1. Introduction
Bacteremia represents a life-threatening medical illness with substantial global health implications, contributing significantly to sepsis-related mortality worldwide. Recent epidemiological analyses from the Global Burden of Disease Study revealed that sepsis accounts for an estimated 48.9 million incident cases annually and results in 11.0 million deaths globally, representing 19.7% of all global deaths [
1]. The clinical outcomes vary significantly by pathogen, with
Staphylococcus aureus bacteremia demonstrating 30-day case-fatality rates of 21% and 90-day mortality ranging from 23% to 39% across different healthcare settings [
2]. Similarly,
Escherichia coli bloodstream infections, despite being among the most common pathogens with incidence rates of 50–60 cases per 100,000 population, carry 30-day case-fatality rates of approximately 10%–15% [
2]. These epidemiological data points underscore the critical need for optimized therapeutic interventions in managing bacteremia.
Providing rapid and appropriate antibiotic treatment is crucial for effectively managing bacteremia and has a significant impact on patient outcomes, regardless of antimicrobial resistance. Extensive healthcare database studies have shown that delays in administering suitable antibiotics increase in-hospital mortality or hospital discharge by 20%, prolong hospital stays by 70%, and raise total inpatient costs by 65%, affecting both resistant and susceptible bacteria [
3]. Patients with multidrug-resistant bloodstream infections face additional challenges; for example, extended-spectrum β-lactamase-producing Enterobacterales have a higher mortality rate (adjusted hazard ratio, 1.63; 95% CI, 1.13–2.35). Third-generation cephalosporin-resistant
E. coli infections led to approximately 8,750 deaths in Europe by 2015 [
2]. Evidence from meta-analyses emphasizes the importance of prompt, appropriate treatment, with molecular rapid diagnostic testing significantly reducing mortality risk (OR 0.66, 95% CI 0.54–0.80) and decreasing the time to effective therapy by approximately 5 hours (95% CI -8.60 to -1.45 hours) [
4].
Integrating artificial intelligence (AI) and machine learning (ML) into clinical decision support systems (CDSS) marks a transformative shift in the management of infectious diseases, significantly improving diagnostic accuracy and treatment precision. These systems have shown notable clinical benefits, including sepsis prediction algorithms that reduce mortality by 30-60% in quasi-experimental studies and 58% in randomized controlled trials. They also enable blood culture collection and antibiotic administration roughly 2.8 hours earlier than traditional methods [
5]. Thanks to AI’s ability to analyze complex clinical data and provide real-time, patient-specific guidance, ML-based CDSS tools are valuable for antimicrobial stewardship, especially in resource-limited settings where infectious disease specialists may be scarce [
5,
6].
Arkstone’s machine learning-based clinical decision support system has shown excellent performance across various internal validation tests. Using methods such as k-fold cross-validation, random subsampling, and holdout validation, the system achieved 100% accuracy in differentiating between trained and untrained single data points in 1,110 tests involving 111 bacterial species and resistance genes [
7]. Further evaluation with 1,401 real lab results from 66 labs in 55 regions confirmed perfect precision and recall (1.0 for both), with no false positives or negatives [
7]. Human-in-the-loop validation also found a 0% significant discrepancy rate compared to clinical guidelines, with only 15.53% minor discrepancies mainly related to antibiotic choices or dosing, confirming the system’s consistency with established infectious disease standards [
7].
Further analysis comparing AI-driven therapeutic suggestions for bacteremia treatment has confirmed the usefulness of molecular-based decision-support systems. The assessment of Arkstone’s OneChoice platform revealed strong agreement between recommendations based solely on molecular data and those incorporating phenotypic susceptibility results (Cohen’s Kappa, 0.80), with guidance provided approximately 29 hours earlier (median, 16.81 versus 46.32 hours) [
8]. For
Escherichia coli bacteremia, the most common pathogen accounting for 41% of cases, the agreement on recommendations was 95%, highlighting the accuracy of AI-assisted antimicrobial decisions [
8].
Despite advances and proven analytical accuracy, the practical utility of these decision support tools in real-world clinical settings remains unclear. This study aimed to explore how specialist physicians perceive the clinical utility of an AI-powered CDSS (OneChoice and OneChoice Fusion). The study focuses on physicians interpretation of the CDSS recommendation, how it affects selection of therapy and whether the CDSS recommendation is consistent with their own clinical judgement in routine practice.
2. Materials and Methods
Study Design and Setting
This study employed a cross-sectional survey design to evaluate the perceived clinical utility of AI-powered CDSS among specialist physicians managing cases of bacteremia. The investigation was conducted at Roe Laboratory, Lima, Peru, and involved healthcare professionals from multiple medical specialties, some with expertise in infectious disease management and others without. The study protocol was designed to assess real-world clinical recommendations by CDSS and determine if physicians presented with the same clinical vignette would recommend the same treatment.
Study Population and Participants
A total of 62 specialist physicians were enrolled in this study using purposive sampling and participated in 90 surveys. Physicians were selected based on their active clinical practice in managing patients with positive blood culture results and their specialization in relevant medical disciplines. All participants possessed clinical experience in interpreting blood culture results and making antimicrobial therapy treatment decisions.
Inclusion criteria encompassed: (1) board-certified specialists in relevant medical disciplines; (2) active clinical practice involving bacteremia management; (3) willingness to participate in the survey evaluation; and (4) familiarity with blood culture interpretation and antimicrobial prescribing practices.
Exclusion criteria included: (1) physicians without active clinical practice; (2) incomplete survey responses; and (3) specialists without experience in antimicrobial therapy decision-making.
Clinical Decision Support Systems Evaluated
Two AI-powered CDSS developed by Arkstone Medical Solutions were evaluated in this study:
OneChoice System: An AI-driven platform that generates therapeutic recommendations based exclusively on molecular diagnostic data along with patient-specific information obtained from blood culture identification panels. The system utilizes ML algorithms trained on extensive real-life clinical databases to provide real-time, pathogen-specific antimicrobial guidance (Supplement 1).
OneChoice Fusion System: An enhanced version of the AI-CDSS that integrates both molecular diagnostic results and conventional phenotypic susceptibility testing data, along with patient-specific information to generate refined therapeutic recommendations. This system combines rapid molecular identification with traditional antimicrobial susceptibility testing profiles to optimize treatment suggestions (Supplement 2).
Data Collection and Survey Methodology
Data collection was conducted using a structured survey instrument administered to participating physicians in accordance with a standardized protocol. Each participant was presented with authentic positive blood culture cases and corresponding AI-CDSS recommendations in a sequential manner:
Phase 1: Participants received initial molecular blood culture identification results accompanied by OneChoice system recommendations for antimicrobial therapy.
Phase 2: Subsequently, conventional susceptibility testing results were provided alongside OneChoice Fusion system recommendations.
The survey instrument evaluated multiple dimensions of clinical utility, including: (1) perceived helpfulness of AI-generated information; (2) impact on therapeutic decision-making processes; (3) concordance between AI recommendations and physician clinical judgment; and (4) implementation of therapy changes based on AI guidance.
Microbiological Characteristics and Pathogen Distribution
The study encompassed a diverse spectrum of bacterial pathogens commonly encountered in cases of clinical bacteremia. The pathogen distribution included Escherichia coli (38.7% of isolates), representing the most frequently identified organism, followed by Pseudomonas aeruginosa (13.3%) and Salmonella typhi (4%). Notably, 30% of bacterial isolates were confirmed as producers of CTX-M extended-spectrum beta-lactamase, reflecting the contemporary antimicrobial resistance patterns encountered in clinical practice.
Molecular Testing: Positive blood culture samples underwent rapid molecular analysis using the FilmArray Blood Culture Identification (BCID) Panel (BioFire Diagnostics, LLC, Salt Lake City, UT, USA) or Xpert® MRSA/SA Blood Culture (Cepheid LLC, Sunnyvale, CA, USA), based on Gram stain results. The assays were conducted according to the manufacturer’s instructions, with specific attention to reagent preparation, sample volume (200 µL), and assay run conditions (temperature and duration).
Phenotypic Testing: Organisms from positive blood cultures were isolated on agar media, including Blood Agar, Chocolate Agar, MacConkey Agar, and Sabouraud Agar. Microbial identification was performed using the MALDI-TOF mass spectrometry system, which was calibrated daily to ensure accuracy. Antimicrobial susceptibility testing (AST) was conducted using the VITEK 2.0 automated system.
Data Analysis
Survey responses were analyzed using descriptive statistics to characterize participant demographics, clinical specialties, and response patterns. Categorical variables were expressed as frequencies and percentages. The utility and impact assessments were quantified through response rate calculations and preference evaluations.
Statistical analyses were performed to determine the proportion of respondents who found AI systems helpful, the percentage reporting that AI facilitated decision-making, the concordance rate between AI recommendations and physician choices, and the frequency of therapy modifications guided by AI.
Ethical Considerations
The Faculty of Health Sciences Ethics Committee at the Universidad Privada de Tacna approved the study protocol. This study was conducted in accordance with the principles outlined in the Declaration of Helsinki for ethical medical research. All survey responses were anonymized to protect participant identity, and participation was entirely voluntary with implied consent obtained through survey completion.
Data and Materials Availability
All survey instruments, data collection protocols, and analytical methodologies employed in this study are available upon reasonable request to facilitate replication and further research. The AI-CDSS evaluated (OneChoice and OneChoice Fusion) are proprietary systems developed by Arkstone Medical Solutions, with technical specifications and algorithmic details subject to intellectual property considerations. Aggregated survey data supporting the study conclusions will be made available through appropriate data-sharing mechanisms while maintaining participant confidentiality.
3. Results
3.1. Participant Demographics and Clinical Characteristics
3.1.1. Specialist Physician Distribution
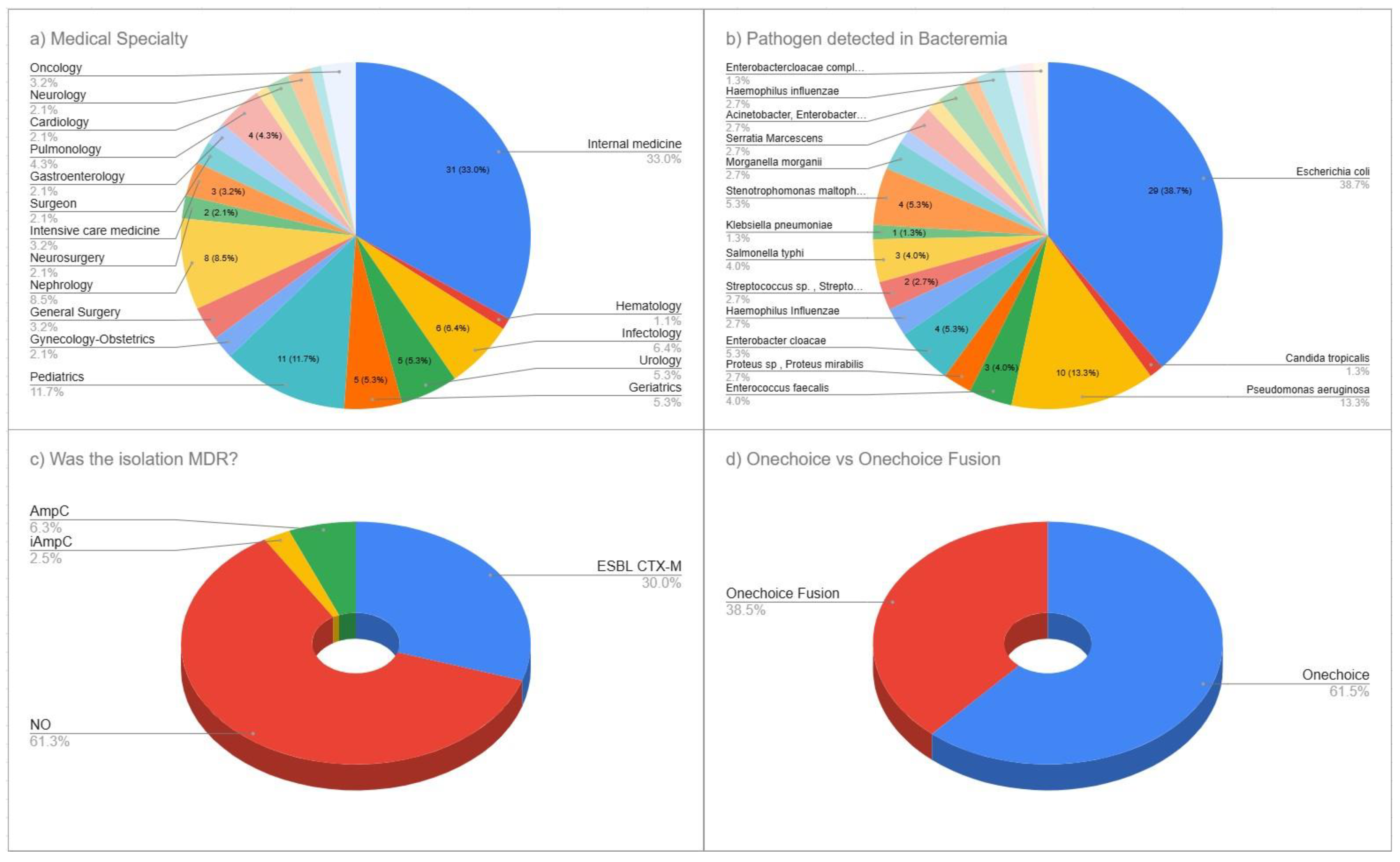
A total of 65 specialist physicians participated in this cross-sectional survey evaluating the AI-powered CDSS, with 90 surveys. Medicine specialists comprised the largest group, at 33.0% (n=31), followed by pediatric specialists at 11.7% (n=11), nephrology specialists at 8.5% (n=8), and infectious disease specialists at 6.4% (n=6). Urology and geriatrics specialists each represented 5.3% (n = 5). Pulmonology specialists made up 4.3% (n=4). Both gastroenterology and intensive care medicine specialists constituted 3.2% (n = 3) each. Additionally, 19.1% (n = 14) of participants came from each of several other specialties, including hematology, neurosurgery, surgical medicine, general surgery, gynecology-obstetrics, cardiology, neurology, oncology, and family and community medicine. This varied specialty participation provided a broad range of clinical expertise relevant to the management of bacteremia (
Figure 1a).
3.1.2. Bacteremia Pathogen Characteristics
The microbiological analysis of positive blood cultures revealed a diverse range of pathogens typical of modern clinical bacteremia.
Escherichia coli was the most common, making up 38.7% (n=29) of all isolates, followed by
Pseudomonas aeruginosa at 13.3% (n=10).
Enterobacter cloacae accounted for 5.3% (n=4), while
Enterococcus faecalis and
Salmonella typhi each constituted 4.0% (n=3). Multiple organisms were found, each at 2.7% (n=2), including Proteus species/
Proteus mirabilis,
Haemophilus influenzae, Streptococcus species
/Streptococcus gallolyticus, and
Morganella morganii. Less common pathogens included
Klebsiella pneumoniae and
Candida tropicalis, each accounting for 1.3% (n = 1). Additional minor pathogens included
Serratia marcescens,
Bacteroides fragilis, Staphylococcus species, the Acinetobacter/
Enterobacter cloacae complex,
Citrobacter freundii, and organisms within the
Enterobacteriaceae complex, reflecting the complexity of microbiological profiles observed in current bacteremia cases (
Figure 1b).
3.1.3. Antimicrobial Resistance Patterns
Analysis of antimicrobial resistance showed a notable prevalence of resistance within the study group. Most isolates (61.3%, n = 55) did not display multidrug resistance however the production of CTX-M extended-spectrum beta-lactamase (ESBL) was seen at 30.0% (n = 27) . AmpC beta-lactamase was found in 6.3% (n = 6) of isolates, and inducible AmpC resistance mechanisms were present in 2.5% (n = 2). This pattern highlights the clinical complexity of managing bacteremia today and suggests that AI-guided treatment recommendations could improve antimicrobial choice for both susceptible and resistant pathogens (
Figure 1c).
3.2. Clinical Utility Assessment of the OneChoice System
3.2.1. Perceived Helpfulness and Decision-Making Impact
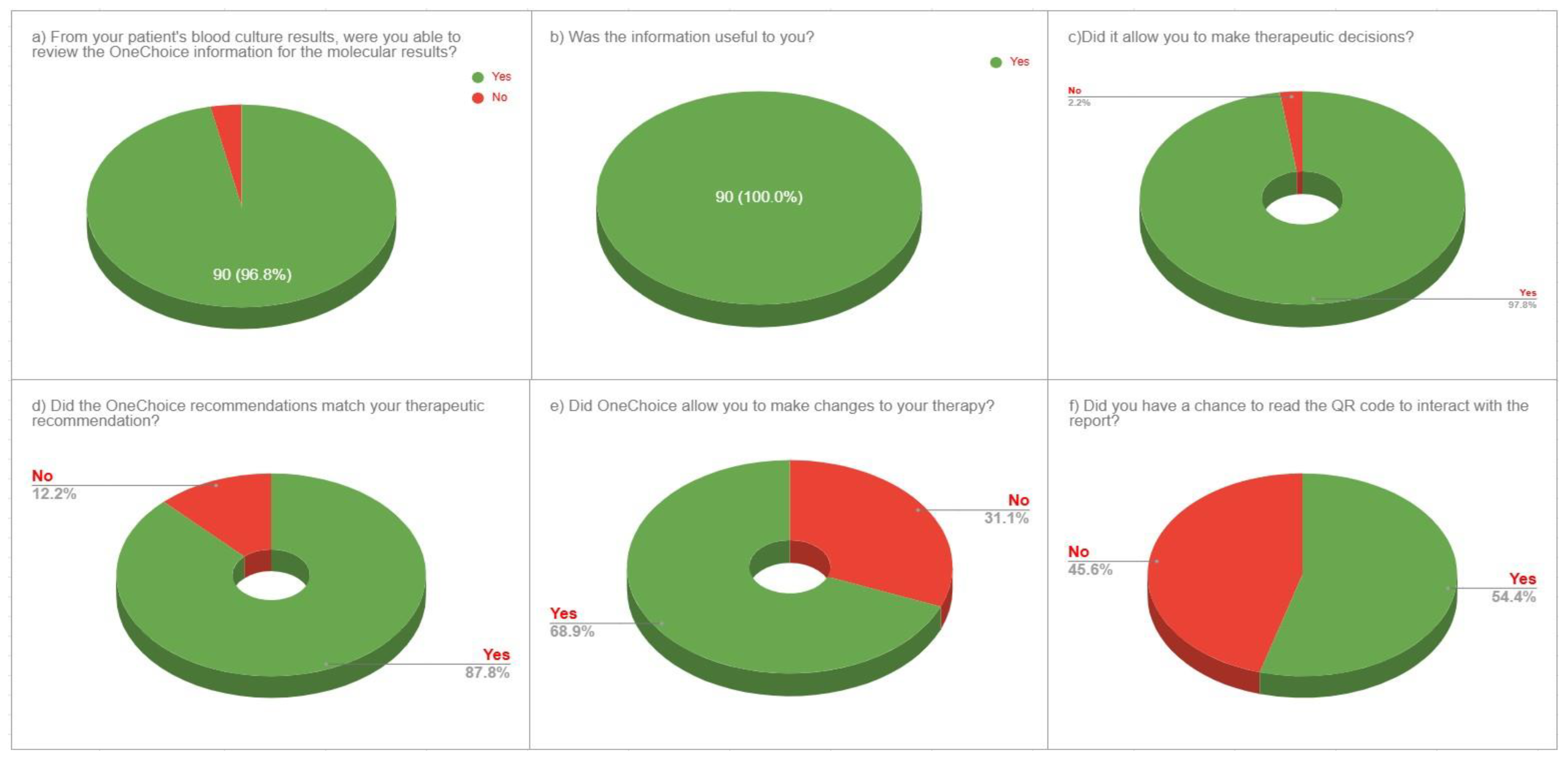
The evaluation of the OneChoice system’s utility showed strong acceptance among physicians. All respondents (100%, n = 90) found the information helpful for clinical decisions. Its influence on therapeutic choices was also notable, with 97.8% (n = 88) of participants stating that it aided their decision-making. Most physicians, 96.7% (n = 87), rated the AI-generated guidance as adequate for a thorough clinical assessment. These results highlight high user satisfaction and perceived clinical value across the entire group of doctors (
Figure 2a-c).
3.2.2. Clinical Concordance and Implementation Patterns
Assessment of clinical concordance between AI recommendations and physician therapeutic preferences showed strong alignment in decision-making. The concordance rate between OneChoice recommendations and physicians’ choices was 87.8% (n = 79), indicating a high level of agreement between AI guidance and clinical judgment. Analysis of implementation revealed a meaningful clinical effect, with 68.9% (n = 62) of cases resulting in treatment changes, according to OneChoice data. Additionally, physician confidence in AI recommendations was evident, with 85.6% (n = 77) of participants willing to follow OneChoice’s advice in routine practice. This demonstrates high trust in the system’s suggestions and a readiness to incorporate AI guidance into standard patient care (
Figure 2d-e).
3.3. Comparative Analysis: OneChoice versus OneChoice Fusion
3.3.1. Digital Engagement and Interactive Features
Physicians adopted the interactive features of AI-CDSS platforms to a moderate degree. Specifically, 54.4% (n = 49) of participants actively used the QR code interaction, which enhanced their engagement with AI-generated reports and recommendations. The other 45.6% (n = 41) relied on core system functions without interactive features, indicating that while digital enhancements can improve the user experience for some clinicians, the essential AI-CDSS functions remain valuable even without interactive options. This suggests the system design effectively supports different levels of technological engagement among healthcare professionals (
Figure 2f).
4. Discussion
The results of this cross-sectional survey provide compelling evidence for the practical application and acceptance of AI-driven CDSS in the management of bacteremia. Results demonstrated near-universal acceptance among physicians and high concordance with their own clinical judgment. The complete recognition (100%) of OneChoice’s clinical value by physicians surpasses the often mixed reception of other healthcare AI applications. Recent systematic reviews of AI-based decision-making systems in healthcare have revealed heterogeneous outcomes across various medical fields, with consistent benefits observed in areas such as depression treatment and pain management. Effects in other clinical sectors, however, remain diverse [
9]. The finding that 97.8% of physicians found therapeutic decision-making easier is consistent with broader evidence on the use of machine learning in infectious diseases, where approximately 40% of systems are developed for intensive care settings and 25% for infectious disease consultations. These systems demonstrate tangible clinical benefits, including sepsis prediction algorithms that have reduced mortality rates by 30-60% in quasi-experimental studies and 58% in randomized controlled trials [
5]. The high congruence rate (87.8%) between OneChoice’s recommendations and physicians therapeutic decisions indicates a strong correlation between AI-generated recommendation and physicians own clinical judgment. In a previous study, we demonstrated the high correlation (80%) between recommendations that relied solely on molecular lab result data and recommendations that incorporated both phenotypic data and molecular data. Therefore, it is easy to postulate that this tool can be used throughout a patient’s clinical course.
The impressive 68.9% implementation rate of treatment changes based on OneChoice’s recommendations underscores a significant clinical impact that extends beyond simple acceptance, demonstrating that AI-driven guidance leads to tangible therapeutic adjustments. This rate is critical when considered in the context of the global challenges that antimicrobial stewardship programs face. In Latin America, data indicates that merely 46% of hospitals in Central and South America have adopted antimicrobial stewardship programs, with major obstacles including a shortage of dedicated pharmacists (63%), the lack of treatment guidelines tailored to local epidemiological data (33%), and insufficient microbiology lab capabilities [
12]. The AI-driven CDSS assessed in our study directly tackles these resource challenges by offering standardized, evidence-based recommendations that can be customized to fit local epidemiological patterns and resistance profiles. This is particularly important given that 30-50% of the population in many Latin American nations depends on underfunded public healthcare systems [
12]. The proven effectiveness of this system is especially pertinent when juxtaposed with findings that show infectious disease consultations, although linked to decreased mortality in gram-negative bloodstream infections (adjusted hazard ratio 0.82, 95% CI 0.77–0.88), display significant variability across hospitals (2.7–76.1%) due to constraints in resources and specialist availability [
10].
These findings hold significant clinical relevance, as highlighted by the current international guidelines from the Surviving Sepsis Campaign. These guidelines clearly acknowledge that machine learning outperforms traditional screening methods like SIRS (AUROC 0.70), MEWS (AUROC 0.50), and SOFA (AUROC 0.78) in identifying sepsis, [
11]. Moreover, the guidelines emphasize the urgent need for prompt antimicrobial treatment, with observational studies indicating that each hour of delay is associated with a 1.04-fold increase in the odds of in-hospital mortality [
11]. The fusion of AI-driven clinical decision support systems with swift molecular diagnostics, as exemplified by the OneChoice platform, which offers therapeutic guidance 29 hours sooner than traditional phenotypic methods [
8], suggests a revolutionary leap forward in managing bacteremia. A 2017 systemic review found that rapid molecular diagnostic testing significantly reduces mortality risk and shortens time to effective therapy by about 5.03 hours [
4]. Conversely, delaying appropriate antibiotic treatment results in a roughly 20% increase in in-hospital mortality, a 70% increase in hospital length of stay, and a 65% increase in total inpatient costs, irrespective of antimicrobial resistance status [
3].
Our research aligns with the growing body of evidence on AI technologies specifically designed to address antimicrobial resistance. Recent studies indicate that AI-powered diagnostic tools can process vast amounts of data with greater precision than humans, enabling quicker, more precise diagnoses through the use of Convolutional Neural Networks (CNNs) for microscopic image analysis and machine learning algorithms for genomic data analysis [
13,
14]. Current machine learning applications in antimicrobial resistance encounter significant hurdles, particularly the scarcity of high-quality, well-annotated, and standardized data, which is critical for training accurate and dependable AI models [
13,
14]. In interpreting our findings, it is important to note several limitations, including the cross-sectional survey design, which offers insights into physicians’ perceptions and self-reported behaviors but does not directly assess patient outcomes or clinical effectiveness in practice. Although the study population is diverse in terms of medical specialties, the research was conducted at a single institution, potentially limiting its applicability to other healthcare environments with varying resources, patient demographics, or resistance patterns.
Furthermore, systematic reviews of AI healthcare applications have shown that only 42% of studies reported adverse events, and none reported an increase in adverse events due to AI interventions, underscoring the need for more thorough safety monitoring in future implementations [
9]. The acceptance and clinical utility of AI-powered CDSS for managing bacteremia, as shown in this study, lay the groundwork for further research and implementation. Future research should focus on prospective clinical trials evaluating patient outcomes, such as mortality rates, hospital stay durations, and antimicrobial stewardship metrics, to complement the encouraging physician acceptance data presented here. Future investigations should focus on tackling the significant challenges in AI applications for antimicrobial resistance, particularly issues related to data quality and standardization that hinder the creation of accurate and reliable AI models [
14].
5. Conclusions
This cross-sectional survey demonstrates exceptional clinical utility and physician acceptance of AI-powered CDSS in bacteremia management, with unanimous recognition (100%) of clinical value and substantial concordance (87.8%) between AI recommendations and physician therapeutic choices. The meaningful implementation of treatment modifications in 68.9% of cases indicates that these systems effectively bridge critical gaps in infectious disease expertise and antimicrobial stewardship resources, particularly relevant given the 30% prevalence of extended-spectrum β-lactamase-producing organisms in our study population. These findings, combined with evidence that AI-powered systems can provide therapeutic guidance approximately 29 hours earlier than conventional approaches while maintaining clinical appropriateness, position machine learning-based clinical decision support as a transformative tool for improving outcomes in bacteremia management.
Author Contributions
Conceptualization, J.C.G.d.l.T. and F.A.; methodology, J.C.G.d.l.T., YH; M.H.-Z.; software, J.C.G.d.l.T., YH .; validation, J.C.G.d.l.T., A.R., C.Ch.L., G.G, E.K and YH.; formal analysis, J.C.G.d.l.T., J.J.L and M.H.-Z.; investigation, J.C.G.d.l.T., J.C, F.A., YH, I.E, E.K, I.S and A.R.; resources, J.C.G.d.l.T., YH, C.Ch.L. JC, RB, IE, I.S, EK, G.G, and M.H.Z.; data curation, J.C.G.d.l.T., J.C, R.B, I.E, G.G, I.S, E.K, S.V.O and C.Ch.L.; writing—original draft preparation, J.C.G.d.l.T., G.G, I.S and C.Ch.L. writing—review and editing, J.C.G.d.l.T., F.A., J.C, R.B, A.R., I.E, E.K, S.V.O, J.S, S.A and M.H.-Z.; visualization, J.C.G.d.l.T., and M.H.-Z.; supervision, F.A. and A.R.; project administration, J.C.G.d.l.T. and F.A.; funding acquisition, F.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and was approved by the Institutional Review Board of the Universidad Privada de Tacna.
Informed Consent Statement
All the participants accept a CI in each survey.
Data Availability Statement
The data analyzed in this manuscript, as well as its definitions, can be requested at any time.
Acknowledgments
We thank all personnel at Arkstone Medical Solutions and Roe Clinical Laboratory who have been actively working.
Conflicts of Interest
Ari Frenkel is Chief Science Officer of Arkstone Medical Solutions, the company that produces the OneChoice report evaluated in this study. JC Gómez de la Torre works as the Director of Molecular Informatics at Arkstone Medical Solutions and as the Medical Director at Roe Lab in Perú. Yoshie Huguchi works at Roe Laboratory. At the same time, Alicia Rendon, Carlos Chavez L., and Miguel Hueda Zavaleta serve as Quality Assurance Managers at Arkstone Medical Solutions. These affiliations may be perceived as potential conflicts of interest. However, the study’s design, data collection, analysis, interpretation, manuscript preparation, and the decision to publish the results were conducted independently, with no undue influence from the authors’ affiliations or roles within the company.
References
- Rudd KE, Johnson SC, Agesa KM, Shackelford KA, Tsoi D, Kievlan DR, et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: analysis for the Global Burden of Disease Study. Lancet. 2020;395(10219):200-211.
- Kern WV, Rieg S. Burden of bacterial bloodstream infection: a brief update on epidemiology and significance of multidrug-resistant pathogens. Clin Microbiol Infect. 2020;26(2):151-157.
- Bonine NG, Berger A, Altincatal A, Wang R, Bhagnani T, Gillard P, Lodise T. Impact of Delayed Appropriate Antibiotic Therapy on Patient Outcomes by Antibiotic Resistance Status From Serious Gram-negative Bacterial Infections. Am J Med Sci. 2019;357(2):103-110.
- Timbrook TT, Morton JB, McConeghy KW, Caffrey AR, Mylonakis E, LaPlante KL. The Effect of Molecular Rapid Diagnostic Testing on Clinical Outcomes in Bloodstream Infections: A Systematic Review and Meta-analysis. Clin Infect Dis. 2017;64(1):15-23.
- Peiffer-Smadja N, Rawson TM, Ahmad R, Buchard A, Georgiou P, Lescure FX, Birgand G, Holmes AH. Machine learning for clinical decision support in infectious diseases: a narrative review of current applications. Clin Microbiol Infect. 2020;26(5):584-595.
- Al Kuwaiti A, Nazer K, Al-Reedy A, Al-Shehri S, Al-Muhanna A, Subbarayalu AV, Al Muhanna D, Al-Muhanna FA. A Review of the Role of Artificial Intelligence in Healthcare. J Pers Med. 2023;13(6):951.
- Frenkel A, Rendon A, Chavez-Lencinas C, Gomez De la Torre JC, MacDermott J, Gross C, et al. Internal Validation of a Machine Learning-Based CDSS for Antimicrobial Stewardship. Life. 2025;15(1):1123.
- Gomez de la Torre JC, Frenkel A, Chavez-Lencinas C, Rendon A, Cáceres JA, Alvarado L, Hueda-Zavaleta M. AI-Based Treatment Recommendations Enhance Speed and Accuracy in Bacteremia Management: A Comparative Study of Molecular and Phenotypic Data. Life. 2025;15(8):864.
- Wilhelm C, Steckelberg A, Rebitschek FG. Benefits and harms associated with the use of AI-related algorithmic decision-making systems by healthcare professionals: a systematic review. BMJ Open. 2023;13(7):e069395.
- Ong SWX, Luo J, Fridman DJ, Lee SM, Johnstone J, Schwartz KL, et al. Association between infectious diseases consultation and mortality in hospitalized patients with Gram-negative bloodstream infection: a retrospective population-wide cohort study. Clin Microbiol Infect. 2024;30(6):789-796.
- Evans L, Rhodes A, Alhazzani W, Antonelli M, Coopersmith CM, French C, et al. Surviving Sepsis Campaign International Guidelines for the Management of Sepsis and Septic Shock 2021. Intensive Care Med. 2021;47(11):1181-1247.
- Fabre V, Cosgrove SE, Secaira C, Tapia Torrez JC, Lessa FC, Patel TS, Quiros R. Antimicrobial stewardship in Latin America: Past, present, and future. Infect Control Hosp Epidemiol. 2022;43(1):78-84.
- Anahtar MN, Yang JH, Kanjilal S. Applications of Machine Learning to the Problem of Antimicrobial Resistance: an Emerging Model for Translational Research. J Clin Microbiol. 2021;59(7):e0126020.
- Branda F, Scarpa F. Implications of Artificial Intelligence in Addressing Antimicrobial Resistance: Innovations, Global Challenges, and Healthcare’s Future. Antibiotics. 2024;13(6):502.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).