Submitted:
05 September 2025
Posted:
05 September 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Bioactive Compounds and Target Predictions
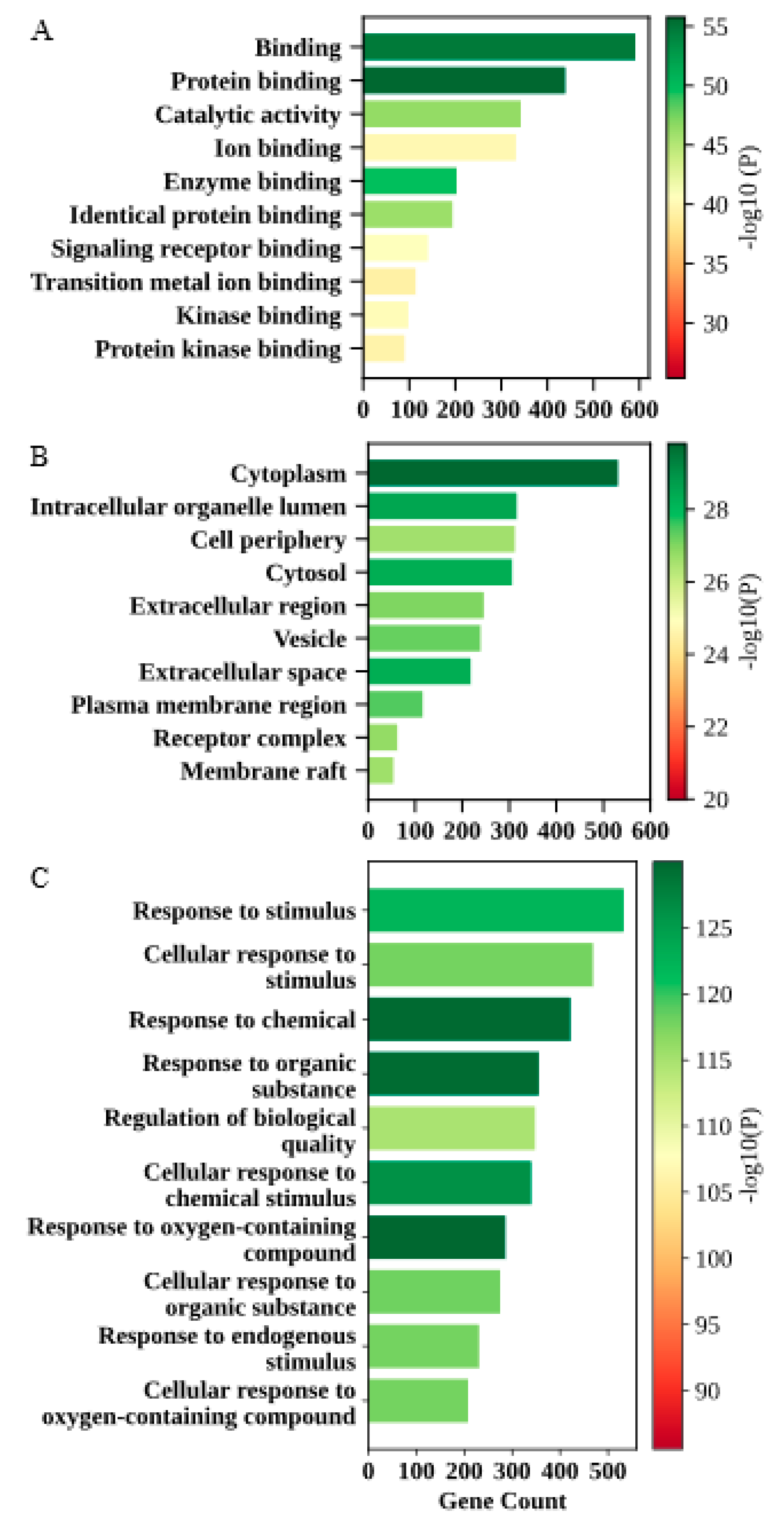
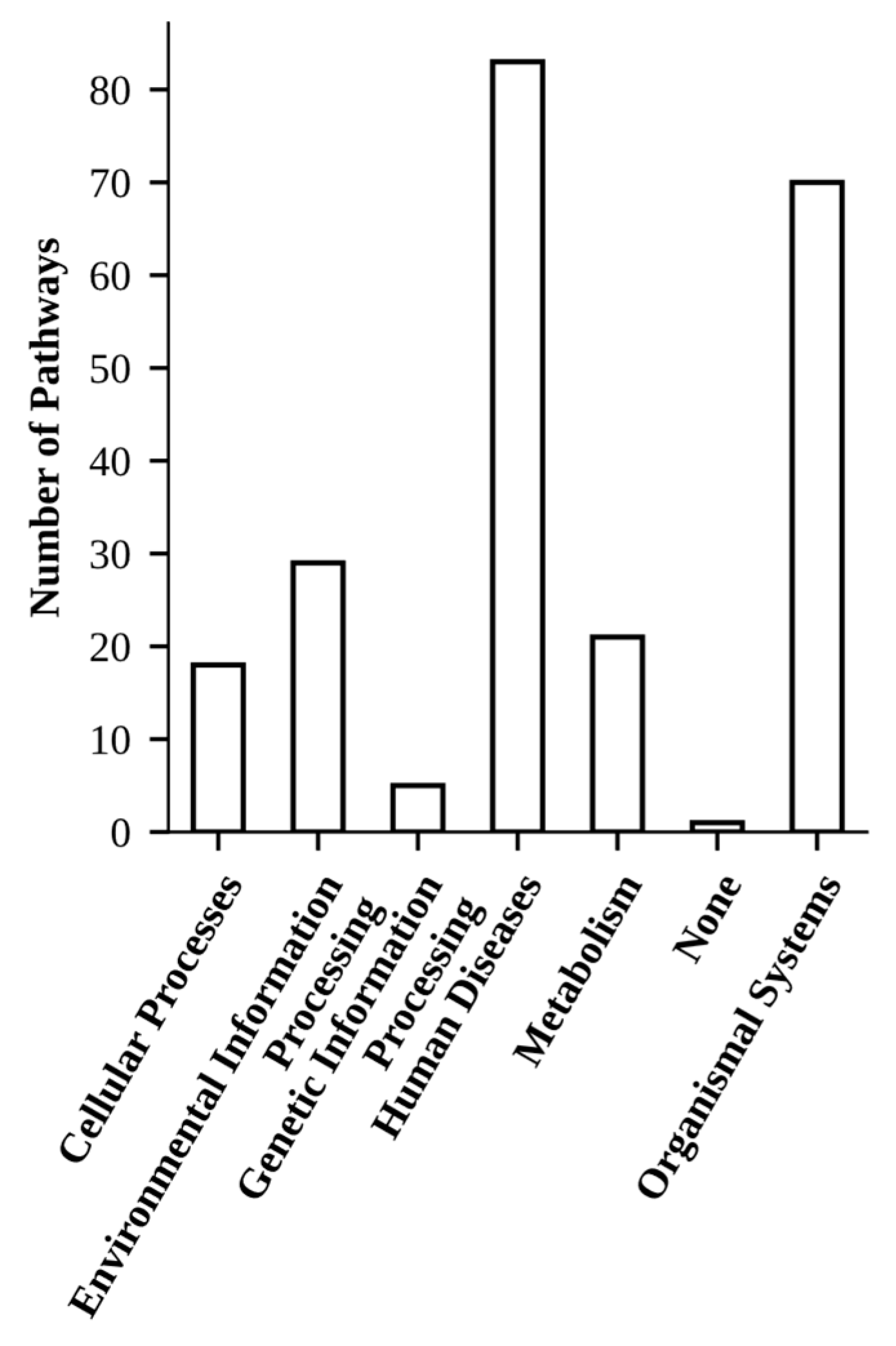
2.2. GO and KEGG Pathway Enrichment
2.3. Community Detection in Target-Pathway-Target Network
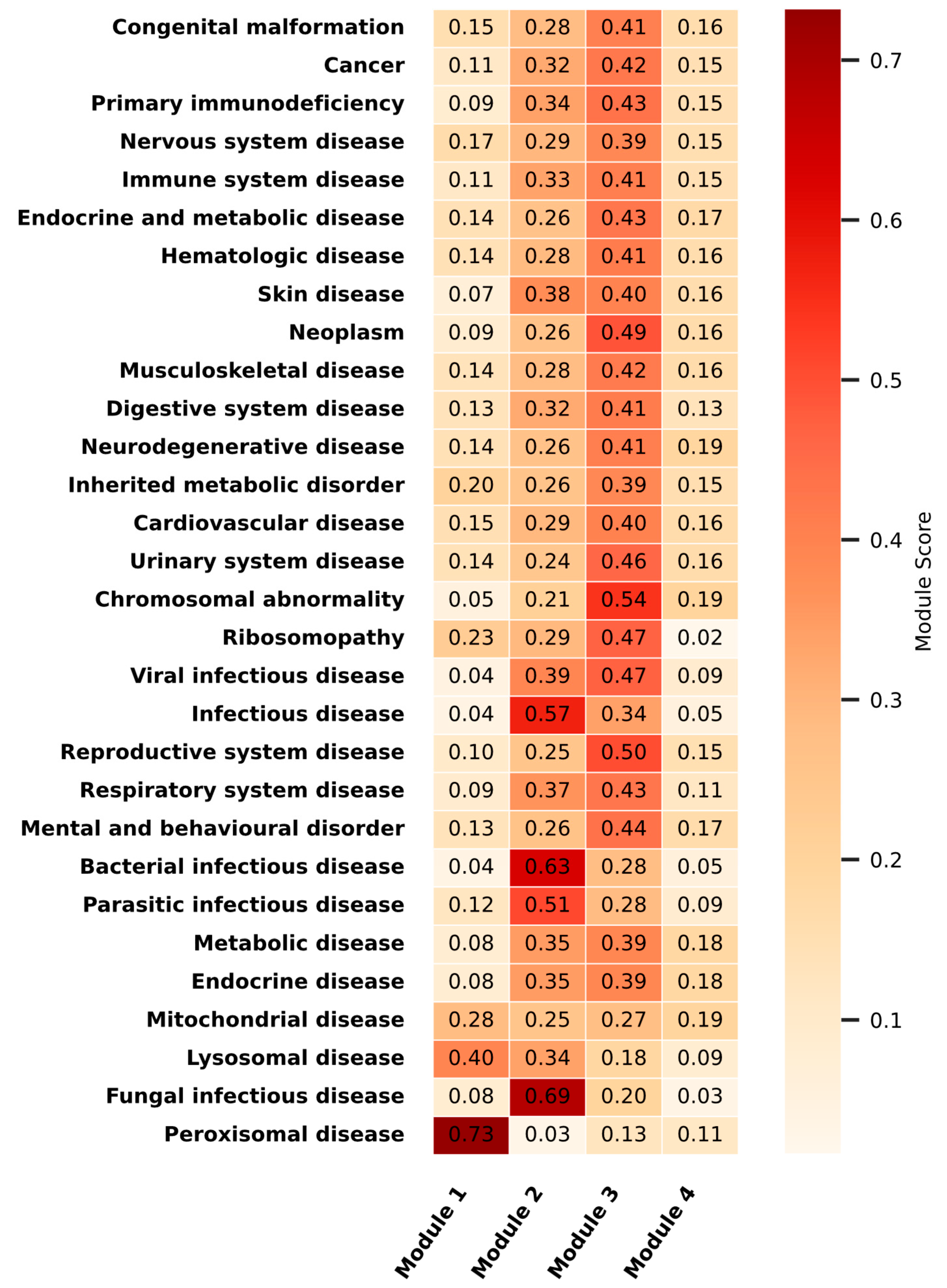
2.4. Disease Association Analysis
3. Materials and Methods
3.1. Compound Acquisition
3.2. Protein Targets Mining
3.3. Functional Enrichment Analysis and Disease Category Mapping
3.4. Network Construction and Module Identification
3.5. Contribution Score Calculation
4. Conclusion
Supplementary Materials
References
- Kwan, K. K. L.; Huang, Y.; Leung, K. W.; Dong, T. T. X.; Tsim, K. W. K. Danggui Buxue Tang, a Chinese herbal decoction containing Astragali Radix and Angelicae Sinensis Radix, modulates mitochondrial bioenergetics in cultured cardiomyoblasts. Front Pharmacol. 2019, 10, 614. [CrossRef]
- Lin, H. Q.; Gong, A. G. W.; Wang, H. Y.; Duan, R.; Dong, T. T. X.; Zhao, K. J.; Tsim, K. W. K. Danggui Buxue Tang (Astragali Radix and Angelicae Sinensis Radix) for menopausal symptoms: A review. J Ethnopharmacol. 2017, 199, 205–10. [CrossRef]
- Gong, A. G.; Zhang, L. M.; Lam, C. T.; Xu, M. L.; Wang, H. Y.; Lin, H.; Dong, T. T.; Tsim, K. W. Polysaccharide of Danggui Buxue Tang, an ancient Chinese herbal decoction, induces expression of pro-inflammatory cytokines possibly via activation of NFKB signaling in cultured raw 264.7 cells. Phytother Res. 2017, 31(2), 274–83. [CrossRef]
- Liu, D.; Mu, Y.; Gao, F.; Zhang, Y.; Shen, Z.; Zhao, Z.; Zhang, P.; Lv, T.; Wang, Y.; Liu, Y. Multi-tissue metabolomics and network pharmacology study on the intervention of Danggui Buxue Decoction in mice with gemcitabine induced myelosuppression. J Ethnopharmacol. 2025, 343, 119498. [CrossRef]
- Guo, R.; Guo, S.; Gao, X.; Wang, H.; Hu, W.; Duan, R.; Dong, T. T. X.; Tsim, K. W. K. Fermentation of Danggui Buxue Tang, an ancient Chinese herbal mixture, together with Lactobacillus plantarum enhances the anti-diabetic functions of herbal product. Chin Med. 2020, 15(1), 98. [CrossRef]
- Huang, Y.; Kwan, K. K. L.; Leung, K. W.; Wang, H.; Kong, X. P.; Dong, T. T. X.; Tsim, K. W. K. The extracts and major compounds derived from Astragali Radix alter mitochondrial bioenergetics in cultured cardiomyocytes: comparison of various polar solvents and compounds. Int J Mol Sci. 2018, 19(6), 1574. [CrossRef]
- Gong, A. G. W.; Huang, V. Y.; Wang, H. Y.; Lin, H. Q.; Dong, T. T. X.; Tsim, K. W. K. Ferulic acid orchestrates anti-oxidative properties of Danggui Buxue Tang, an ancient herbal decoction: elucidation by chemical knock-out approach. PLOS ONE. 2016, 11(11), e0165486. [CrossRef]
- Li, C.; Zhu, F.; Wang, S.; Wang, J.; Wu, B. Danggui Buxue Decoction ameliorates inflammatory bowel disease by improving inflammation and rebuilding intestinal mucosal barrier. Evid Based Complement Alternat Med. 2021, 2021, 1–12. [CrossRef]
- Shi, X. Q.; Yue, S. J.; Tang, Y. P.; Chen, Y. Y.; Zhou, G. S.; Zhang, J.; Zhu, Z. H.; Liu, P.; Duan, J. A. A network pharmacology approach to investigate the blood enriching mechanism of Danggui Buxue Decoction. J Ethnopharmacol. 2019, 235, 227–42. [CrossRef]
- Hua, Y. L.; Ma, Q.; Yuan, Z. W.; Zhang, X. S.; Yao, W. L.; Ji, P.; Hu, J. J.; Wei, Y. M. A novel approach based on metabolomics coupled with network pharmacology to explain the effect mechanisms of Danggui Buxue Tang in anaemia. Chin J Nat Med. 2019, 17(4), 275–90. [CrossRef]
- Liu, Y.; Ju, Y.; Qin, X. Studies on the compatibility mechanism and material basis of Danggui Buxue Decoction against anemia mice using metabonomics and network pharmacology. J Pharm Pharmacol. 2021, 73(6), 767–77. [CrossRef]
- Yu, B.; Lv, G.; Li, Z.; Li, Y.; Xu, H. Utilizing bioinformatics technology to explore the potential mechanism of Danggui Buxue Decoction against NSCLC. Dis Markers. 2022, 2022, 5296830. [CrossRef]
- Zhu, D.; Li, S.; Xu, L.; Ren, X.; Wang, S.; Chen, J.; Zhao, E.; Zheng, Z. Investigation of the molecular mechanism of Danggui Buxue Tang in treating lung cancer using network pharmacology and molecular docking techniques. Nat Prod Res. 2025, 39(11), 3312–5. [CrossRef]
- Liu, W. J.; Ma, S. B.; Li, J. X.; Fan, B. S.; Du, Y.; Xu, Z. H.; Li, X. Q.; Cao, W.; Tang, Y. P. Explore the key targets and mechanism of Danggui Buxue Decoction against ulcerative colitis: Network pharmacology and experimental validation. J Ethnopharmacol. 2025, 344, 119580. [CrossRef]
- Xu, H.; Zhang, T.; He, L.; Yuan, M.; Yuan, X.; Wang, S. Exploring the mechanism of Danggui Buxue Decoction in regulating atherosclerotic disease network based on integrated pharmacological methods. Biosci Rep. 2021, 41(10), BSR20211429. [CrossRef]
- Zheng, G.; Cao, X.; Jing, Y.; Wang, L.; Yan, R.; Ji, Y.; Zhang, Y.; Li, H.; Wang, Y.; Shi, Y.; Yu, Y.; Xiong, Q. An integrative approach for mechanistic insights into the atherosclerotic plaque-stabilizing properties of Danggui Buxue Decoction. J Ethnopharmacol. 2025, 343, 119450. [CrossRef]
- Shen, C.; Chen, Q.; Chen, S.; Lin, Y. Mechanism of Danggui Buxue Decoction in the treatment of myocardial infarction based on network pharmacology and experimental identification. Heliyon. 2024, 10(8), e29360. [CrossRef]
- Fan, Q.; Liu, X.; Zhang, Y.; Kang, W.; Si, S.; Zhang, H. Integration of metabolomics and network pharmacology technology to explain the effect mechanisms of Danggui Buxue Decoction in vascular dementia. Biomed Chromatogr. 2024, 38(4), e5822. [CrossRef]
- Zuo, H.; Zhang, Q.; Su, S.; Chen, Q.; Yang, F.; Hu, Y. A network pharmacology-based approach to analyse potential targets of traditional herbal formulas: An example of Yu Ping Feng Decoction. Sci Rep. 2018, 8(1), 11418. [CrossRef]
- Chen, J.; Teng, D.; Wu, Z.; Li, W.; Feng, Y.; Tang, Y.; Liu, G. Insights into the molecular mechanisms of Liuwei Dihuang Decoction via network pharmacology. Chem Res Toxicol. 2021, 34(1), 91–102. [CrossRef]
- He, Y.; Sun, M. M.; Zhang, G. G.; Yang, J.; Chen, K. S.; Xu, W. W.; Li, B. Targeting PI3K/AKT signal transduction for cancer therapy. Signal Transduct Target Ther. 2021, 6(1), 425. [CrossRef]
- Deng, R. ming; Zhou, J. The role of PI3K/AKT signaling pathway in myocardial ischemia-reperfusion injury. Int Immunopharmacol. 2023, 123, 110714. [CrossRef]
- Yu, H.; Zhang, W. L.; Ding, X.; Zheng, K. Y. Z.; Ho, C. M.; Tsim, K. W. K.; Lee, Y. K. Optimizing combinations of flavonoids deriving from Astragali Radix in activating the regulatory element of erythropoietin by a feedback system control scheme. Evid Based Complement Alternat Med. 2013, 2013, 1–10. [CrossRef]
- De Andrade, C. M.; De Sá, M. F. S.; Toloi, M. R. T. Effects of phytoestrogens derived from soy bean on expression of adhesion molecules on HUVEC. Climacteric. 2012, 15(2), 186–94. [CrossRef]
- Liu, H.; Yang, H.; Qin, Z.; Chen, Y.; Yu, H.; Li, W.; Zhu, X.; Cai, J.; Chen, J.; Zhang, M. Exploration of the Danggui Buxue Decoction mechanism regulating the balance of ESR and AR in the TP53-AKT signaling pathway in the prevention and treatment of POF. Evid Based Complement Alternat Med. 2021, 2021, 1–16. [CrossRef]
- Lou, X.; Ma, Y.; Deng, J.; Lv, Y.; Li, R.; Shang, M.; Zhang, Q.; Zhang, X.; Hou, T. Exploring the mechanism of Danggui Buxue Decoction against acute renal insufficiency using network pharmacology and molecular docking. Mol Cell Biomech. 2024, 21(3), 388. [CrossRef]
- Bo, H.; He, J.; Wang, X.; Du, R.; Bei, H.; Chen, J.; Wang, J.; Wu, F.; Zhang, W.; Chen, Q. Danggui Buxue Tang promotes the adhesion and migration of bone marrow stromal cells via the focal adhesion pathway in vitro. J Ethnopharmacol. 2019, 231, 90–7. [CrossRef]
- Zhang, Z. T.; Qi, Y.; Chen, P.; Chen, L.; Jiang, Y.; Fan, Z.; Guan, H.; Bai, L.; Liu, J.; Zhao, D.; Yan, G. Dang-Gui-Bu-Xue decoction against diabetic nephropathy via modulating the carbonyl compounds metabolic profile and AGEs/RAGE pathway. Phytomedicine. 2024, 135, 156104. [CrossRef]
- Sun, L.; Yang, Z.; Zhao, W.; Chen, Q.; Bai, H.; Wang, S.; Yang, L.; Bi, C.; Shi, Y.; Liu, Y. Integrated lipidomics, transcriptomics and network pharmacology analysis to reveal the mechanisms of Danggui Buxue Decoction in the treatment of diabetic nephropathy in type 2 diabetes mellitus. J Ethnopharmacol. 2022, 283, 114699. [CrossRef]
- Hu, G.; Yang, P.; Zeng, Y.; Zhang, S.; Song, J. Danggui Buxue Decoction promotes angiogenesis by up-regulation of VEGFR1/2 expressions and down-regulation of sVEGFR1/2 expression in myocardial infarction rat. J Chin Med Assoc. 2018, 81(1), 37–46. [CrossRef]
- Liu, H.; Lian, L.; Hou, L.; Liu, C.; Ren, J.; Qiao, Y.; Wen, S.; Li, Q. Herb pair of Huangqi-Danggui exerts anti-tumor immunity to breast cancer by upregulating PIK3R1. Anim Models Exp Med. 2024, 7(3), 234–58. [CrossRef]
- Zhang, Y.; Li, X.; Shi, Y.; Chen, T.; Xu, Z.; Wang, P.; Yu, M.; Chen, W.; Li, B.; Jing, Z.; Jiang, H.; Fu, L.; Gao, W.; Jiang, Y.; Du, X.; Gong, Z.; Zhu, W.; Yang, H.; Xu, H. ETCM v2.0: An update with comprehensive resource and rich annotations for traditional Chinese medicine. Acta Pharm Sin B. 2023, 13(6), 2559–71. [CrossRef]
- Gao, K.; Liu, L.; Lei, S.; Li, Z.; Huo, P.; Wang, Z.; Dong, L.; Deng, W.; Bu, D.; Zeng, X.; Li, C.; Zhao, Y.; Zhang, W.; Wang, W.; Wu, Y. HERB 2.0: an updated database integrating clinical and experimental evidence for traditional Chinese medicine. Nucleic Acids Res. 2025, 53(D1), D1404–14. [CrossRef]
- Singhal, A.; Cao, S.; Churas, C.; Pratt, D.; Fortunato, S.; Zheng, F.; Ideker, T. Multiscale community detection in Cytoscape. PLOS Comput Biol. 2020, 16(10), e1008239. [CrossRef]


| PubChem CID | Compound name | OBa (%) | DLb |
|---|---|---|---|
| 64971 | Mairin | 55.38 | 0.78 |
| 5318869 | Jaranol | 50.83 | 0.29 |
| 73299 | Hederagenin | 36.91 | 0.75 |
| 15976101 | (3S,8S,9S,10R,13R,14S,17R)-10,13-dimethyl-17-[(2R,5S)-5-propan-2-yloctan-2-yl]-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-3-ol | 36.23 | 0.78 |
| 5281654 | Isorhamnetin | 49.6 | 0.31 |
| 15689655 | 3,9-di-O-methylnissolin | 53.74 | 0.48 |
| Not Found | 5’-hydroxyiso-muronulatol-2’,5’-di-O-glucoside | 41.72 | 0.69 |
| 15689652 | 7-O-methylisomucronulatol | 74.69 | 0.3 |
| 86566448 | 9,10-dimethoxypterocarpan-3-O-β-D-glucoside | 36.74 | 0.92 |
| 14077830 | (6aR,11aR)-9,10-dimethoxy-6a,11a-dihydro-6H-benzofurano [3,2-c]chromen-3-ol | 64.26 | 0.42 |
| 108213 | Bifendate | 31.1 | 0.67 |
| 5280378 | Formononetin | 69.67 | 0.21 |
| 160767 | Isoflavanone | 109.99 | 0.3 |
| 5280448 | Calycosin | 47.75 | 0.24 |
| 5280863 | Kaempferol | 41.88 | 0.24 |
| 6037 | FA | 68.96 | 0.71 |
| 10380176 | (3R)-3-(2-hydroxy-3,4-dimethoxyphenyl)chroman-7-ol | 67.67 | 0.26 |
| 15689653 | Isomucronulatol-7,2’-di-O-glucosiole | 49.28 | 0.62 |
| 5316760 | 1,7-Dihydroxy-3,9-dimethoxypterocarpene | 39.05 | 0.48 |
| 5280343 | Quercetin | 46.43 | 0.28 |
| 222284 | Beta-sitosterol | 36.91 | 0.75 |
| 5280794 | Stigmasterol | 43.83 | 0.76 |
| Modulea | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Top Frequent Pathway identifier | hsa01100 | hsa04621 | hsa05200 | hsa04080 |
| Pathway name | Metabolic pathways | NOD-like receptor signaling pathway | Pathways in cancer | Neuroactive ligand-receptor interaction |
| Repeated times | 6555 | 703 | 3403 | 1081 |
| Total pathways in module | 8328 | 16064 | 25471 | 3732 |
| Number of unique pathways in module | 86 | 145 | 178 | 106 |
| Relevant diseasea | Disease category |
|---|---|
| Ulcerative colitis, chronic non-healing ulcers, Rheumatoid arthritis | Immune system disease |
| Menstrual anemia, myelosuppression, chemotherapy-induced bone marrow suppression, blood deficiency, anemia | Hematologic disease |
| Non-small-cell lung cancer, breast cancer, lung cancer, metastatic colon cancer | Cancer |
| Atherosclerosis, Myocardial infarction, coronary heart disease (CHD) | Cardiovascular disease |
| Diabetic nephropathy (DN) | Metabolic disease; endocrine disease; Urinary system disease |
| Parkinson’s disease, vascular dementia | Neurodegenerative disease |
| Age-related macular degeneration | Nervous system disease |
| Non-proliferative diabetic retinopathy | Endocrine and metabolic disease; Nervous system disease |
| Idiopathic pulmonary fibrosis | Respiratory system disease |
| Premature ovarian failure | Reproductive system disease |
| Disease Categorya | Target Genes |
|---|---|
| Nervous system disease | GSK3B, AKT1, SRC, BAX, CCND1, MAPK3, MAPK1, MTOR, EGFR, RAF1, PIK3R1, PIK3CD, PIK3CA, PLCG1, PRKCB, PRKCA, TP53 |
| Immune system disease | IGF1, AKT1, SRC, BAX, CCND1, MAPK3, MAPK1, MTOR, EGFR, RAF1, PIK3R1, PIK3CD, PIK3CA, PLCG1, PRKCB, PRKCA, TP53 |
| Hematologic disease | GSK3B, AKT1, SRC, BAX, CCND1, MAPK3, MAPK1, MTOR, CDKN1A, EGFR, RAF1, PIK3R1, PIK3CD, PIK3CA, PRKCB, PRKCA, TP53 |
| Neurodegenerative disease | IGF1, GSK3B, AKT1, SRC, BAX, MAPK3, MAPK1, MTOR, EGFR, RAF1, PIK3R1, PIK3CD, PIK3CA, PLCG1, PRKCB, PRKCA, TP53 |
| Cardiovascular disease | IGF1, GSK3B, AKT1, SRC, CCND1, MAPK3, MAPK1, MTOR, EGFR, RAF1, PIK3R1, PIK3CD, PIK3CA, PLCG1, PRKCB, PRKCA, TP53 |
| Urinary system disease | IGF1, GSK3B, AKT1, SRC, CCND1, MAPK3, MAPK1, EGFR, RAF1, PTK2, PIK3R1, PIK3CD, PIK3CA, PLCG1, PRKCB, PRKCA, TP53 |
| Cancer | GSK3B, AKT1, SRC, BAX, CCND1, MAPK3, MAPK1, MTOR, EGFR, RAF1, PIK3R1, PIK3CD, PIK3CA, PLCG1, PRKCB, PRKCA, TP53 |
| Gene Symbola | Uniport ID | Relevant Compound (source herb) b |
| AKT1 | P31749 | Formononetin (AR), Stigmasterol (AS), Kaempferol (AR), Quercetin (AR), Calycosin (AR) |
| EGFR | P00533 | Formononetin (AR), Quercetin (AR), Jaranol (AR), Isorhamnetin (AR), Calycosin (AR) |
| MAPK1 | P28482 | Calycosin (AR), Mairin (AR), Quercetin (AR) |
| MAPK3 | P27361 | Calycosin (AR), Quercetin (AR) |
| PIK3CA | Calycosin (AR), Formononetin (AR), Quercetin (AR) | |
| PIK3CD | P42336 | Calycosin (AR) |
| PIK3R1 | P27986 | Quercetin (AR) |
| PRKCA | P17252 | Beta-sitosterol (ASR), Quercetin (AR) |
| PRKCB | P05771 | Mairin (AR), Quercetin (AR) |
| RAF1 | P04049 | Quercetin (AR) |
| SRC | P12931 | Calycosin (AR), Quercetin (AR) |
| TP53 | P04637 | Formononetin (AR), Quercetin (AR) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





