Submitted:
17 October 2025
Posted:
17 October 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
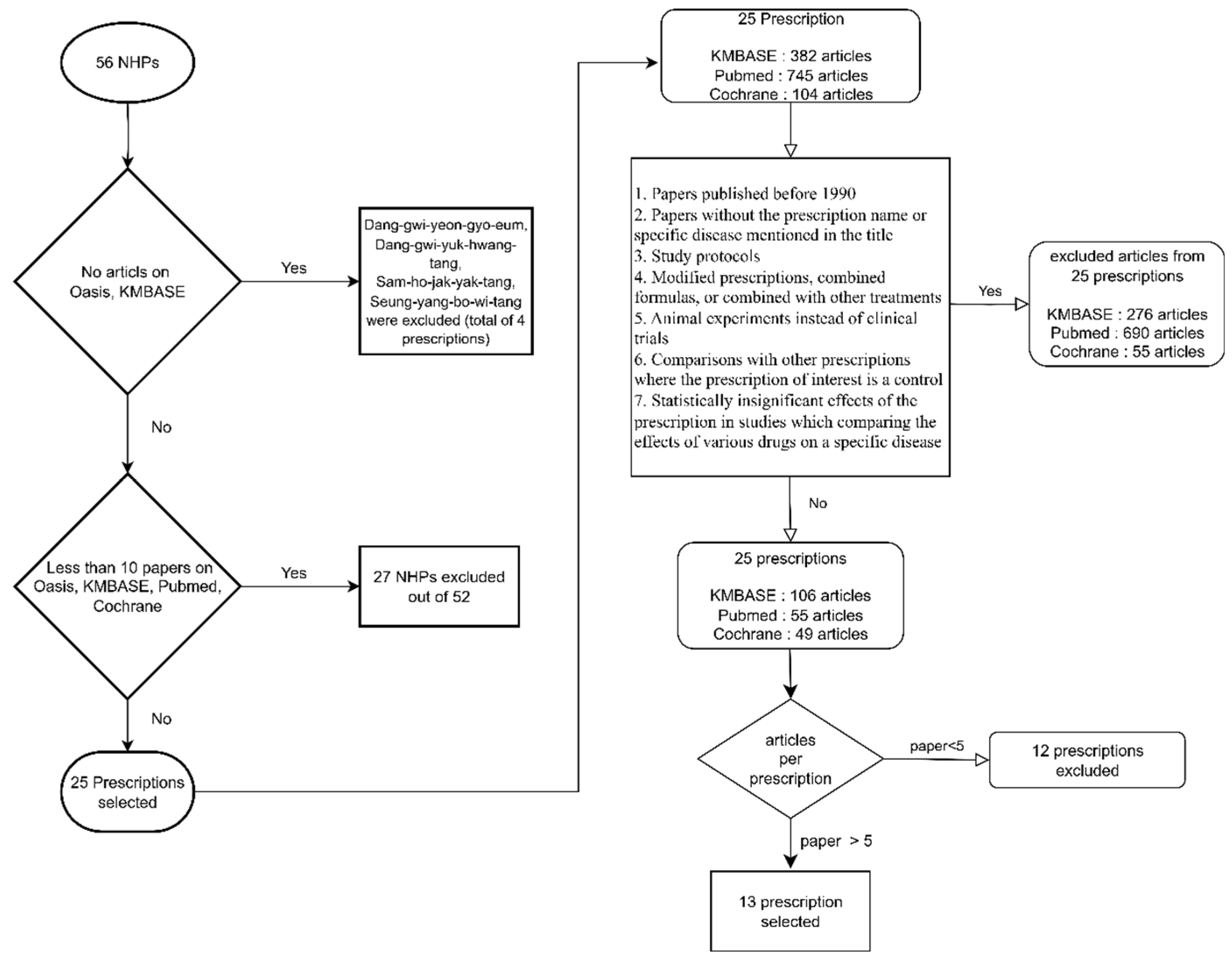
2.1. Search Method
2.2. Criteria for Excluding Prescriptions (Figure 1)
2.3. Criteria for Excluding Papers (Figure 1)
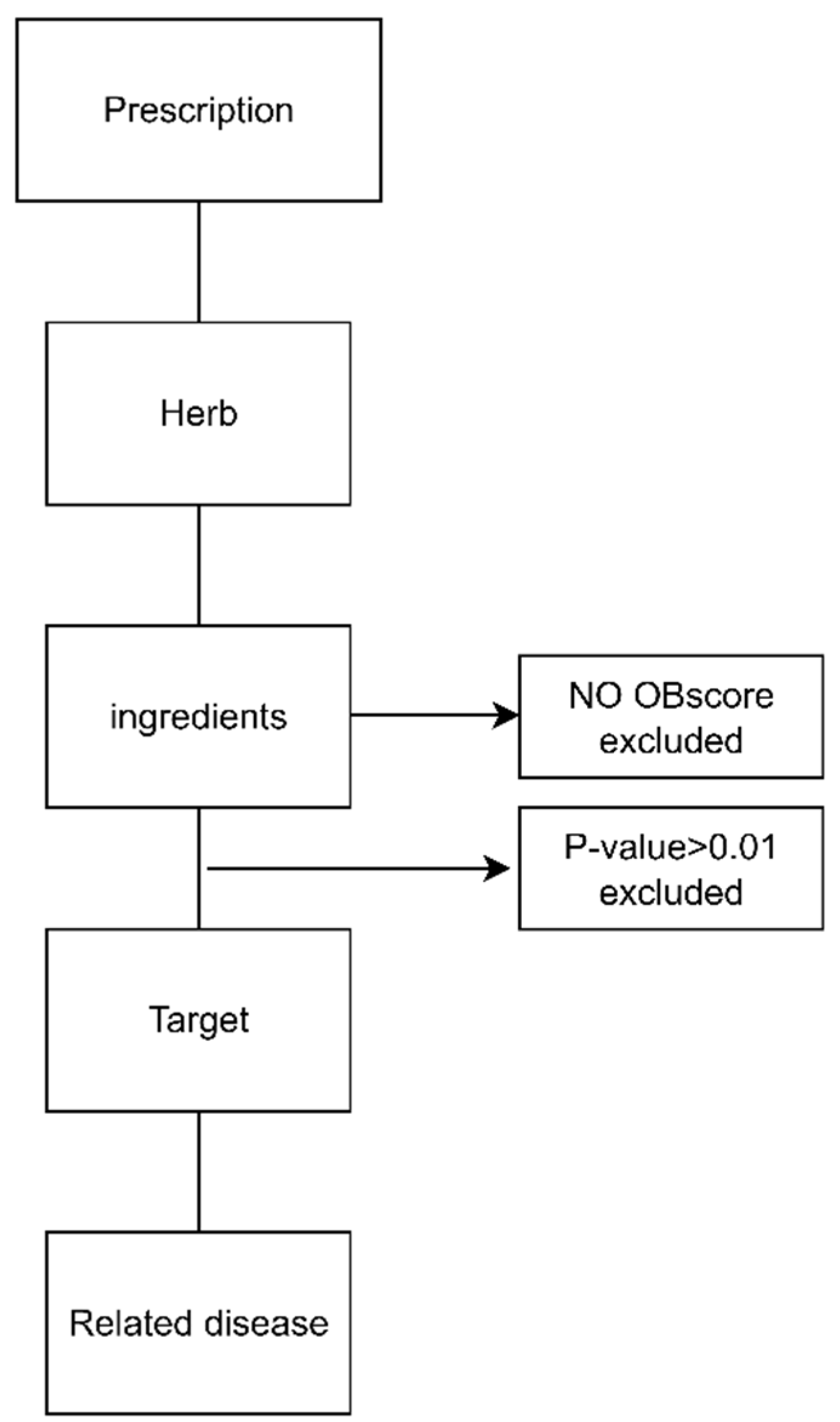
2.4. Network Construction of NHPs
2.5. Pathway Analysis of NHPs
2.6. Comparative Analysis Between the Main Indications of NHPs and the Diseases Associated with Clinical Research and Databases
3. Results
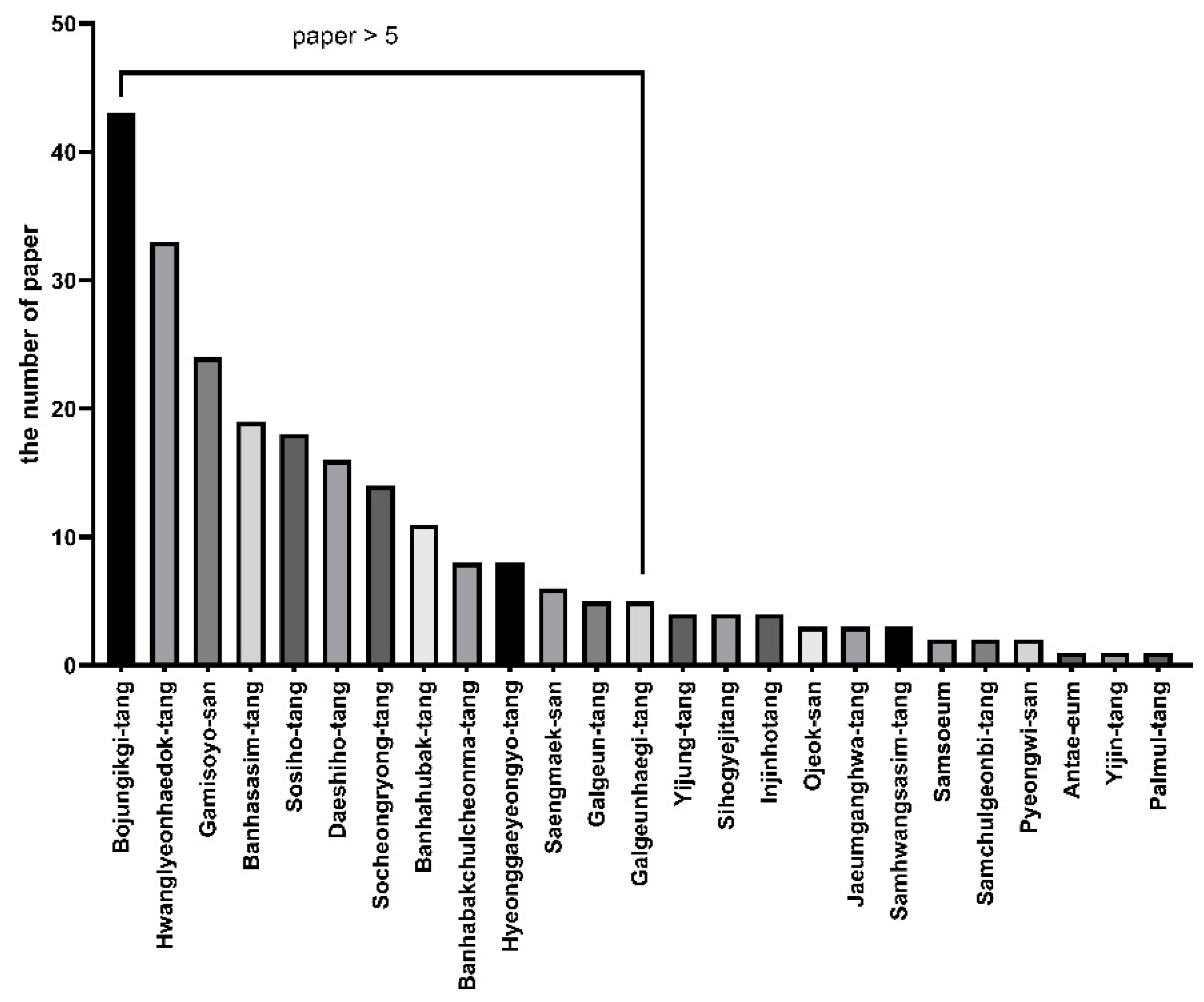
3.1. Main Indications of NHPs
3.2. Selection of Clinically Relevant NHPs for Comparative Analysis
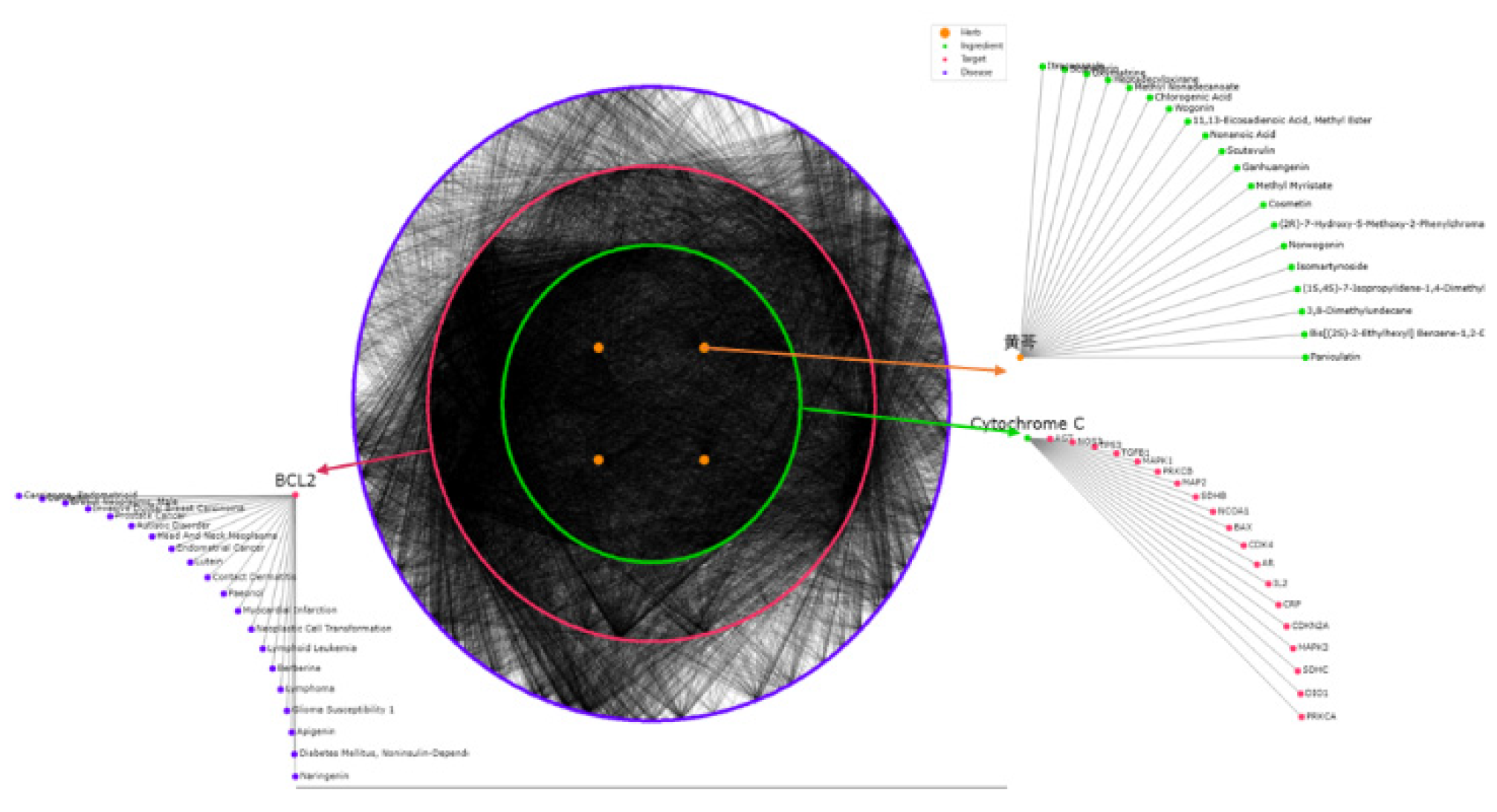
3.3. Herb-Based Reconstruction of Prescription Networks
3.4. Comparative Analysis of the Main Indications, Networks, and Clinical Research Concordance of NHPs
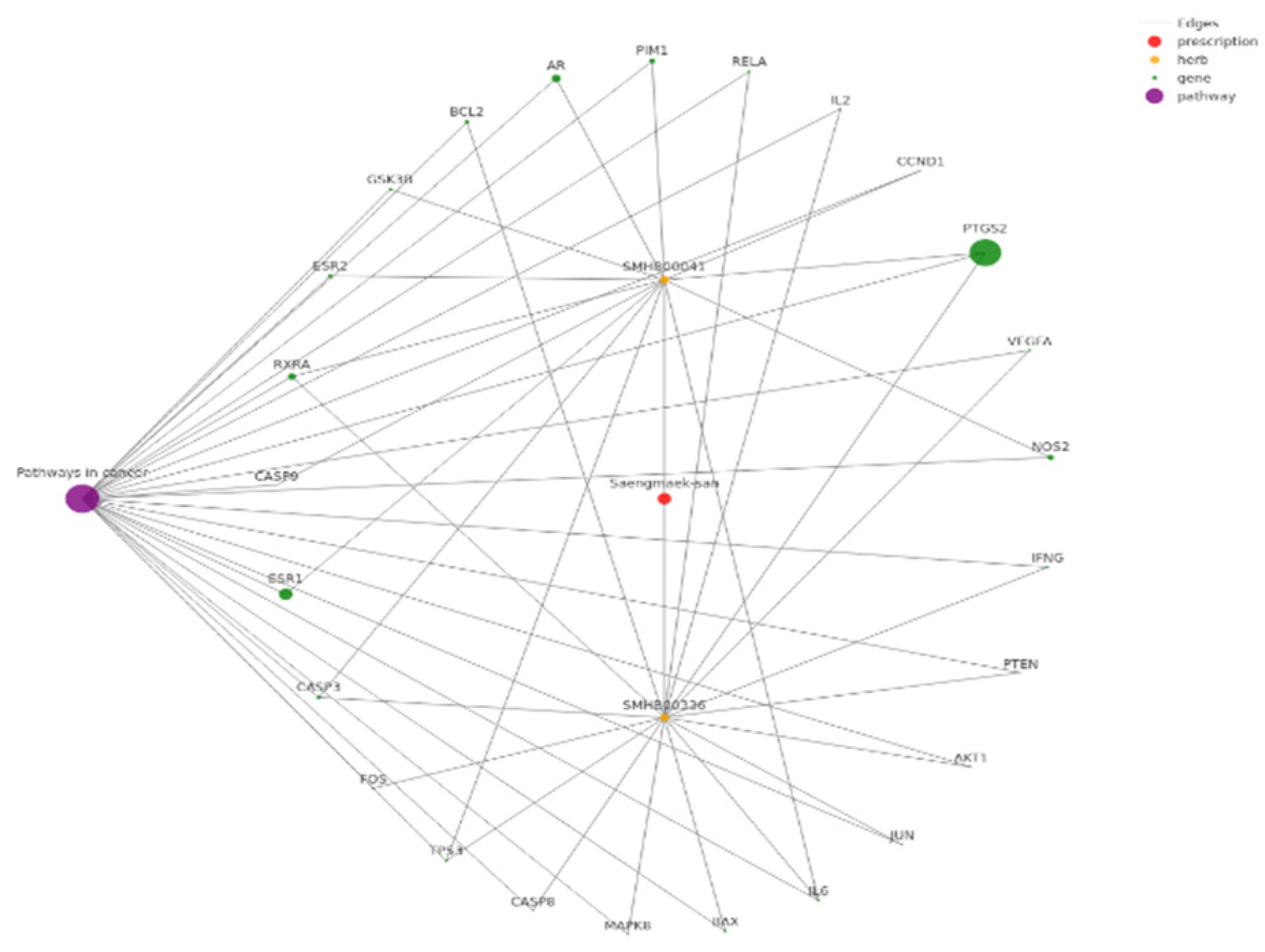
3.5. Molecular Mechanism-Based Analysis of Network-Inferred Prescription-Disease Association
4. Discussion
Limitations of the Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NHPs | National Health Insurance herbal Prescriptions |
| TCM | Traditional Chinese Medicine |
| TKM | Traditional Korean medicine |
| OASIS | Oriental Medicine Advanced Searching Integrated System |
| KMBASE | Korean Medical database |
| OB | Oral bioavailability |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| HIV | Human Immunodeficiency Virus |
| AGE | Advanced Glycation End-product |
| RAGE | Receptor for Advanced Glycation End-product |
| HIF-1 | Hypoxia-Inducible Factor-1 |
| NF-κB | Nuclear Factor kappa-light-chain-enhancer of activated B cells |
| GERD | Gastroesophageal Reflux Disease |
| KCI | Korean Citation Index |
References
- Wu, Y.; Zhang, F.; Yang, K.; Liu, Y.; Zhou, M.; Zhao, Y.; et al. SymMap: an integrative database of traditional Chinese medicine enhanced by symptom mapping. Nucleic Acids Res. 2019, 47, D1110–D1117.
- Tran, M.N.; Baek, S.J.; Jun, H.J.; Lee, S. Identifying target organ location of Radix Achyranthis Bidentatae: a bioinformatics approach on active compounds and genes. Front Pharmacol. 2023, 14, 1187896.
- Park, S.; Kim, Y.J.; Bae, G.S.; Lee, J.; Kim, S.Y.; Choi, J.Y.; et al. Network analysis using the established database (K-herb Network) on herbal medicines used in clinical research on heart failure. The Journal of Internal Korean Medicine. 2023, 44, 313.
- Oriental Medicine Advanced Searching Integrated System. K Herb Network. Available online: https://oasis.kiom.re.kr/kmedi/main.jsp (accessed on 28 January 2025).
- Oriental Medicine Advanced Searching Integrated System. Prescription search. Available online: https://oasis.kiom.re.kr/oasis/pres/prbaseSearch.jsp?srch_menu_nix=1GydHbs0 (accessed on 28 January 2025).
- Streamlit Inc. Streamlit: Turn Python scripts into beautiful ML tools. Available online: https://streamlit.io/ (accessed on 17 July 2025).
- Ministry of Health and Welfare of Korea. Indications and dosages for herbal formulas covered by health insurance. Available online: https://www.mohw.go.kr/board.es?mid=a10409020000&bid=0026&act=view&list_no=1481950&tag=&nPage=1 (accessed on 7 May 2025).
- Kang, E.; Ryu, H.; Kim, Y.; Kim, K. A case report of patient with type 2 diabetes mellitus treated with Galgeun-tang. The Journal of Internal Korean Medicine. 2017, 38, 541–546.
- Oh, J.; Shim, H.; Cha, J.; Kim, J.; Yoo, H.; Lee, Y.; et al. The anti-obesity effects of Bangpungtongseong-san and Daesiho-tang: a study protocol of randomized, double-blinded clinical trial. J Korean Med Obes Res. 2020, 20, 138–149.
- Oh, S.; Jung, W.; Song, M.; Kim, E.; Jeong, H.; Park, K.; et al. A retrospective study of the safety and effect of co-administration of glucose-lowering medication and Bojungikgi-tang on blood glucose level in patients with type 2 diabetes mellitus. The Journal of Internal Korean Medicine. 2023, 44, 354–365.
- Sul, M.C.; Kim, D.S.; Kim, D.H.; Shin, D.H. Usage of Bojungikgi-tang to maintain and treat asthmatic patients. The Journal of Internal Korean Medicine. 2011, 32, 497–503.
- Li, K.Z. Observation of Jiawei Shengmai San on 30 patients with type 2 diabetes accompanied somatic neuropathy. Information on traditional chinese medicine. 2003, 20, 45–46.
- Baek, J.; Shin, S.; Shin, D.; Cho, C. Efficacy and safety of the antidiabetic effect of Hwangryunhaedok-tang for type 2 diabetes mellitus patients without complications: a systematic review and meta-analysis. The Journal of Internal Korean Medicine. 2021, 42, 605–621.
- Clarrett, D.M.; Hachem, C. Gastroesophageal reflux disease (GERD). Mo Med. 2018, 115, 214–218.
- Cui, C.Y.; Liu, X.; Peng, M.H.; Yu, Y.; Zhang, J.; Li, W.; et al. Identification of key candidate genes and biological pathways in neuropathic pain. Comput Biol Med. 2022, 150, 106135.
- Li, Y.; Miao, P.; Li, F.; Zhou, J.; Wang, Q.; Zhao, L.; et al. An association study of clock genes with major depressive disorder. J Affect Disord. 2023, 341, 147–153.
- Li, Q.; Bai, J.; Ma, Y.; Zhang, Y.; Liu, X.; Chen, Y.; et al. Pharmacometabolomics and mass spectrometry imaging approach to reveal the neurochemical mechanisms of Polygala tenuifolia. J Pharm Anal. 2024, 14, 100973.
- Kondoh, T.; Mallick, H.N.; Torii, K. Activation of the gut-brain axis by dietary glutamate and physiologic significance in energy homeostasis. Am J Clin Nutr. 2009, 90, 832S–837S.
- Shang, Y.; Smith, S.; Hu, X. Role of Notch signaling in regulating innate immunity and inflammation in health and disease. Protein Cell 2016, 7, 159–174.
- Badmus, O.O.; Hillhouse, S.A.; Anderson, C.D.; Ghosh, S.; Rahman, S.M.; Imig, J.D. Molecular mechanisms of metabolic associated fatty liver disease (MAFLD): functional analysis of lipid metabolism pathways. Clin Sci (Lond) 2022, 136, 1347–1366.
- National Research Foundation of Korea. Korea Citation Index. Available online: https://www.kci.go.kr/kciportal/main.kci?locale=en (accessed on 28 January 2025).
- Chu, H.; Kim, Y.; Min, B.K. A guide to writing a case report according to CARE guideline. Journal of Korean Medical Society of Acupotomology 2021, 5, 41–45.
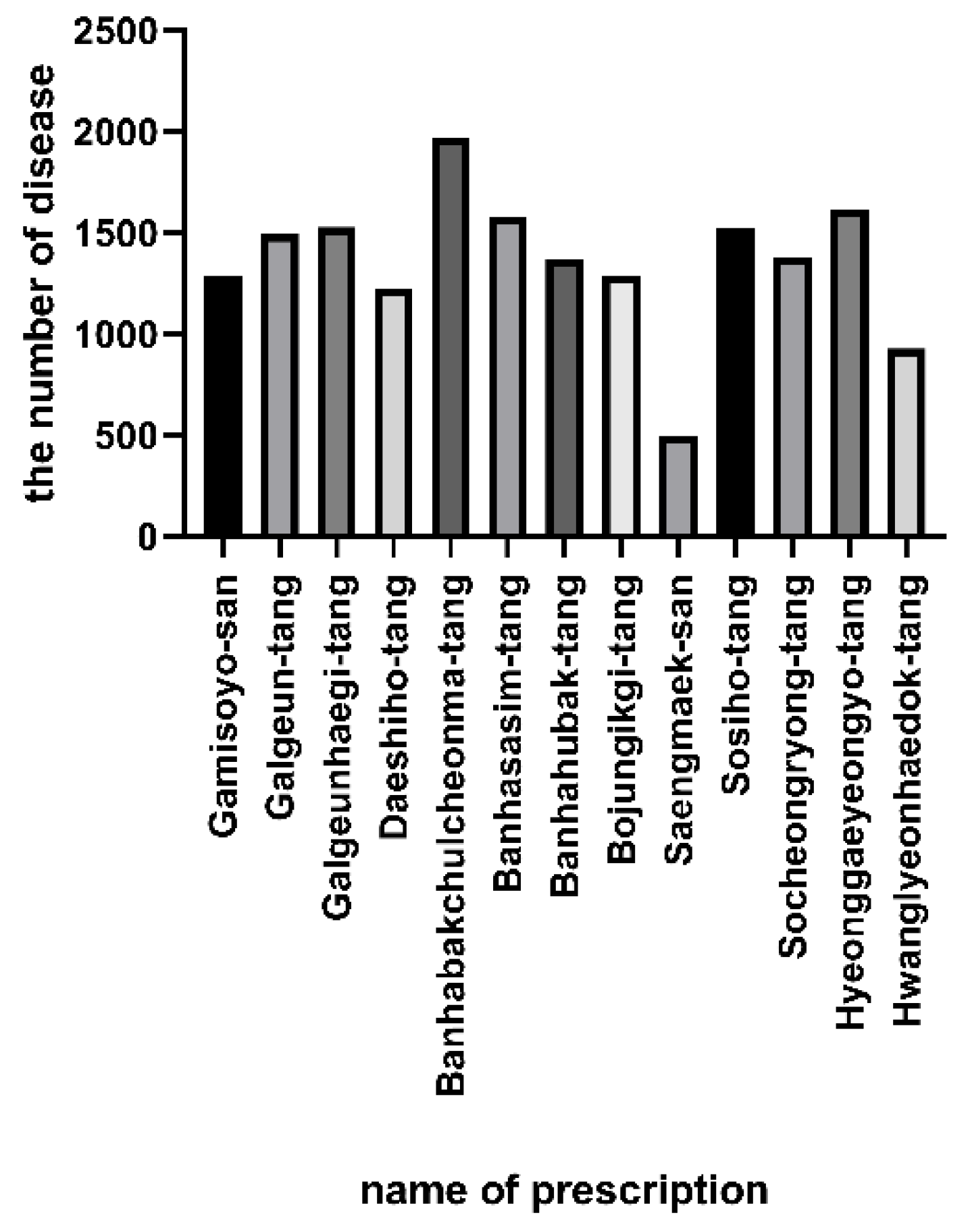

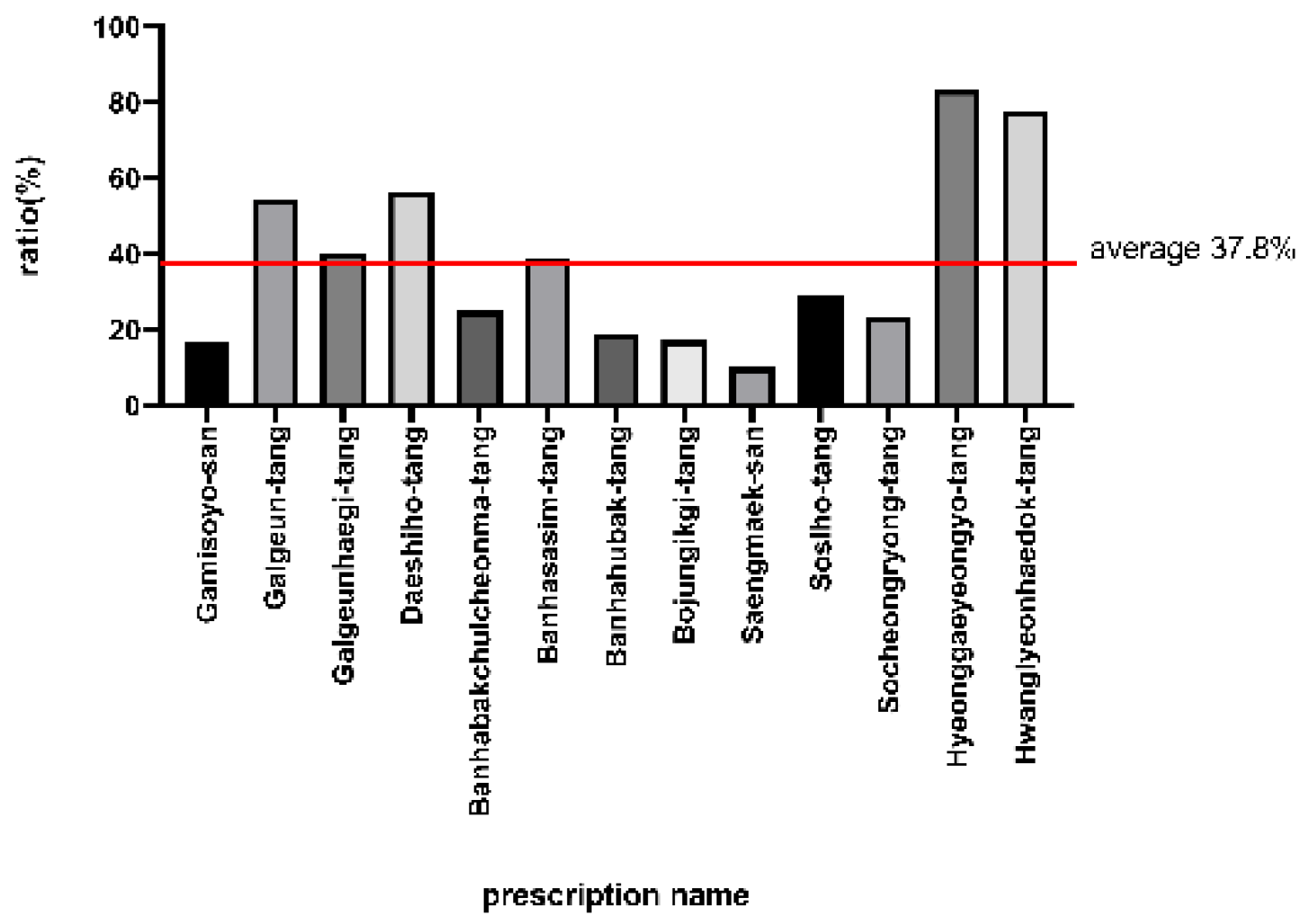
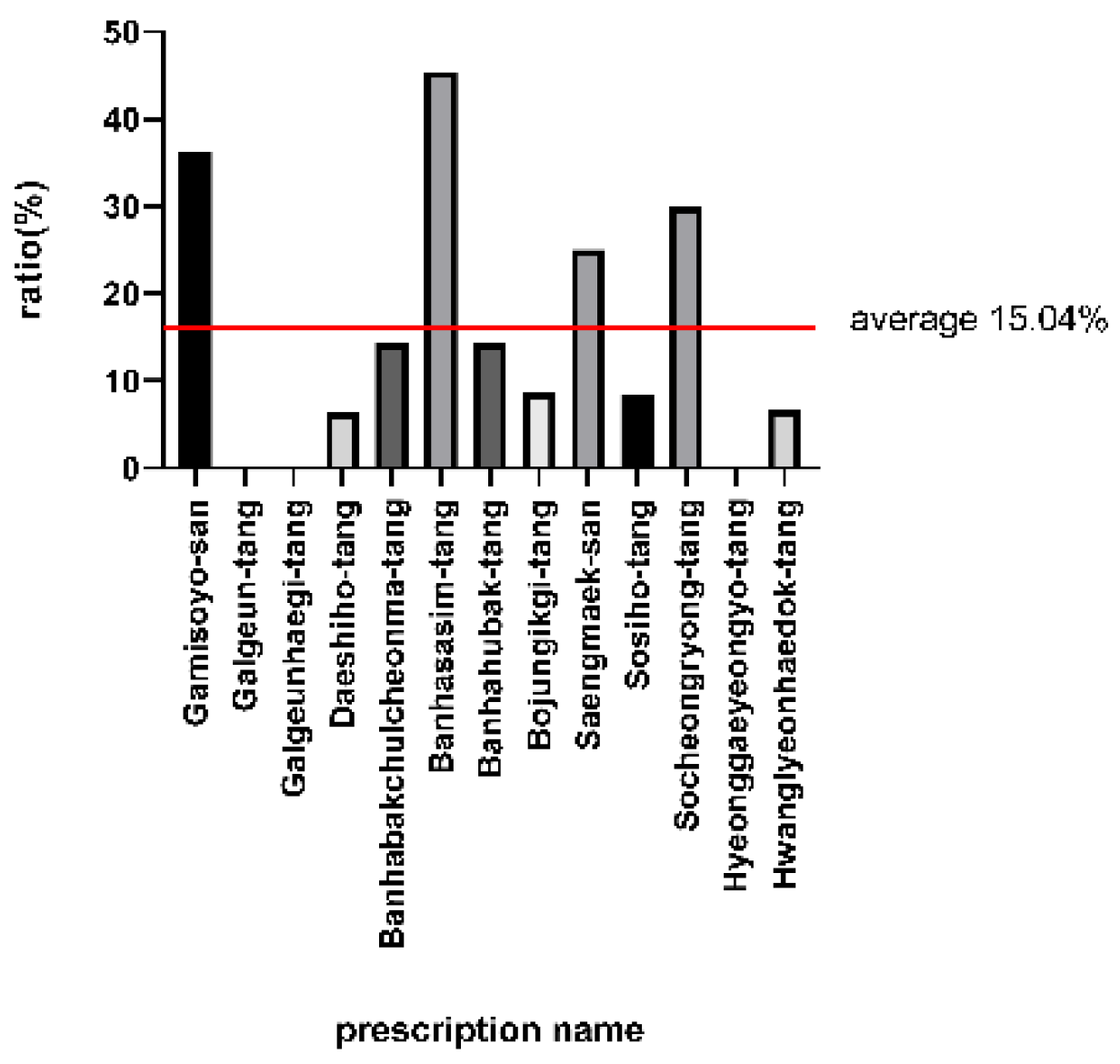
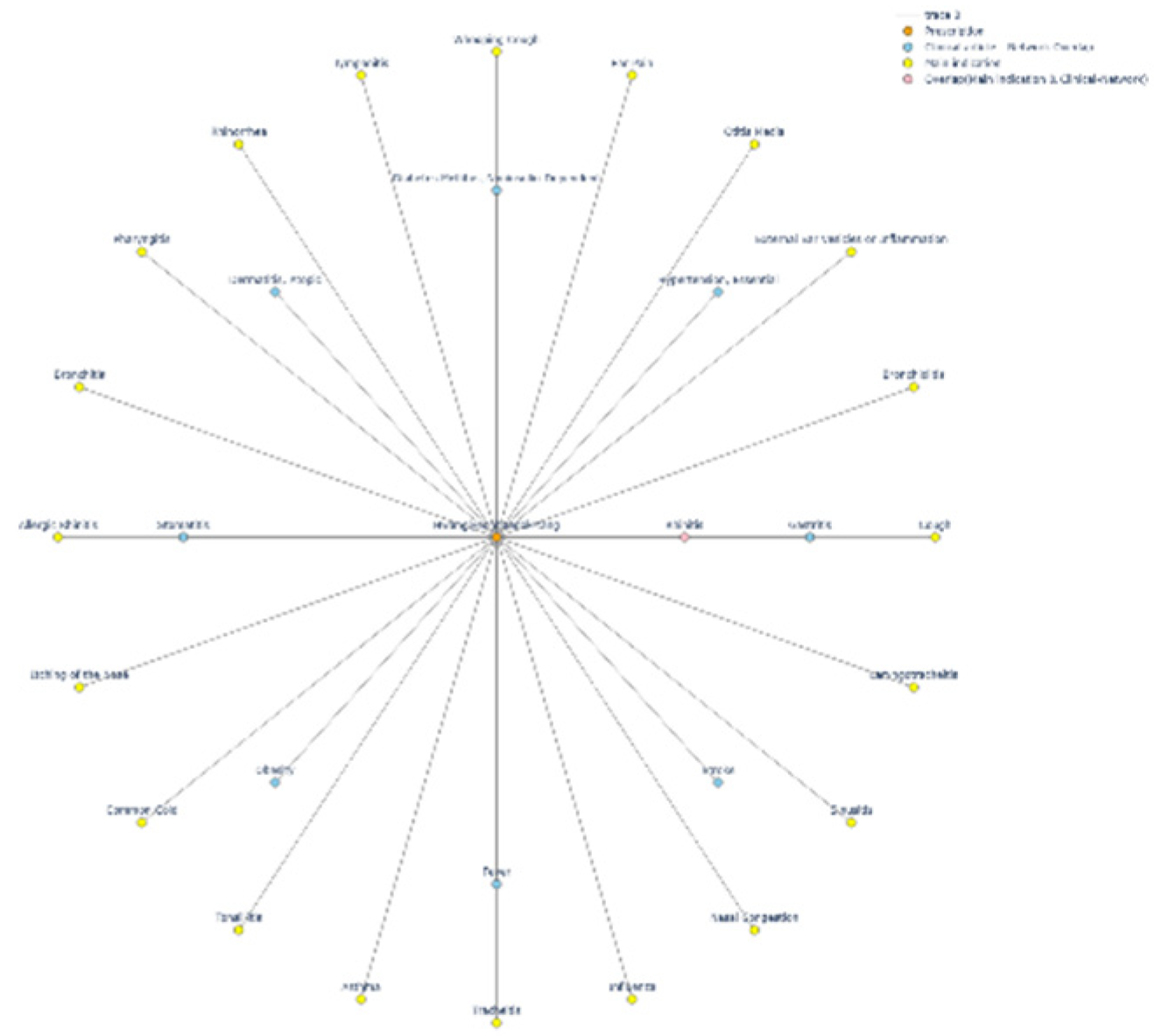
| Prescription | Disease |
| Gamisoyo-san | Lung Cancer, Carcinoma of Lung. Lung Neoplasms, Schizophrenia, Non-Small Cell Lung Carcinoma. Diabetes Mellitus, Noninsulin-Dependent, Ovarian Neoplasm, Arthritis, Rheumatoid, Leukemia, Adenocarcinoma, Liver Cirrhosis, Experimental, Asthma, Aneurysm, Myocardial Infarction, Prostatic Neoplasms, Spinal Cord Ischemia, Diabetes Mellitus, Experimental, Malaria, Hepatoblastoma, Disorder of Eye |
| Galgeun-tang | Leukemia, Glioma, Lung Cancer, Kidney Neoplasm, Carcinoma of Lung, Lung Neoplasms, Adenocarcinoma, Non-Small Cell Lung Carcinoma, Schizophrenia, Melanoma, Prostatic Neoplasms, Astrocytoma, Hepatocellular Carcinoma, Myeloid Leukemia, Liver Carcinoma, Diabetes Mellitus, Non-Insulin-Dependenta, Liver Cirrhosis, Experimental, Depressive Disorder, Lymphoproliferative Disorders, Ovarian Neoplasm |
| Galgeunhaegi-tang | Leukemia, Glioma, Lung Cancer, Adenocarcinoma, Lung Neoplasms, Carcinoma of Lung, Schizophrenia, Prostatic Neoplasms, Non-Small Cell Lung Carcinoma, Mammary Neoplasms, Diabetes Mellitus, Noninsulin-Dependent, Hepatocellular Carcinoma, Colorectal Neoplasms, Astrocytoma, Diabetes Mellitus, Experimental, Myeloid Leukemia, Liver Cirrhosis, Experimental, Hypertensive Disease, Kidney Neoplasm, Asthma |
| Daeshiho-tang | Adenocarcinoma, Lung Cancer, Carcinoma of Lung, Non-Small Cell Lung Carcinoma, Schizophrenia, Mammary Neoplasms, Glioma, Diabetes Mellitus, Noninsulin-Dependent, Hepatocellular Carcinoma, Prostatic Neoplasms, Liver Carcinoma, Ovarian Neoplasm, Malignant Neoplasm of Breast, Leukemia, Esophageal Neoplasms, Colorectal Neoplasms, Liver Cirrhosis, Experimental, Colorectal Cancer, Obesity b†, Malaria |
| Banhabakchulcheonma-tang | Schizophrenia, Diabetes Mellitus, Non-Insulin-Dependent, Mammary Neoplasms, Lung Cancer, Ovarian Neoplasm, Liver Cirrhosis, Experimental, Bipolar Disorder, Liver Carcinoma, Non-Small Cell Lung Carcinoma, Colorectal Neoplasms, Carcinoma of Lung, Hepatocellular, Carcinoma, Inflammatory Bowel Diseases, Alzheimer’s Disease, Asthma, Alcoholic, Intoxication, Chronic, Prostatic Neoplasms, Colorectal Cancer, Major Depressive Disorder, Lung Neoplasms |
| Banhasasim-tang | Mammary Neoplasms, Prostatic Neoplasms, Schizophrenia, Lung Cancer, Carcinoma of Lung, Hepatocellular Carcinoma, Liver Carcinoma, Animal Mammary Neoplasms, Leukemia, Liver Cirrhosis, Experimental, Diabetes Mellitus, Non-Insulin-Dependent, Colorectal Neoplasms, Ovarian Neoplasm, Non-Small Cell Lung Carcinoma, Malignant Neoplasm of Breast, Colitis (Main), Alzheimer Disease, Inflammatory Bowel Diseases, Depressive Disorder, Colorectal Cancer |
| Banhahubak-tang | Mammary Neoplasms, Schizophrenia, Leukemia, Lung Cancer, Diabetes Mellitus, Noninsulin-Dependent, Liver Cirrhosis, Experimental, Carcinoma of Lung, Lung Neoplasms, Alzheimer'S Disease, Astrocytoma, Major Depressive Disorder, Animal Mammary Neoplasms, Prostatic Neoplasms, Liver Carcinoma, Depressive Disorder, Melanoma, Diabetes Mellitus, Experimental, Alcoholic Intoxication, Chronic, Kidney Neoplasm, Colorectal Neoplasms |
| Bojungikgi-tang | Lung Cancer, Carcinoma of Lung, Schizophrenia, Prostatic Neoplasms, Diabetes Mellitus, Noninsulin-Dependentc, Ovarian Neoplasm, Bipolar Disorder, Asthmad, Liver Carcinoma, Non-Small Cell Lung Carcinoma, Liver Cirrhosis, Experimental, Lung Neoplasms, Hypertensive Disease, Mammary Neoplasms, Arthritis, Rheumatoid, Myocardial Infarction, Ovarian Cancer, Colorectal Neoplasms, Autistic Disorder, Adult Primary Hepatocellular Carcinoma |
| Saengmaek-san | Mammary Neoplasms, Prostatic Neoplasms, Renal Cell Carcinoma, Bipolar Disorder, Malaria, Disorder of Eye, Diabetes Mellitus, Noninsulin-Dependente, Induced Malaria, Insulin Resistance, Malaria, Cerebral, Ewings Sarcoma-Primitive Neuroectodermal Tumor (Primitive neuro-ectodermal tumor), Ewings Sarcoma, Arthritis, Rheumatoid, Mood Disorders, Schizophrenia, Asthma, Obesity, Nervous System Disorder, Ovarian Neoplasm, Mental Retardation |
| Sosiho-tang | Mammary Neoplasms, Prostatic Neoplasms, Lung Cancer, Schizophrenia, Carcinoma of Lung, Leukemia, Diabetes Mellitus, Non-Insulin-Dependent, Hepatocellular Carcinoma, Ovarian Neoplasm, Liver Carcinoma, Non-Small Cell Lung Carcinoma, Lung Neoplasms, Animal Mammary Neoplasms, Liver Cirrhosis, Experimental, Glioma, Colorectal Neoplasms, Alzheimer’s Disease, Disorder Of Eye, Obesity, Malignant Neoplasm of Breast |
| Socheongryong-tang | Schizophrenia, Lung Cancer, Carcinoma of Lung, Non-Small Cell Lung Carcinoma, Hepatocellular Carcinoma, Liver Carcinoma, Liver Cirrhosis, Experimental, Myocardial Infarction, Diabetes Mellitus, Non-Insulin-Dependent, Prostatic Neoplasms, Mammary Neoplasms, Bipolar Disorder, Esophageal Neoplasms, Alzheimer’s Disease, Depressive Disorder, Diabetes Mellitus, Experimental, Hypertensive Disease, Glioma, Ovarian Neoplasm, Leukemia |
| Hyeonggaeyeongyo-tang | Schizophrenia, Lung Cancer, Carcinoma of Lung, Diabetes Mellitus, Non-Insulin-Dependent, Prostatic Neoplasms, Non-Small Cell Lung Carcinoma, Asthma, Lung Neoplasms, Adenocarcinoma, Hypertensive Disease, Colorectal Neoplasms, Liver Cirrhosis, Experimental, Mammary Neoplasms, Diabetes Mellitus, Experimental, Colorectal Cancer, Hepatocellular Carcinoma, Myocardial Infarction, Arthritis, Rheumatoid, Bipolar Disorder, Ovarian Neoplasm |
| Hwanglyeonhaedok-tang | Mammary Neoplasms, Leukemia, Schizophrenia, Prostatic Neoplasms, Malignant Neoplasm of Breast, Breast Cancer, Colorectal Neoplasms, Glioma Susceptibility 1, Colorectal Cancer, Liver Cirrhosis, Experimental, Glioblastoma, Asthma†, Inflammatory Bowel Diseases, Lung Neoplasms, Non-Small Cell Lung Carcinoma, Hepatocellular Carcinoma, Liver Carcinoma, Hypertensive Disease, Prostate Cancer, Diabetes Mellitus, Noninsulin-Dependentf |
| Prescription | Network-Clinical Paper Concordance |
| Gamisoyo-san | Depressive Disorder(6A7Z) Mastitis (GB21) |
| Galgeun-tang | Diabetes Mellitus (Non-Insulin-Dependent) (5A11) |
| Galgeunhaegi-tang | Fever (MG26) Trigeminal Neuropathy(8B82) Erysipelas (1B70.0) Hanta Viral Infections(1D62) |
| Daeshiho-tang | Cholelithiasis (DC11) Dyslipidemias(5C8Z) Acute Cholecystitis (DC12) Cholecystitis (DC12) Hyperlipidemia(5C80) Obesity(5B81) |
| Banhabakchulcheonma-tang | Obesity(5B81) Cerebral Hemorrhage(8B00) Hemiplegia (MB53) |
| Banhasasim-tang | Diabetes Mellitus, Noninsulin-Dependent(5A11) Gastrointestinal Diseases (DE2Z) Gastroesophageal Reflux Disease (DA22) Dyspepsia And Other Specified Disorders of Function of Stomach (MD92) Gastritis, Atrophic (DA42) Irritable Bowel Syndrome (DD91) |
| Banhahubak-tang | Mental Depression(6E20/6A70) Gastroesophageal Reflux Disease (DA22) Panic Disorder(6B01) Sleep Apnea(7A41) Mental Depression(6E20/6A70) |
| Bojungikgi-tang | Diabetes Mellitus, Noninsulin-Dependent(5A11) Asthma (CA23) Myocardial Infarction (BA41) Stroke(8B20) Cerebrovascular Disorders(8B2Z) Fever (MG26) Anemia(3A9Z) Dermatitis, Atopic (EA80) Chronic Fatigue Syndrome(8E49) Otitis Media (AB0Z) Irritable Bowel Syndrome (DD91) Gastric Cancer(2B72) Male Infertility (GB04) Recurrent Urinary Tract Infection (GC04) Allergic Rhinitis (Disorder)(SC90) |
| Saengmaek-san | Diabetes Mellitus, Noninsulin-Dependent(5A11) Myocarditis (BC42) |
| Sosiho-tang | Parkinson Disease(8A00) Chronic Fatigue Syndrome(8E49) Dermatitis, Atopic (EA80) Hepatitis, Chronic (DB97.2) Burning Mouth Syndrome (DA0F.0) Hepatitis C, Chronic (1E51.1) Hepatitis C (1E51.1) Hepatitis B, Chronic (1E51.0) |
| Socheongryong-tang | Asthma (CA23.32) Gastrointestinal Diseases (DE2Z) Dermatitis, Atopic (EA80) Allergic Rhinitis (Disorder)(SC90) Urticaria (EB05) |
| Hyeonggaeyeongyo-tang | Allergic Rhinitis (Disorder)(SC90) Bronchiectasis (CA24) |
| Hwanglyeonhaedok-tang | Diabetes Mellitus, Noninsulin-Dependent(5A11) Stroke(8B20) Obesity(5B81) Hypertension, Essential (BA00) Fever (MG26) Gastritis (DA42) Dermatitis, Atopic (EA80) Stomatitis (DA01) Rhinitis (CA09) |
| Prescription | Disease | Matched Pathways |
| Banhabakchulcheonma-tang | Schizophrenia | Dopaminergic synapse; ErbB signaling pathway; Glutamatergic synapse; Neuroactive ligand-receptor interaction; Serotonergic synapse |
| Banhabakchulcheonma-tang | Diabetes Mellitus, Non-Insulin-Dependent | Cell cycle; Insulin secretion; PPAR signaling pathway; Pancreatic secretion; Protein processing in endoplasmic reticulum; TGF-beta signaling pathway; Wnt signaling pathway; p53 signaling pathway |
| Banhabakchulcheonma-tang | Mammary Neoplasms | Breast cancer; PI3K-Akt signaling pathway |
| Banhabakchulcheonma-tang | Lung Cancer (small cell) | Apoptosis; Small cell lung cancer |
| Banhabakchulcheonma-tang | Lung Cancer (non-small cell) | Calcium signaling pathway; Non-small cell lung cancer |
| Banhabakchulcheonma-tang | Ovarian Cancer | PI3K-Akt signaling pathway; Thyroid hormone signaling pathway |
| Gamisoyo-san | Lung Cancer (small cell) | Apoptosis; Small cell lung cancer |
| Gamisoyo-san | Non-Small Cell Lung Carcinoma | Calcium signaling pathway; ErbB signaling pathway; Neuroactive ligand-receptor interaction; Non-small cell lung cancer |
| Gamisoyo-san | Schizophrenia | Dopaminergic synapse; Glutamatergic synapse; PPAR signaling pathway; Pancreatic secretion; Protein processing in endoplasmic reticulum; Serotonergic synapse; Wnt signaling pathway |
| Gamisoyo-san | Diabetes Mellitus, Noninsulin-Dependent | Cell cycle; Insulin secretion; TGF-beta signaling pathway; p53 signaling pathway |
| Galgeun-tang | Leukemia (Acute myeloid leukemia) | Acute myeloid leukemia; Efferocytosis |
| Galgeun-tang | Leukemia (chronic myelogenous leukemia) | Chronic myeloid leukemia |
| Galgeun-tang | Leukemia (chronic lymphocytic leukemia) | - |
| Galgeun-tang | Glioma | Glioma |
| Galgeun-tang | Non-Small Cell Lung Carcinoma | Calcium signaling pathway; ErbB signaling pathway; Neuroactive ligand-receptor interaction; Non-small cell lung cancer |
| Galgeun-tang | Lung Cancer (small cell) | Apoptosis; Small cell lung cancer |
| Galgeunhaegi-tang | Leukemia (Acute myeloid leukemia) | Acute myeloid leukemia; Efferocytosis |
| Galgeunhaegi-tang | Leukemia (chronic myelogenous leukemia) | Chronic myeloid leukemia |
| Galgeunhaegi-tang | Leukemia (chronic lymphocytic leukemia) | - |
| Galgeunhaegi-tang | Glioma | Glioma |
| Galgeunhaegi-tang | Non-Small Cell Lung Carcinoma | Calcium signaling pathway; ErbB signaling pathway; Neuroactive ligand-receptor interaction; Non-small cell lung cancer |
| Galgeunhaegi-tang | Lung Cancer (small cell) | Apoptosis; Small cell lung cancer |
| Daeshiho-tang | Non-Small Cell Lung Carcinoma | Calcium signaling pathway; ErbB signaling pathway; Neuroactive ligand-receptor interaction; Non-small cell lung cancer |
| Daeshiho-tang | Lung Cancer (small cell) | Apoptosis; Small cell lung cancer |
| Daeshiho-tang | Non-Small Cell Lung Carcinoma | Calcium signaling pathway; ErbB signaling pathway; Neuroactive ligand-receptor interaction; Non-small cell lung cancer |
| Daeshiho-tang | Schizophrenia | Dopaminergic synapse; Glutamatergic synapse; Serotonergic synapse |
| Banhasasim-tang | Mammary Neoplasms | Breast cancer; PI3K-Akt signaling pathway |
| Banhasasim-tang | Prostatic Neoplasms | Prostate cancer |
| Banhasasim-tang | Schizophrenia | Dopaminergic synapse; Glutamatergic synapse; Serotonergic synapse |
| Banhasasim-tang | Non-Small Cell Lung Carcinoma | Calcium signaling pathway; ErbB signaling pathway; Neuroactive ligand-receptor interaction; Non-small cell lung cancer |
| Banhasasim-tang | Lung Cancer (small cell) | Apoptosis; Small cell lung cancer |
| Banhahubak-tang | Mammary Neoplasms | Breast cancer; ErbB signaling pathway; Neuroactive ligand-receptor interaction; PI3K-Akt signaling pathway |
| Banhahubak-tang | Schizophrenia | Dopaminergic synapse; Glutamatergic synapse; Serotonergic synapse |
| Banhahubak-tang | Leukemia (Acute myeloid leukemia) | Acute myeloid leukemia; Efferocytosis |
| Banhahubak-tang | Leukemia (chronic myelogenous leukemia) | Chronic myeloid leukemia |
| Banhahubak-tang | Leukemia (chronic lymphocytic leukemia) | - |
| Banhahubak-tang | Non-Small Cell Lung Carcinoma | Calcium signaling pathway; ErbB signaling pathway; Neuroactive ligand-receptor interaction; Non-small cell lung cancer |
| Banhahubak-tang | Lung Cancer (small cell) | Apoptosis; Small cell lung cancer |
| Banhahubak-tang | Diabetes Mellitus, Noninsulin-Dependent | Cell cycle; Insulin secretion; TGF-beta signaling pathway; p53 signaling pathway |
| Bojungikgi-tang | Non-Small Cell Lung Carcinoma | Calcium signaling pathway; ErbB signaling pathway; Neuroactive ligand-receptor interaction; Non-small cell lung cancer |
| Bojungikgi-tang | Lung Cancer (small cell) | Apoptosis; Small cell lung cancer |
| Bojungikgi-tang | Schizophrenia | Dopaminergic synapse; Glutamatergic synapse; Serotonergic synapse |
| Bojungikgi-tang | Prostatic Neoplasms | Prostate cancer |
| Bojungikgi-tang | Diabetes Mellitus, Noninsulin-Dependent | Cell cycle; Insulin secretion; TGF-beta signaling pathway; p53 signaling pathway |
| Saengmaek-san | Mammary Neoplasms | Breast cancer; PI3K-Akt signaling pathway |
| Saengmaek-san | Prostatic Neoplasms | Prostate cancer |
| Saengmaek-san | Renal Cell Carcinoma | Renal cell carcinoma |
| Saengmaek-san | Malaria | Malaria |
| Sosiho-tang | Mammary Neoplasms | Breast cancer; PI3K-Akt signaling pathway |
| Sosiho-tang | Prostatic Neoplasms | Prostate cancer |
| Sosiho-tang | Non-Small Cell Lung Carcinoma | Calcium signaling pathway; ErbB signaling pathway; Neuroactive ligand-receptor interaction; Non-small cell lung cancer |
| Sosiho-tang | Lung Cancer (small cell) | Apoptosis; Small cell lung cancer |
| Sosiho-tang | Schizophrenia | Dopaminergic synapse; Glutamatergic synapse; Serotonergic synapse |
| Socheongryong-tang | Schizophrenia | - |
| Socheongryong-tang | Non-Small Cell Lung Carcinoma | Neuroactive ligand-receptor interaction |
| Socheongryong-tang | Lung Cancer (small cell) | Apoptosis; Small cell lung cancer |
| Socheongryong-tang | Hepatocellular Carcinoma | PI3K-Akt signaling pathway |
| Hyeonggaeyeongyo-tang | Schizophrenia | Dopaminergic synapse; Glutamatergic synapse; Serotonergic synapse |
| Hyeonggaeyeongyo-tang | Non-Small Cell Lung Carcinoma | Calcium signaling pathway; ErbB signaling pathway; Neuroactive ligand-receptor interaction; Non-small cell lung cancer |
| Hyeonggaeyeongyo-tang | Lung Cancer (small cell) | Apoptosis; Small cell lung cancer |
| Hyeonggaeyeongyo-tang | Diabetes Mellitus, Non-Insulin-Dependent | Cell cycle; Insulin secretion; TGF-beta signaling pathway; p53 signaling pathway |
| Hyeonggaeyeongyo-tang | Prostatic Neoplasms | Prostate cancer |
| Hwanglyeonhaedok-tang | Mammary Neoplasms | Breast cancer; PI3K-Akt signaling pathway |
| Hwanglyeonhaedok-tang | Leukemia (Acute myeloid leukemia) | Acute myeloid leukemia; Efferocytosis |
| Hwanglyeonhaedok-tang | Leukemia (chronic myelogenous leukemia) | Chronic myeloid leukemia |
| Hwanglyeonhaedok-tang | Leukemia (chronic lymphocytic leukemia) | - |
| Hwanglyeonhaedok-tang | Schizophrenia | Dopaminergic synapse; Glutamatergic synapse; Serotonergic synapse |
| Hwanglyeonhaedok-tang | Prostatic Neoplasms | Prostate cancer |
| Hwanglyeonhaedok-tang | Malignant Neoplasm of Breast | Breast cancer; PI3K-Akt signaling pathway |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





