Africa. With further development and validation, the system holds strong potential for improving radiographic interpretation, especially in resource-limited settings.
Author Contributions
Conceptualization, E.U.U., S.O.P., and A.O.A.; Methodology, E.U.U., S.O.P., and A.O.A.; Software, E.U.U.; Validation, S.A.E., F.I.O., and N.O.E.; Investigation, E.V.U., F.I.O., and A.E.E.; Data Curation, E.V.U., B.E.A., and A.E.E.; Writing—Original Draft Preparation, E.U.U.; Writing—Review and Editing, A.O.A., S.A.E., F.I.O., N.O.E., B.E.A., and G.K.; Visualization, E.U.U. and G.K.; Supervision, S.A.E., A.O.A., and G.K.; Project Administration, E.U.U. and S.O.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Ethical approval for this study was obtained from the Health Research Ethics Committee of the University of Calabar Teaching Hospital and the Doren Specialist Hospital Health Research Ethics Committee. Approval letters are available upon reasonable request.
Informed Consent Statement
Patient consent was waived due to retrospective anonymized data.
Data Availability Statement
Data supporting the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
AI Tools Declaration: Artificial intelligence tools (e.g., ChatGPT by OpenAI) were used solely to assist with language editing, formatting, and improving the clarity of the manuscript. No generative AI was used for data analysis, model development, or content creation.
Abbreviations
The following abbreviations are used in this manuscript:
| AI |
Artificial Intelligence |
| AUC |
Area Under the Curve |
| CNN |
Convolutional Neural Network |
| CTR |
Cardiothoracic Ratio |
| DICOM |
Digital Imaging and Communications in Medicine |
| DL |
Deep Learning |
| IRB |
Institutional Review Board |
| PA |
Posteroanterior |
| ROC |
Receiver Operating Characteristic |
| U-Net |
U-shaped Convolutional Network |
| X-ray |
X-radiation |
References
- Truszkiewicz, K.; Poręba, R.; Gać, P. Radiological cardiothoracic ratio in evidence based medicine. J. Clin. Med. 2021, 10, 2016. [CrossRef]
- Grotenhuis, H.B.; Zhou, C.; Tomlinson, G.; Isaac, K.V.; Seed, M.; Grosse-Wortmann, L.; Yoo, S.J. Cardiothoracic ratio on chest radiograph in pediatric heart disease: correlation with heart volumes at magnetic resonance imaging. Pediatr. Radiol. 2015, 45, 1616–1623. [CrossRef]
- Sahin, H.; Chowdhry, D.N.; Olsen, A.; Nemer, O.; Wahl, L. Is there any diagnostic value of anteroposterior chest radiography in predicting cardiac chamber enlargement? Int. J. Cardiovasc. Imaging 2019, 35, 195–206. [CrossRef]
- Anjuna, R.; Paulius, S.; Manuel, G.G.; Audra, B.; Jurate, N.; Monika, R. Diagnostic value of cardiothoracic ratio in patients with non-ischemic cardiomyopathy: comparison to cardiovascular magnetic resonance imaging. Curr. Probl. Diagn. Radiol. 2024, 53, 353–358. [CrossRef]
- Chamveha, I.; Promwiset, T.; Tongdee, T.; Saiviroonporn, P.; Chaisangmongkon, W. Automated cardiothoracic ratio calculation and cardiomegaly detection using deep learning approach. arXiv 2020, arXiv:2002.07468. [CrossRef]
- Gupte, T.; Niljikar, M.; Gawali, M.; Kulkarni, V.; Kharat, A.; Pant, A. Deep learning models for calculation of cardiothoracic ratio from chest radiographs for assisted diagnosis of cardiomegaly. arXiv 2021, arXiv:2101.07606. [CrossRef]
- Ajmera, P.; Kharat, A.; Gupte, T.; Pant, R.; Kulkarni, V.; Duddalwar, V.; Lamghare, P. Observer performance evaluation of the feasibility of a deep learning model to detect cardiomegaly on chest radiographs. Acta Radiol. Open 2022, 11, 20584601221107345. [CrossRef]
- Chou, H.-H.; Lin, J.-Y.; Shen, G.-T.; Huang, C.-Y. Validation of an Automated Cardiothoracic Ratio Calculation for Hemodialysis Patients. Diagnostics 2023, 13, 1376. [CrossRef]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. ChestX-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. Proc. IEEE Conf. Comput. Vis. Pattern Recognit. 2017, 2017, 3462–3471. [CrossRef]
- Ajmera, P.; Kharat, A.; Gupte, T.; Pant, R.; Kulkarni, V.; Duddalwar, V.; Lamghare, P. Observer performance evaluation of the feasibility of a deep learning model to detect cardiomegaly on chest radiographs. Acta Radiol. Open 2022, 11, 20584601221107345. [CrossRef]
- Tang, Y.-X.; Tang, Y.-B.; Peng, Y.; Yan, K.; Harrison, A.P.; Bagheri, M.; Summers, R.M. Automated abnormality classification of chest radiographs using deep convolutional neural networks. NPJ Digit. Med. 2020, 3, 70. [CrossRef]
- Arora, R.; Banerjee, I.; Soni, A.; Sivasankaran, S.; Aggarwal, S.; Mahajan, V.; Arora, A. Developing AI for low-resource settings: a case study on a real-time offline model for tuberculosis detection. BMJ Health Care Inform. 2022, 29, e100496. [CrossRef]
- Ayeni, F.A.; Adedeji, A.T.; Agwuna, E.S.; Omoregbe, N.; Ojo, O.; Fakunle, O.O. Assessing the readiness of low-resource hospitals for artificial intelligence integration in Sub-Saharan Africa. Health Technol. 2023, 13, 241–251. [CrossRef]
- Kazeminia, S.; Khaki, M.; Asadi, A.; Jolfaei, A.; Ghassemi, N.; Parsaei, H.; Rajaraman, S. Deep learning for medical image analysis in low-resource settings: a review. Comput. Biol. Med. 2023, 158, 106874. [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention (MICCAI); Springer: Cham, Switzerland, 2015; Vol. 9351, pp. 234–241. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).