Submitted:
11 October 2025
Posted:
15 October 2025
Read the latest preprint version here
Abstract
Keywords:
1. Introduction
2. Biogenesis and Functional Mechanisms of microRNAs
3. Exosomes and Exosomal microRNAs: Biogenesis and Selective Packaging

4. Physiological Roles of Exosomal miRNAs
5. Pathological Roles of Exosomal miRNAs in Diseases
6. Liver Cancer: Pathogenesis and Clinical Challenges
7. Roles of Exosomal microRNAs in Liver Cancer
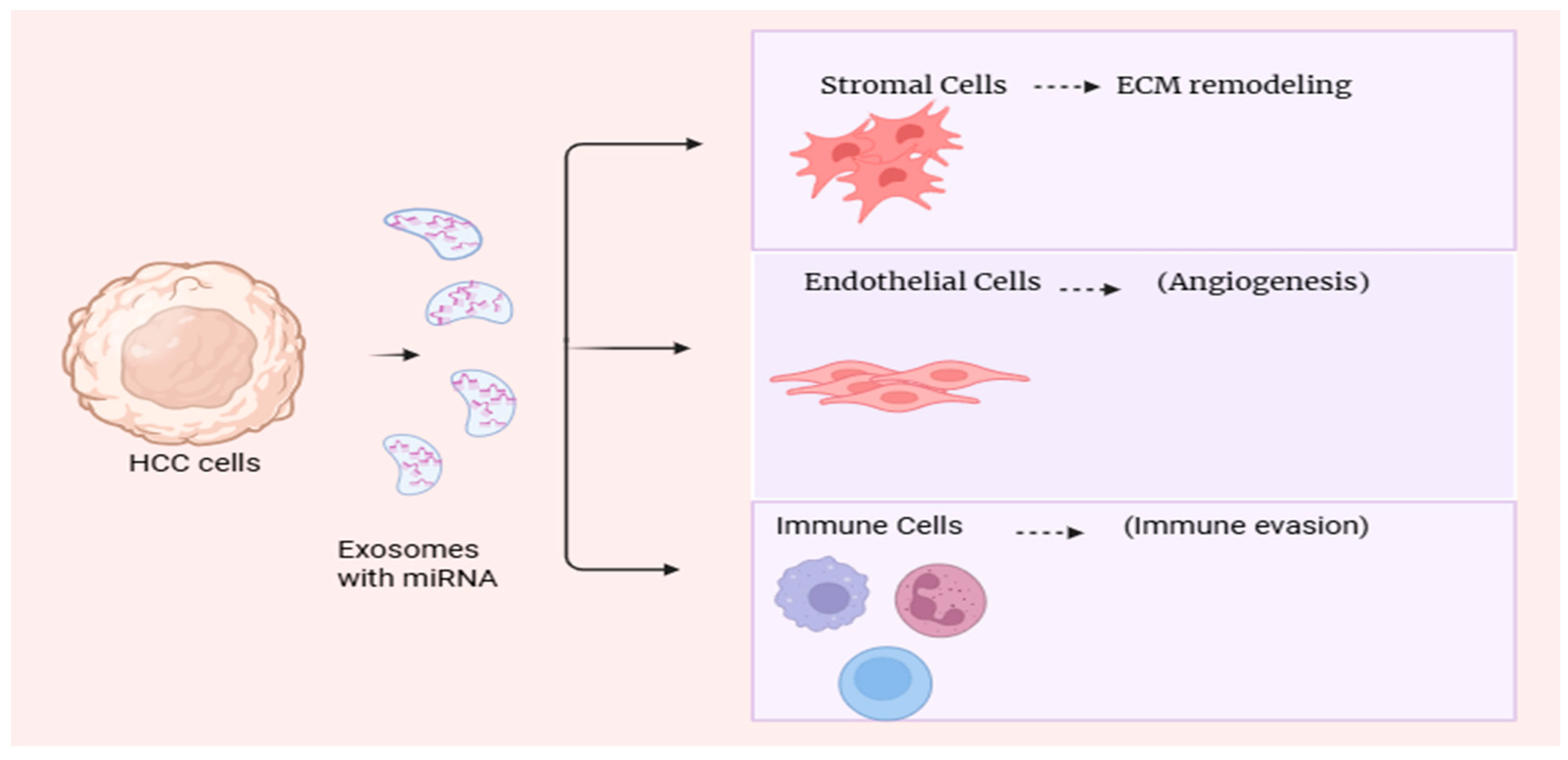
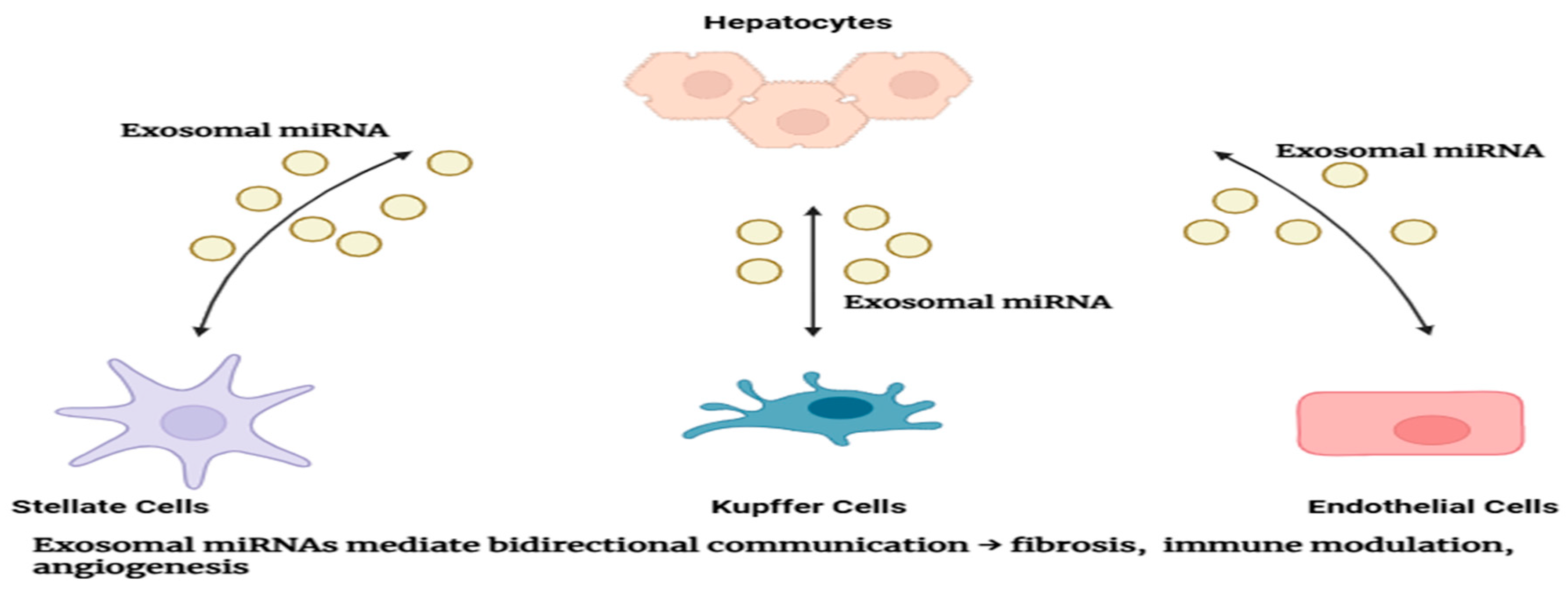
8. Therapeutic Implications and Potential
9. Emerging Technologies and Future Directions
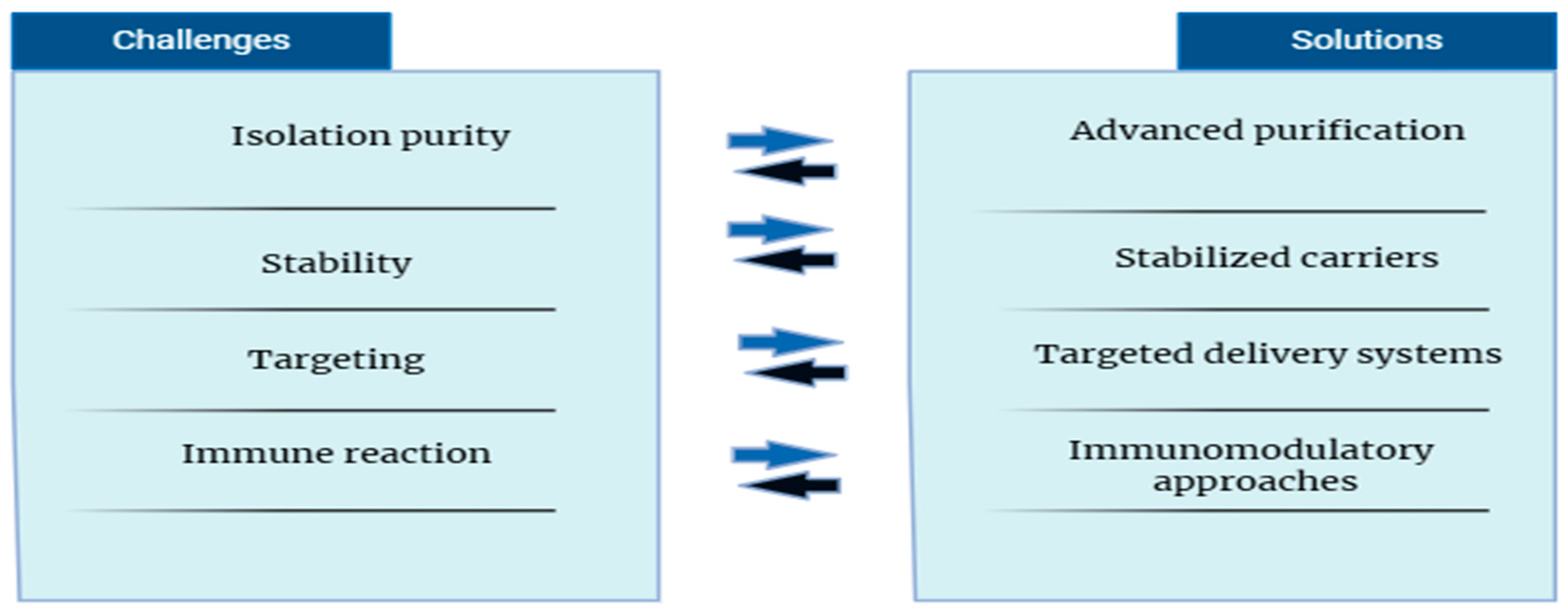
10. Conclusions
Author Contributions
Acknowledgments
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Llovet, J.M.; Zucman-Rossi, J.; Pikarsky, E.; Sangro, B.; Schwartz, M.; Sherman, M. , et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016, 2, 16018. [Google Scholar] [CrossRef] [PubMed]
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–14. [Google Scholar] [CrossRef] [PubMed]
- Trevisani, F.; D'Intino, P.E.; Morselli-Labate, A.M.; Mazzella, G.; Accogli, E.; Caraceni, P.; Domenicali, M.; De Notariis, S.; Roda, E.; Bernardi, M. Serum α-fetoprotein for diagnosis of hepatocellular carcinoma in patients with chronic liver disease: influence of HBsAg and anti-HCV status. J. Hepatol. 2001, 34, 570–575. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004, 116, 281–97. [Google Scholar] [CrossRef] [PubMed]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006, 6, 857–66. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Kogure T, Yan IK, Lin WL, Patel T. Extracellular Vesicle–Mediated Transfer of a Liver-Specific MicroRNA, miR-122, Regulates Hepatic Lipid Metabolism. Hepatology. 2011, 54, 1164–74. [Google Scholar]
- He, C.; Zheng, S.; Luo, Y.; Wang, B. Exosome Theranostics: Biology and Translational Medicine. Theranostics 2018, 8, 237–255. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.-H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef] [PubMed]
- Denli, A.M.; Tops, B.B.J.; Plasterk, R.H.A.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Lund, E.; Güttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear Export of MicroRNA Precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.S.; Hutvágner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the Assembly of the RNAi Enzyme Complex. Cell 2003, 115, 199–208. [Google Scholar] [CrossRef]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006, 6, 259–69. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Gutiérrez-Vázquez, C.; Sánchez-Cabo, F.; Pérez-Hernández, D.; Vázquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [PubMed]
- Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, Schekman R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife. 2016, 5, e19276. [Google Scholar] [CrossRef] [PubMed]
- Guduric-Fuchs, J.; O’connor, A.; Camp, B.; O'Neill, C.L.; Medina, R.J.; A Simpson, D. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genom. 2012, 13, 357–357. [Google Scholar] [CrossRef]
- Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011, 108, 5003–8. [Google Scholar] [CrossRef]
- Montecalvo, A.; Larregina, A.T.; Shufesky, W.J.; Beer Stolz, D.; Sullivan, M.L.G.; Karlsson, J.M.; Baty, C.J.; Gibson, G.A.; Erdos, G.; Wang, Z.; et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 2012, 119, 756–766. [Google Scholar] [CrossRef]
- Mittelbrunn, M.; Sánchez-Madrid, F. Intercellular communication: diverse structures for exchange of genetic information. Nat. Rev. Mol. Cell Biol. 2012, 13, 328–335. [Google Scholar] [CrossRef]
- Bruno, S.; Grange, C.; Deregibus, M.C.; Calogero, R.A.; Saviozzi, S.; Collino, F.; Morando, L.; Busca, A.; Falda, M.; Bussolati, B.; et al. Mesenchymal Stem Cell-Derived Microvesicles Protect Against Acute Tubular Injury. J. Am. Soc. Nephrol. 2009, 20, 1053–1067. [Google Scholar] [CrossRef]
- Chen L, Zhang S, Wang J, et al. Exosomes derived from hepatocellular carcinoma cells induce activation of hepatic stellate cells through transferring miR-21. Cancer Sci. 2018, 109, 1965–76. [Google Scholar]
- Roderburg C, Luedde T. The role of the microRNA-29 family in liver fibrosis and hepatocellular carcinoma. J Hepatol. 2014, 61, 507–8. [Google Scholar]
- T, T.; M, M.; J, D.; M, K.; M, S.; C, W.; T, N.R.; J, W.; R, G.-M.; S, G.; et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Yearb. Paediatr. Endocrinol. 2018. [Google Scholar] [CrossRef]
- Xu, R.; Rai, A.; Chen, M.; Suwakulsiri, W.; Greening, D.; Simpson, R. Extracellular vesicles in cancer — implications for future improvements in cancer care. [CrossRef]
- Verma SK, Baliyan S, Patil V, et al. Role of extracellular vesicles in liver fibrosis: a concise review. Int J Mol Sci. 2020;21(21):7746.
- Zhang, L.; Yu, D. Exosomes in cancer development, metastasis, and immunity. Biochim. Biophys. Acta (BBA) Rev. Cancer 2019, 1871, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Li, Y.; Pan, Y.; Lan, X.; Song, F.; Sun, J.; Zhou, K.; Liu, X.; Ren, X.; Wang, F.; et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat. Commun. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Mark, M.T.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Chen WX, Liu XM, Lv MM, et al. Exosomal miR-21 regulates the sensitivity of breast cancer cells to doxorubicin by targeting PTEN. J Cell Mol Med. 2018, 22, 5385–97. [Google Scholar]
- Schwarzenbach, H.; Nishida, N.; Calin, G.A.; Pantel, K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 2014, 11, 145–156. [Google Scholar] [CrossRef]
- El-Serag, H.B. Hepatocellular carcinoma. N Engl J Med. 2011, 365, 1118–27. [Google Scholar] [CrossRef]
- Villanueva, A. Hepatocellular carcinoma. N Engl J Med. 2019, 380, 1450–62. [Google Scholar] [CrossRef]
- Llovet, J.M.; Ricci, S.; Mazzaferro, V. , et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008, 359, 378–90. [Google Scholar] [CrossRef]
- Sugimachi, K.; Matsumura, T.; Hirata, H.; Uchi, R.; Ueda, M.; Ueo, H.; Shinden, Y.; Iguchi, T.; Eguchi, H.; Shirabe, K.; et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br. J. Cancer 2015, 112, 532–538. [Google Scholar] [CrossRef]
- Lin XJ, Gao W, Wan J, et al. Serum exosomal miR-122 and miR-148a are promising biomarkers for early diagnosis of hepatocellular carcinoma. J Cancer. 2019, 10, 4582–9. [Google Scholar]
- Zhang X, Yang J, Li L, et al. A novel panel of serum exosomal microRNAs for early diagnosis of hepatocellular carcinoma. J Cell Biochem. 2019, 120, 17322–30. [Google Scholar]
- Lou, G.; Chen, Z.; Zheng, M.; Liu, Y. Mesenchymal stem cell-derived exosomes as a new therapeutic strategy for liver diseases. Exp. Mol. Med. 2017, 49, e346–e346. [Google Scholar] [CrossRef] [PubMed]
- Shi M, Zhang Z, Xu R, et al. Exosomal miR-103a-3p promotes hepatocellular carcinoma metastasis by targeting SFRP4 and activating Wnt/β-catenin signaling. Mol Ther Nucleic Acids. 2020, 22, 1–15. [Google Scholar]
- Wang B, Yao K, Huuskes BM, Shen HH, Zhuang J, Godson C. Exosomes from M2 macrophages promote angiogenesis in hepatocellular carcinoma by transferring miR-21. J Exp Clin Cancer Res. 2018, 37, 132. [Google Scholar]
- Xu H, Ma Q, Liu W, et al. Exosomal miR-199a-3p promotes sorafenib resistance in hepatocellular carcinoma. Mol Cancer. 2020, 19, 148. [Google Scholar]
- Qu, L.; Ding, J.; Chen, C.; Wu, Z.-J.; Liu, B.; Gao, Y.; Chen, W.; Liu, F.; Sun, W.; Li, X.-F.; et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell 2016, 29, 653–668. [Google Scholar] [CrossRef]
- Kannan M, Kaur G, Haque SJ. Fibrosis and hepatocellular carcinoma: molecular connections and therapeutic targets. Front Pharmacol. 2019, 10, 994. [Google Scholar]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Liang G, Zhu Y, Ali DJ, Tian T, Chen X. Engineered exosomes for targeted drug delivery. Theranostics. 2021, 11, 3183–95. [Google Scholar] [CrossRef]
- Essandoh K, Li Y, Huo J, Fan GC. Exosomes as a nanocarrier for gene therapy: Progress and challenges. Nanomedicine. 2015, 11, 3219–32. [Google Scholar]
- Luan, X.; Sansanaphongpricha, K.; Myers, I.; Chen, H.; Yuan, H.; Sun, D. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol. Sin. 2017, 38, 754–763. [Google Scholar] [CrossRef]
- ClinicalTrials.gov [Internet]. Bethesda (MD): National Library of Medicine (US). Identifier NCT03608631, Exosomes in liver cancer (HCC) diagnosis and therapy; 2020 May 1 [cited 2025 Oct 3]. Available online: https://clinicaltrials.gov/ct2/show/NCT03608631 (accessed on 3 October 2025).
- Zhang P, Zhou X, He M, Shang Y, Tetlow AL, Godwin AK, et al. Ultrasensitive microfluidic analysis of circulating exosomes using a nanostructured graphene oxide–polyethyleneimine coating. Lab Chip. 2016, 16, 3033–42. [Google Scholar] [CrossRef] [PubMed]
- Nordin, J.Z.; Lee, Y.; Vader, P.; Mäger, I.; Johansson, H.J.; Heusermann, W.; Wiklander, O.P.B.; Hällbrink, M.; Seow, Y.; Bultema, J.J.; et al. Ultrafiltration with size-exclusion liquid chromatography for high yield isolation of extracellular vesicles preserving intact biophysical and functional properties. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 879–883. [Google Scholar] [CrossRef]
- Kalluri, R. The biology and function of exosomes in cancer. J. Clin. Investig. 2016, 126, 1208–1215. [Google Scholar] [CrossRef]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
| Step | Key Molecules/Proteins | Description | Relevance to Exosomal Packaging | Reference(s) |
|---|---|---|---|---|
| Transcription | RNA Polymerase II | Primary miRNA (pri-miRNA) synthesis | Initial step; source of all miRNAs | [5,11] |
| Nuclear processing | Drosha, DGCR8 | Processing pri-miRNA to precursor miRNA (pre-miRNA) | Generates pre-miRNA for export | [12] |
| Nuclear export | Exportin-5 | Transports pre-miRNA to cytoplasm | Enables cytoplasmic processing | [13] |
| Cytoplasmic processing | Dicer | Converts pre-miRNA into mature miRNA duplex | Produces mature miRNAs, ready for function | [14] |
| RISC loading | Argonaute proteins (Ago2) | Assembly into RNA-induced silencing complex (RISC) | Guides miRNA targeting; selective exosomal sorting | [15,24] |
| Exosomal sorting | hnRNPA2B1, YBX1 | RNA-binding proteins mediate selective packaging | Determines miRNA export via exosomes | [21,22] |
| miRNA | Sample Type | Diagnostic/Prognostic Utility | Sensitivity/Specificity (if available) | Reference(s) |
|---|---|---|---|---|
| miR-21 | Serum exosomes | Early diagnosis, poor prognosis marker | Sensitivity ~85%, Specificity ~80% | [28,36] |
| miR-122 | Plasma exosomes | Early detection biomarker | Sensitivity ~90%, Specificity ~85% | [9,42] |
| miR-148a | Serum exosomes | Predicts recurrence after treatment | Data limited | [42,43] |
| miR-221 | Serum exosomes | Associated with aggressive tumor behavior | Data limited | [7] |
| miR-199a-3p | Serum exosomes | Predicts resistance to sorafenib therapy | Data limited | [47] |
| miRNA | Expression Pattern | Target Genes/Pathways | Functional Role in HCC | Reference(s) |
|---|---|---|---|---|
| miR-21 | Upregulated | PTEN, PDCD4 | Promotes proliferation, invasion | [28,46] |
| miR-122 | Downregulated | Cyclin G1, ADAM17 | Tumor suppressor, regulates metabolism | [9,42] |
| miR-199a | Downregulated | mTOR, c-Met | Suppresses tumor growth | [47] |
| miR-221 | Upregulated | CDKN1B, PTEN | Enhances proliferation and survival | [7] |
| miR-25-3p | Upregulated | Notch signaling pathway | Promotes metastasis | [34] |
| miR-148a | Downregulated | DNMT1 | Tumor suppressor | [42,43] |
| miR-103a | Upregulated | SFRP4, Wnt/β-catenin | Promotes metastasis and EMT | [45] |
| Therapeutic Strategy | Target miRNA(s) | Mode of Delivery | Preclinical/Clinical Status | Outcomes/Notes | Reference(s) |
|---|---|---|---|---|---|
| miRNA mimics | miR-122, miR-199a | Lipid nanoparticles, exosomes | Preclinical | Suppression of tumor growth in vivo | [44,45] |
| Anti-miRNA oligonucleotides | miR-21, miR-221 | Systemic administration | Preclinical | Reduced tumor proliferation and metastasis | [28,46] |
| Exosome-based drug delivery | Various miRNAs | Engineered exosomes | Early clinical/preclinical | Improved targeting, reduced off-target effects | [51,52,53] |
| Combination therapies | miRNAs + Sorafenib | Co-delivery via nanoparticles | Preclinical | Overcomes drug resistance | [47,48] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





