Submitted:
28 October 2025
Posted:
29 October 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
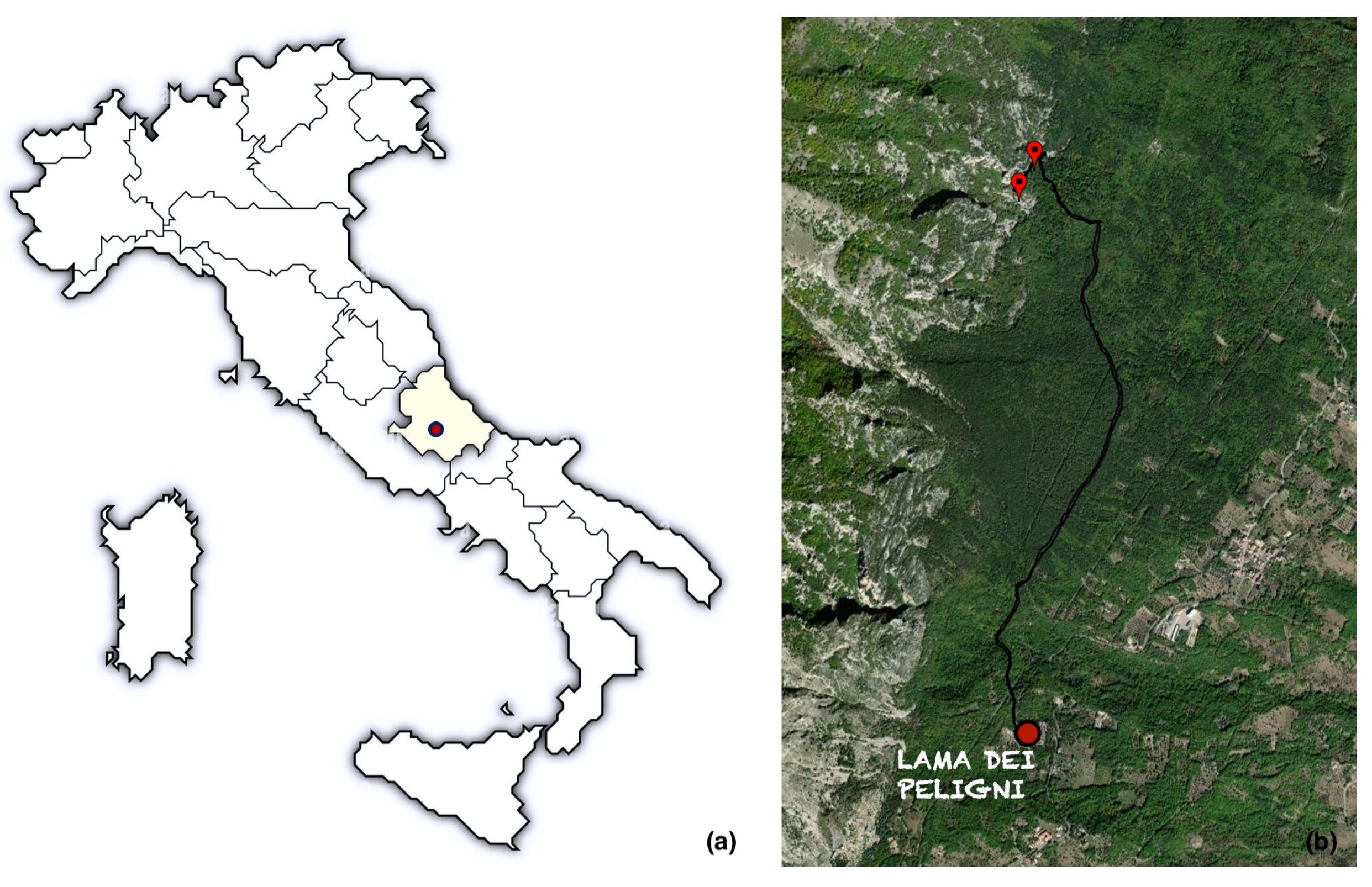
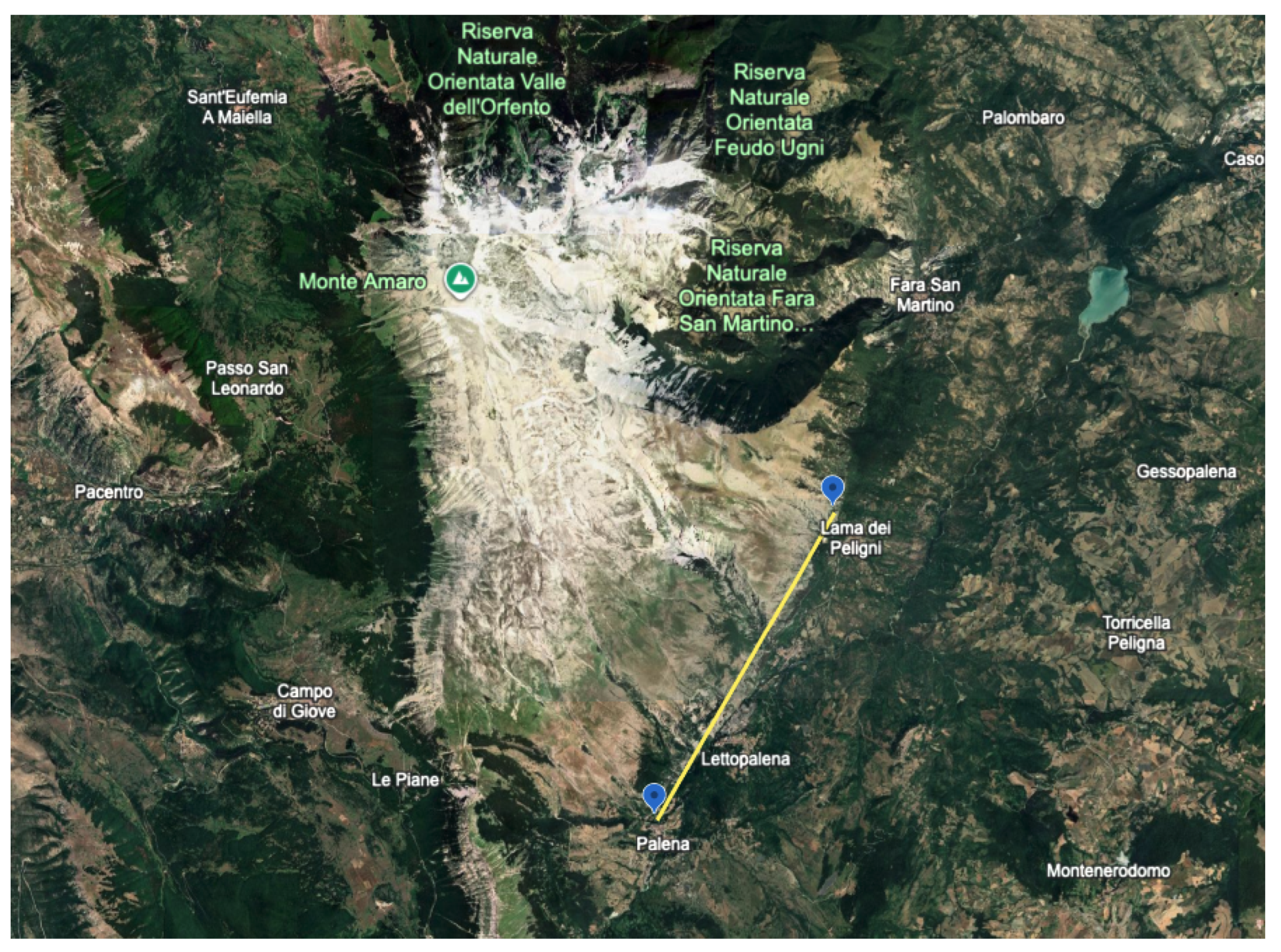
2.1. Research Design
2.2. Atmospheric Analysis
2.3. Laboratory Tests
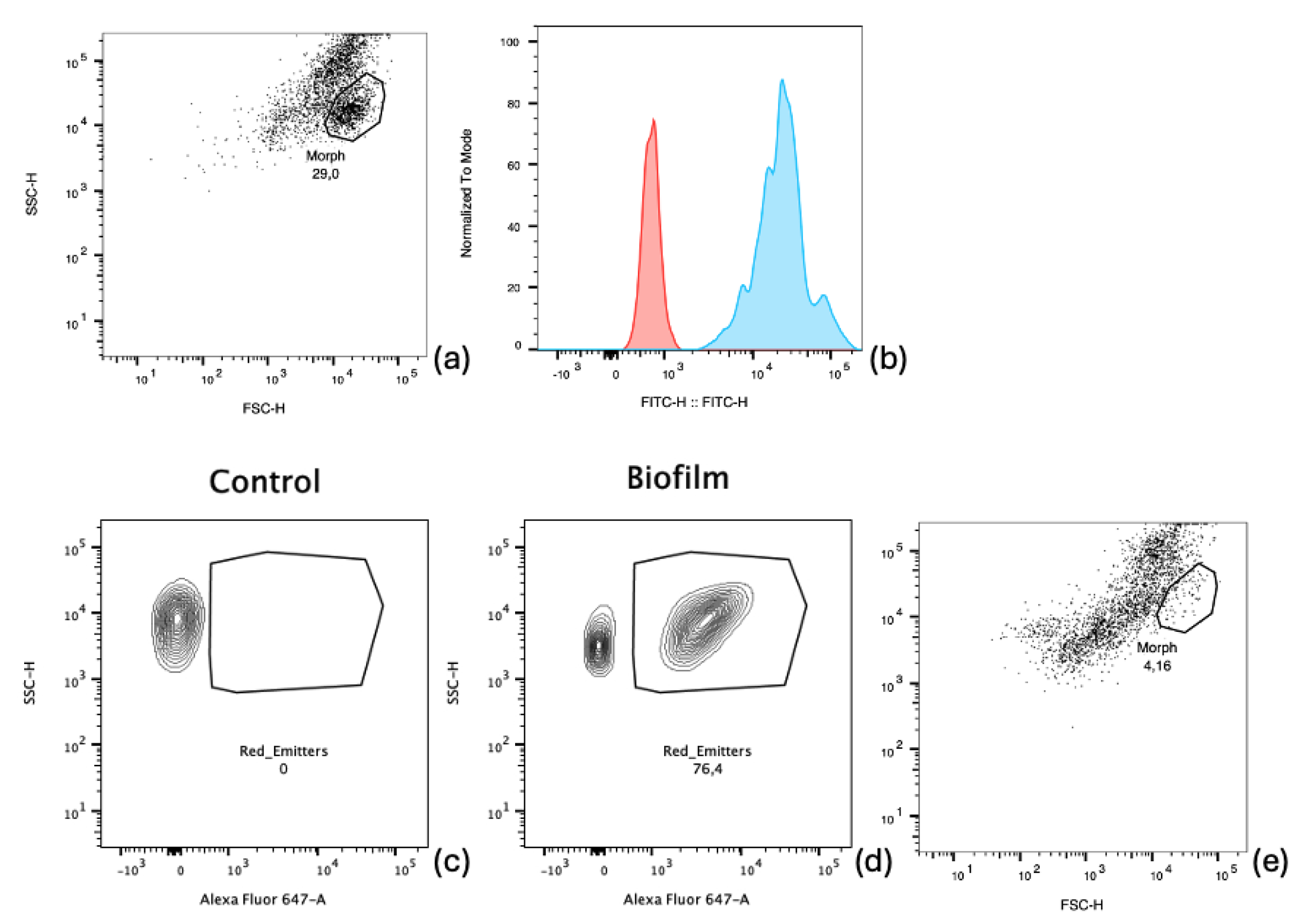
2.3.1. Cytometry Analysis
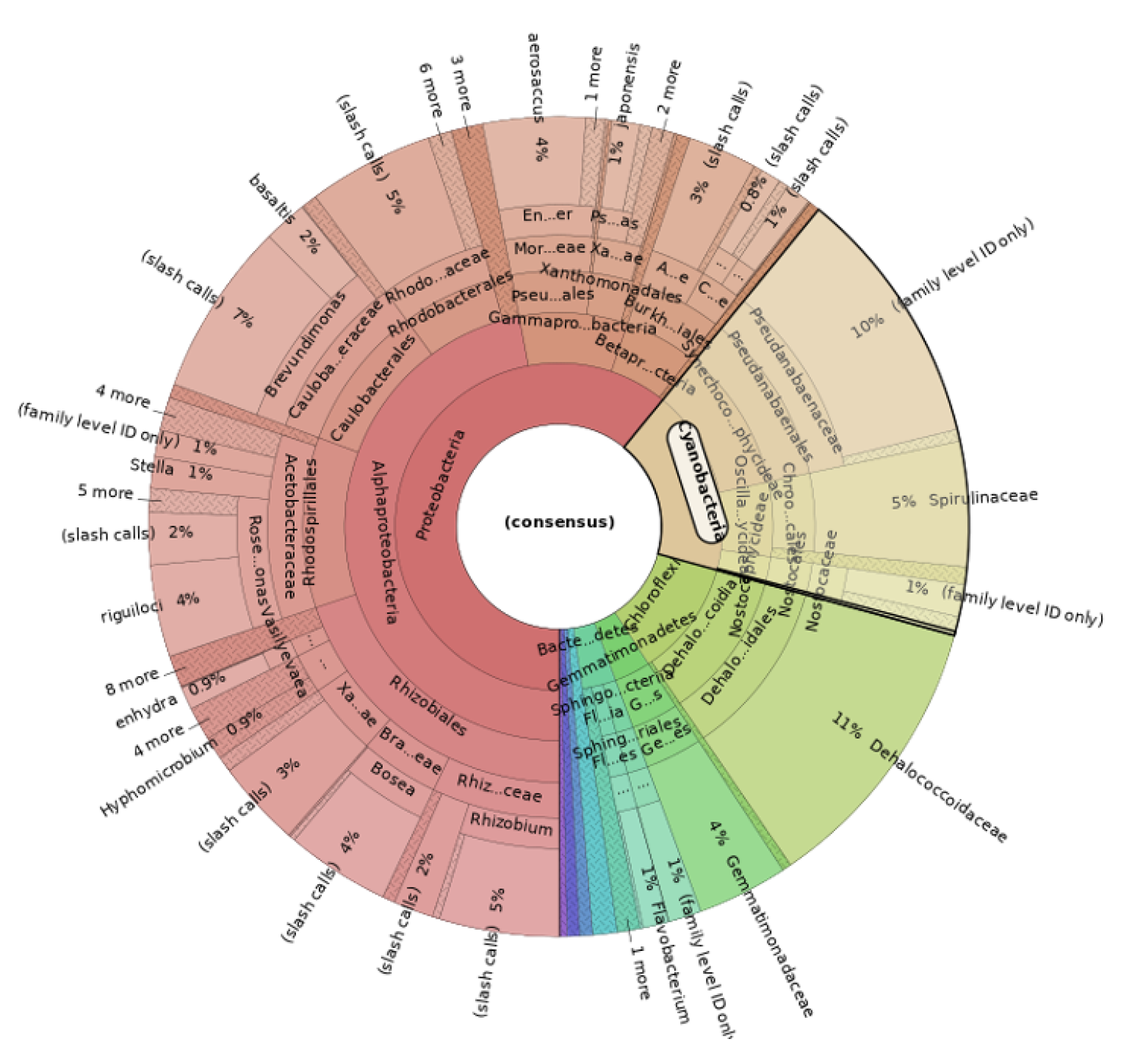
2.3.2. Genetics Analysis
2.3.3. Inductively Coupled Plasma—Mass Spectrometry (ICP-MS) Analysis
2.3.4. Shotgun Proteomics Analysis
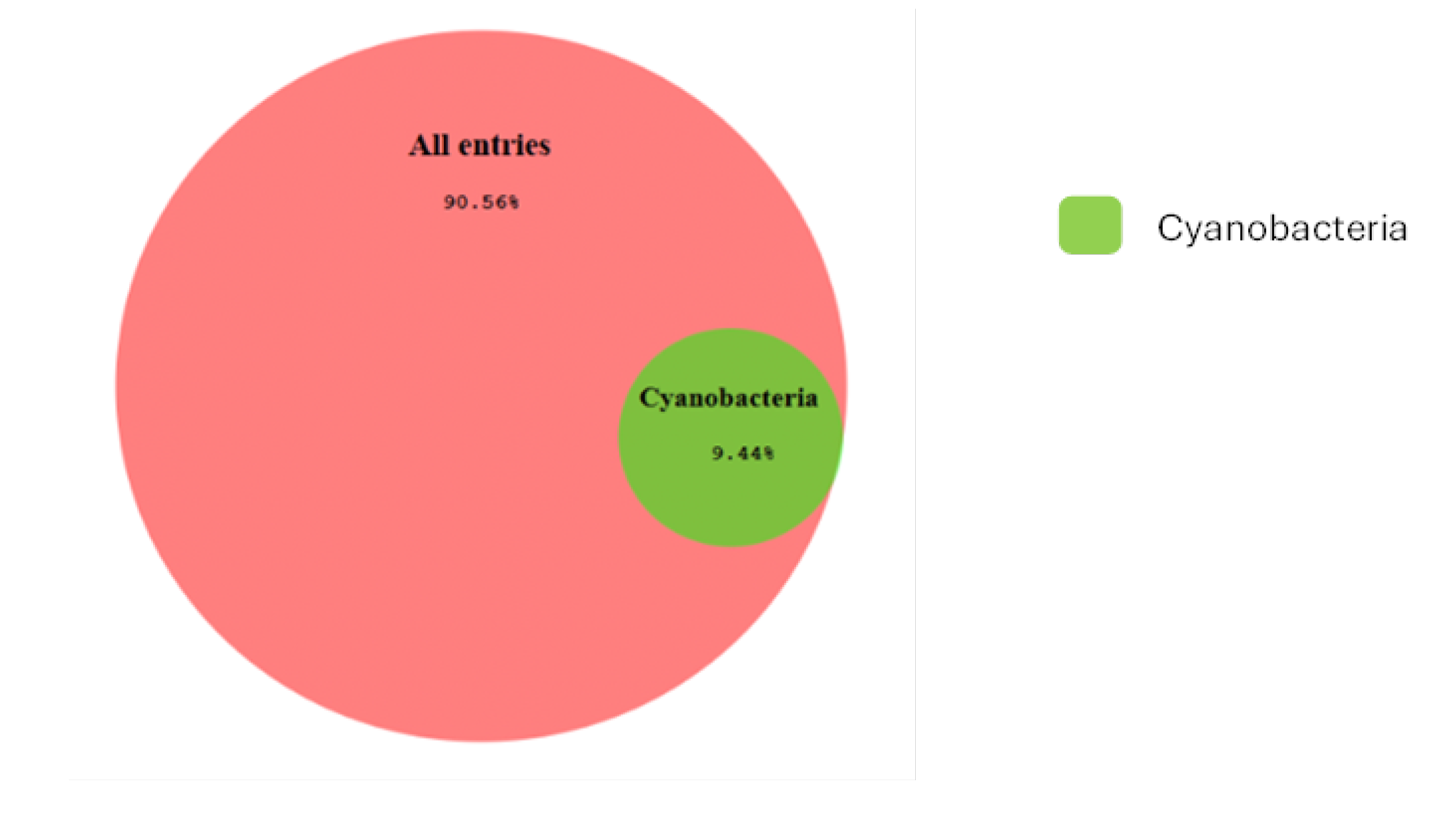
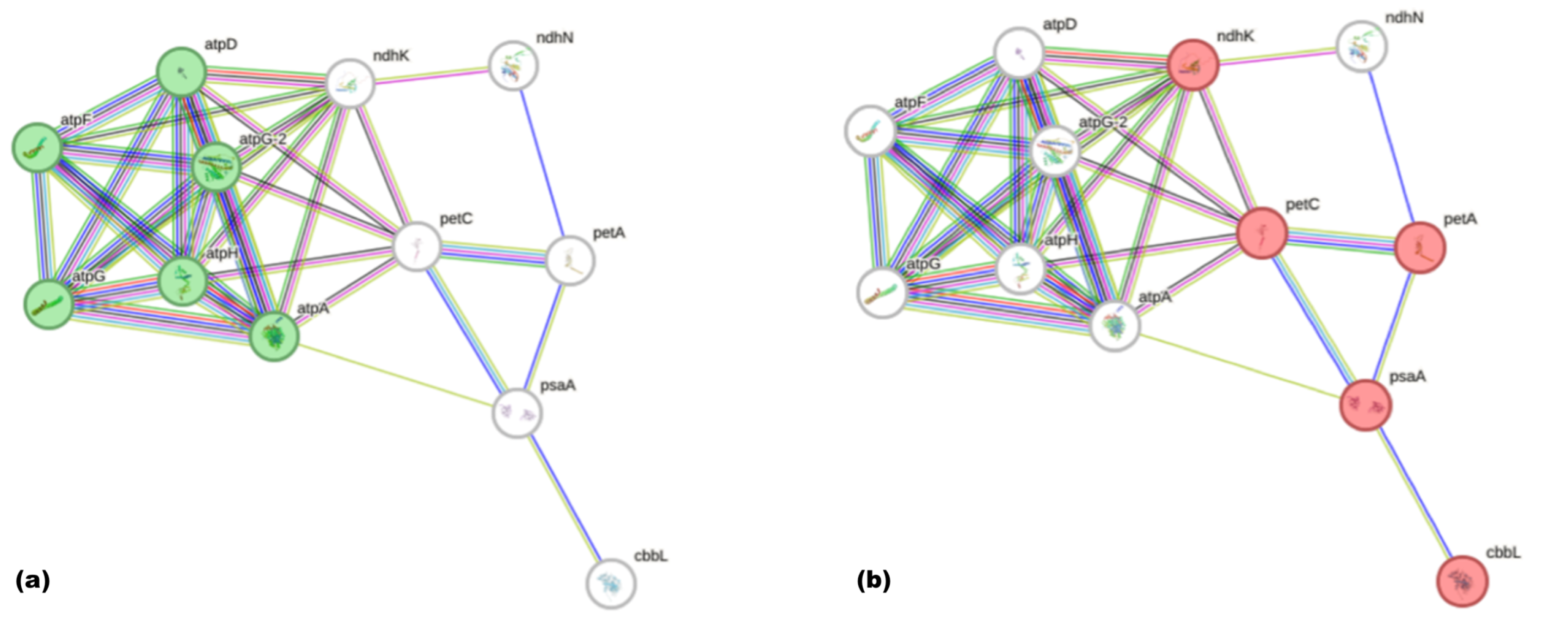
3. Results
4. Discussion
5. Conclusions
Author Contributions
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Palmerini, G. L’arte rupestre nel Parco Nazionale della Maiella. Ricerche storiche, indagini in corso e nuove prospettive; Ente Parco Nazionale della Maiella, 2023.
- Mattioli, T. L’arte rupestre in Italia centrale: Umbria, Lazio, Abruzzo; Vol. 4, Tommaso Mattioli, 2007.
- Palmerini, G.; Beck, L.; Di Martino, L.; Gallet, X.; Lebon, M.; Manzi, A.; Nicoud, E.; Mariano, A.; Villa, V. # MaiellaRockArtProject: Nuove ricerche sull’arte rupestre dell’Appennino abruzzese. In Proceedings of the XXVIII Valcamonica Symposium: ROCK-ART, A HUMAN HERITAGE. Centro Camuno di Studi Preistorici, 2021.
- Di Fraia, T. Le nuove scoperte di arte rupestre in Abruzzo: verso un’interpretazione sistemica. L’RTE RUPESTRE, 2015. [Google Scholar]
- Mattioli, T. Landscape analysis of a sample of rock-art sites in Central Italy. In Proceedings of the Layers of perception: Proceedings of the 35th International Conference on Computer Applications and Quantitative Methods in Archaeology (CAA), 2008, pp. 342–43.
- Nuhoglu, Y.; Oguz, E.; Uslu, H.; Ozbek, A.; Ipekoglu, B.; Ocak, I.; Hasenekoglu, I. The accelerating effects of the microorganisms on biodeterioration of stone monuments under air pollution and continental-cold climatic conditions in Erzurum, Turkey. Science of the total environment 2006, 364, 272–283. [Google Scholar] [CrossRef]
- Kumar, R.; Kumar, A.V. Biodeterioration of stone in tropical environments. An overview. The Getty Conservation Institute, Los Angeles, Calif, 1999. [Google Scholar]
- Ortega-Morales, B.O.; Gaylarde, C.C.; Englert, G.E.; Gaylarde, P.M. Analysis of salt-containing biofilms on limestone buildings of the Mayan culture at Edzna, Mexico. Geomicrobiology Journal 2005, 22, 261–268. [Google Scholar] [CrossRef]
- Tomaselli, L.; Lamenti, G.; Bosco, M.; Tiano, P. Biodiversity of photosynthetic micro-organisms dwelling on stone monuments. International Biodeterioration & Biodegradation 2000, 46, 251–258. [Google Scholar] [CrossRef]
- Gaylarde, P.M.; Gaylarde, C.C. Algae and cyanobacteria on painted buildings in Latin America. International Biodeterioration & Biodegradation 2000, 46, 93–97. [Google Scholar] [CrossRef]
- Crispim, C.A.; Gaylarde, C. Cyanobacteria and biodeterioration of cultural heritage: a review. Microbial ecology 2005, 49, 1–9. [Google Scholar] [CrossRef]
- Zanardini, E.; Abbruscato, P.; Ghedini, N.; Realini, M.; Sorlini, C. Influence of atmospheric pollutants on the biodeterioration of stone. International biodeterioration & biodegradation 2000, 45, 35–42. [Google Scholar]
- Warscheid, T.; Braams, J. Biodeterioration of stone: a review. International Biodeterioration & Biodegradation 2000, 46, 343–368. [Google Scholar] [CrossRef]
- Scheerer, S.; Ortega-Morales, O.; Gaylarde, C. Microbial deterioration of stone monuments—an updated overview. Advances in applied microbiology 2009, 66, 97–139. [Google Scholar] [PubMed]
- Ortega-Morales, O.; Montero-Muñoz, J.L.; Neto, J.A.B.; Beech, I.B.; Sunner, J.; Gaylarde, C. Deterioration and microbial colonization of cultural heritage stone buildings in polluted and unpolluted tropical and subtropical climates: A meta-analysis. International Biodeterioration & Biodegradation 2019, 143, 104734. [Google Scholar] [CrossRef]
- Louati, M.; Ennis, N.J.; Ghodhbane-Gtari, F.; Hezbri, K.; Sevigny, J.L.; Fahnestock, M.F.; Cherif-Silini, H.; Bryce, J.G.; Tisa, L.S.; Gtari, M. Elucidating the ecological networks in stone-dwelling microbiomes. Environmental microbiology 2020, 22, 1467–1480. [Google Scholar] [CrossRef]
- Sesana, E.; Gagnon, A.S.; Ciantelli, C.; Cassar, J.; Hughes, J.J. Climate change impacts on cultural heritage: A literature review. Wiley Interdisciplinary Reviews: Climate Change 2021, 12, e710. [Google Scholar] [CrossRef]
- Gu, J.D.; Katayama, Y. Microbiota and biochemical processes involved in biodeterioration of cultural heritage and protection. Microorganisms in the deterioration and preservation of Cultural Heritage 2021, 37, 37–58. [Google Scholar]
- Macedo, M.F.; Miller, A.Z.; Dionísio, A.; Saiz-Jimenez, C. Biodiversity of cyanobacteria and green algae on monuments in the Mediterranean Basin: an overview. Microbiology 2009, 155, 3476–3490. [Google Scholar] [CrossRef]
- Urzì, C.; Realini, M. Colour changes of Notos calcareous sandstone as related to its colonisation by microorganisms. International Biodeterioration & Biodegradation 1998, 42, 45–54. [Google Scholar] [CrossRef]
- Gambino, M.; Sanmartín, P.; Longoni, M.; Villa, F.; Mitchell, R.; Cappitelli, F. Surface colour: An overlooked aspect in the study of cyanobacterial biofilm formation. Science of the Total Environment 2019, 659, 342–353. [Google Scholar] [CrossRef]
- Ortega-Morales, O.; Guezennec, J.; Hernandez-Duque, G.; Gaylarde, C.C.; Gaylarde, P.M. Phototrophic biofilms on ancient Mayan buildings in Yucatan, Mexico. Current Microbiology 2000, 40, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Gorbushina, A.A. Life on the rocks. Environmental microbiology 2007, 9, 1613–1631. [Google Scholar] [CrossRef] [PubMed]
- Guillitte, O. Bioreceptivity: a new concept for building ecology studies. Science of the total environment 1995, 167, 215–220. [Google Scholar] [CrossRef]
- Sanmartín, P.; Miller, A.; Prieto, B.; Viles, H.A. Revisiting and reanalysing the concept of bioreceptivity 25 years on. Science of the total environment 2021, 770, 145314. [Google Scholar] [CrossRef]
- Miller, A.; Sanmartín, P.; Pereira-Pardo, L.; Dionísio, A.; Sáiz-Jiménez, C.; Macedo, M.; Prieto, B. Bioreceptivity of building stones: A review. Science of the total environment 2012, 426, 1–12. [Google Scholar] [CrossRef]
- Viles, H.; Ahmad, H. Architectural controls on the bioreceptivity of sandstone to green algal colonization. ECBSM2016. In Proceedings of the European Conference on Biodeterioration of Stone Monuments-Second Edition. Cergy-Pontoise, France. November 17–18, 2016, 2016.
- Cattò, C.; Mu, A.; Moreau, J.W.; Wang, N.; Cappitelli, F.; Strugnell, R. Biofilm colonization of stone materials from an Australian outdoor sculpture: importance of geometry and exposure. Journal of Environmental Management 2023, 339, 117948. [Google Scholar] [CrossRef]
- Trovão, J.; Portugal, A. The impact of stone position and location on the microbiome of a marble statue. The Microbe 2024, 2, 100040. [Google Scholar] [CrossRef]
- Ohkubo, S.; Miyashita, H. A niche for cyanobacteria producing chlorophyll f within a microbial mat. The ISME Journal 2017, 11, 2368–2378. [Google Scholar] [CrossRef]
- Involvement of Cyanobacterial Phytochromes in Growth Under Different Light Qualitities and Quantities¶.
- Liu, X.; Qian, Y.; Wu, F.; Wang, Y.; Wang, W.; Gu, J.D. Biofilms on stone monuments: biodeterioration or bioprotection? Trends in Microbiology 2022, 30, 816–819. [Google Scholar] [CrossRef]
- Favero-Longo, S.E.; Viles, H.A. A review of the nature, role and control of lithobionts on stone cultural heritage: weighing-up and managing biodeterioration and bioprotection. World Journal of Microbiology and Biotechnology 2020, 36, 100. [Google Scholar] [CrossRef]
- Pinna, D. Coping with biological growth on stone heritage objects: methods, products, applications, and perspectives; Apple academic press, 2017.
- Cappitelli, F.; Cattò, C.; Villa, F. The control of cultural heritage microbial deterioration. Microorganisms 2020, 8, 1542. [Google Scholar] [CrossRef]
- Macchia, A.; Aureli, H.; Prestileo, F.; Ortenzi, F.; Sellathurai, S.; Docci, A.; Cerafogli, E.; Colasanti, I.A.; Ricca, M.; La Russa, M.F. In-situ comparative study of eucalyptus, basil, cloves, thyme, pine tree, and tea tree essential oil biocide efficacy. Methods and Protocols 2022, 5, 37. [Google Scholar] [CrossRef]
- Decho, A. W. a. Microbial indicators of environmental change, 2010.
- C. Gaylarde, C. Influence of environment on microbial colonization of historic stone buildings with emphasis on cyanobacteria. Heritage 2020, 3, 1469–1482. [Google Scholar] [CrossRef]
- May, E.; Papida, S.; Abdulla, H.; Tayler, S.; Dewedar, A. Comparative studies of microbial communities on stone monuments in temperate and semi-arid climates. In Of microbes and art: the role of microbial communities in the degradation and protection of cultural heritage; Springer, 2000; pp. 49–62.
- Perez-Monserrat, E.M.; Varas-Muriel, M.J.; Alvarez De Buergo, M.; Fort, R. Black layers of decay and color patterns on heritage limestone as markers of environmental change. Geosciences 2016, 6, 4. [Google Scholar] [CrossRef]
- Chen, X.; Bai, F.; Huang, J.; Lu, Y.; Wu, Y.; Yu, J.; Bai, S. The organisms on rock cultural heritages: growth and weathering. Geoheritage 2021, 13, 56. [Google Scholar] [CrossRef]
- Ding, X.; Lan, W.; Yan, A.; Li, Y.; Katayama, Y.; Gu, J.D. Microbiome characteristics and the key biochemical reactions identified on stone world cultural heritage under different climate conditions. Journal of Environmental Management 2022, 302, 114041. [Google Scholar] [CrossRef]
- Wang, C.; Wang, L.; Bai, C.; Wang, M.; Ma, T.; Ma, H.; Zhang, G.; Wang, W.; Guo, Z.; Sun, Y.; et al. Identification of the key factors influencing biodeterioration of open-air cultural heritage in the temperate climate zone of China. International Biodeterioration & Biodegradation 2025, 196, 105954. [Google Scholar]
- Pointing, S. B. a. Microbial growth patterns linked to climatic factors, 2009.
- Viles, H.A.; Cutler, N.A. Global environmental change and the biology of heritage structures. Global Change Biology 2012, 18, 2406–2418. [Google Scholar] [CrossRef]
- Traversetti, L.; Bartoli, F.; Caneva, G. Wind-driven rain as a bioclimatic factor affecting the biological colonization at the archaeological site of Pompeii, Italy. International biodeterioration & biodegradation 2018, 134, 31–38. [Google Scholar]
- Orr, S.A.; Cassar, M. Exposure indices of extreme wind-driven rain events for built heritage. Atmosphere 2020, 11, 163. [Google Scholar] [CrossRef]
- Albertano, P. Cyanobacterial biofilms in monuments and caves. In Ecology of cyanobacteria II: their diversity in space and time; Springer, 2012; pp. 317–343.
- Chiaudani, A.; Flamminii, F.; Consalvo, A.; Bellocci, M.; Pizzi, A.; Passamonti, C.; Cichelli, A. Rare Earth Element Variability in Italian Extra Virgin Olive Oils from Abruzzo Region. Foods 2024, 13. [Google Scholar] [CrossRef]
- Potenza, F.; Cufaro, M.C.; Di Biase, L.; Panella, V.; Di Campli, A.; Ruggieri, A.G.; Dufrusine, B.; Restelli, E.; Pietrangelo, L.; Protasi, F.; et al. Proteomic analysis of marinesco–sjogren syndrome fibroblasts indicates pro-survival metabolic adaptation to SIL1 loss. International Journal of Molecular Sciences 2021, 22, 12449. [Google Scholar] [CrossRef]
- Ucci, A.; Giacchi, L.; Cufaro, M.C.; Puri, C.; Ciocca, M.; Di Ferdinando, F.; Del Boccio, P.; Cappariello, A.; Rucci, N. Human osteosarcoma cell secretome impairs neonatal mouse calvarial osteogenic cells functions and modifies the nanoparticles-derived protein profile. Life Sciences 2025, p. 123837.



| Instrument parameters | |
|---|---|
| Nebulizer | Babington |
| Torch | Quartz glass torch |
| Spray chamber | Scott double-pass type at 2 ∘C |
| Sample cone Nickel | 1.00 mm aperture |
| Skimmer cone Nickel | 0.40 mm aperture |
| Plasma mode | Normal plasma |
| RF power (W) | 1550 |
| RF matching (V) | 1.8 |
| Sample depth (mm) | 10 |
| Nebulizer gas (L ) | 1.03 |
| Nebulizer pump (rps) | 0.1 |
| Plasma gas (L ) | 15 |
| Sampling period (s) | 0.3 |
| Repetitions 3 | 3 |
| Sample uptake rate (mL ) | 0.4 |
| Integration time (s) | 0.1 |
| MOSS | BLACKENED MOSS | SOIL | ROCK SURFACE |
|---|---|---|---|
| Co = 6.6 ± 0.2 | Co = 2.9 ± 1 | Co = 12.9 ± 5.7 | Co = 3.8 ± 2.7 |
| Cr = 2.2 ± 2.9 | Cr = 5.6 ± 2.2 | Cr = 1.5 ± 6.5 | Cr = 11.7 ± 4.7 |
| Al = 713.3*± 8.4 | Al = 3329.3*± 0.7 | Al = 435.2*± 1.7 | Al = 1532.3*± 2.4 |
| Ag = N/A** | Ag = 0.23 ± 4.7 | Ag = N/A** | Ag = N/A** |
| As = 4.4 ± 5.1 | As = 2.01 ± 1.9 | As = 4.8 ± 0.6 | As = 12.6 ± 4.2 |
| Cd = 0.13 ± 3.4 | Cd = 5.4 ± 1.7 | Cd = 0.18 ± 10.9 | Cd = 624.2* ± 1.3 |
| Cu = 16.9 ± 6.7 | Cu = 19.3 ± 2.3 | Cu = 16.2 ± 1 | Cu = 42.2 ± 3.1 |
| Mn = 25.2 ± 7.2 | Mn = 115.5* ± 0.6 | Mn = 17.7 ± 1.7 | Mn = 188.4* ± 2.7 |
| Ni = 56.3 ± 0.1 | Ni = 12.9 ± 0.9 | Ni = 107 ± 6.5 | Ni = 41.4 ± 8.7 |
| Pb = 1.9 ± 8.6 | Pb = 255.6 ± 4.9 | Pb = 1.4 ± 1 | Pb = 24.8 ± 6.9 |
| Se = 1.8 ± 13.4 | Se = 11.5 ± 7.2 | Se = 3.2 ± 16 | Se = 18 ± 6.9 |
| V = 11.7 ± 7 | V = 6.9 ± 0.8 | V = 12.1 ± 0.5 | V = 23.31 ± 1.6 |
| Zn = 17.9 ± 7.1 | Zn = 54.7 ± 1.7 | Zn = 22.2 ± 1.1 | Zn = 82.9 ± 3.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





