Submitted:
07 October 2025
Posted:
08 October 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
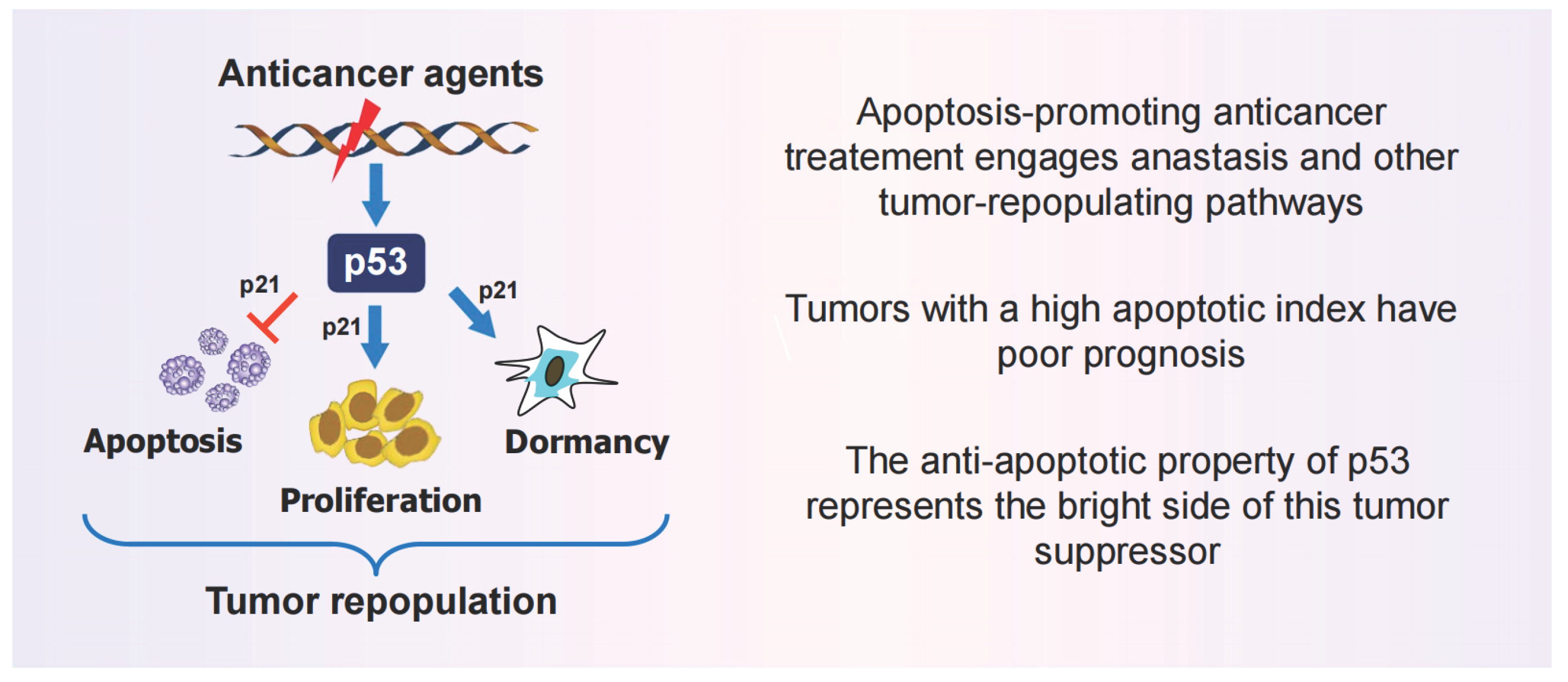
2. Is Apoptosis a Tumor Suppression Mechanism?
2.1. Precision Oncology Targeting Apoptosis: Reality or False Promises?
2.2. The Apoptosis-Anastasis Tumor-Repopulating Pathway
2.3. Other Apoptosis-Related Tumor-Repopulating Pathways
2.4. Increased Apoptosis in Solid Tumors is linked to an Unfavorable Clinical Outcome
2.5. Take Home Messages
3. Apoptotic Cancer Cells Promote Tumor Diversity and Heterogeneity
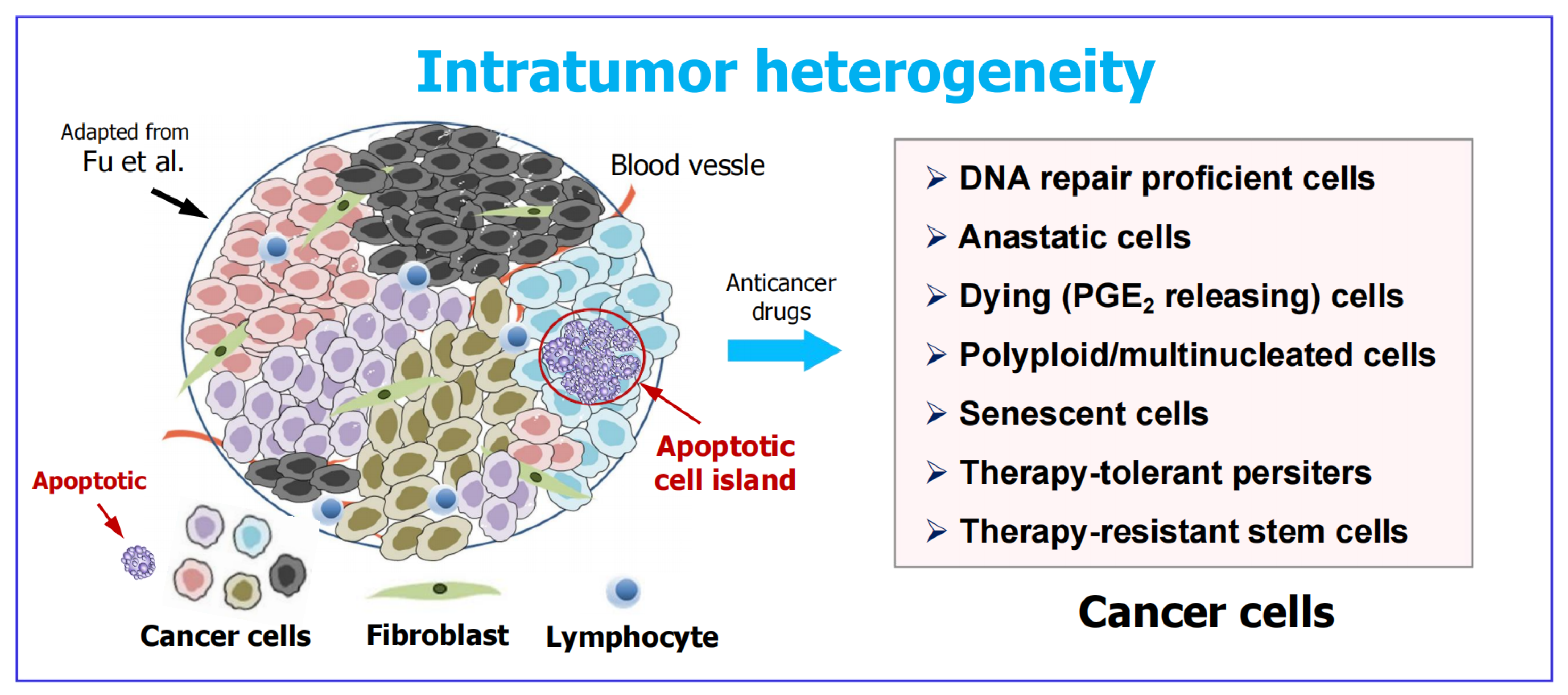
4. Intratumor Heterogeneity: A Well Established (Yet Widely Overlooked) Obstacle in Cancer Therapy
- Is cancer cell dormancy a greater threat in managing solid tumors or treacherous apoptosis (encompassing anastasis)? Probably the former is a bigger fish to fry based on reasons discussed previously [3,89,90]. For example, judging from tissue culture studies, clinically relevant anticancer exposure (radiation, drugs) triggers cancer cell dormancy but rarely engages regulated cell death [3,89,90,91]. This observation gives credence to the emerging trend of deintensification in cytotoxic cancer therapy [92], which would be expected to minimize the occurrence of side effects as well as regulated cell death and other tumor-repopulating events [4].
- Given that intratumor heterogeneity was well established over two decades ago [83], why did it take so long for most cancer research community members to appreciate its impact on resistance and relapse? Who knows! Perhaps “in the quest for the next cancer cure, few researchers bother to look back at the graveyard of failed medicines to figure out what went wrong” [93].
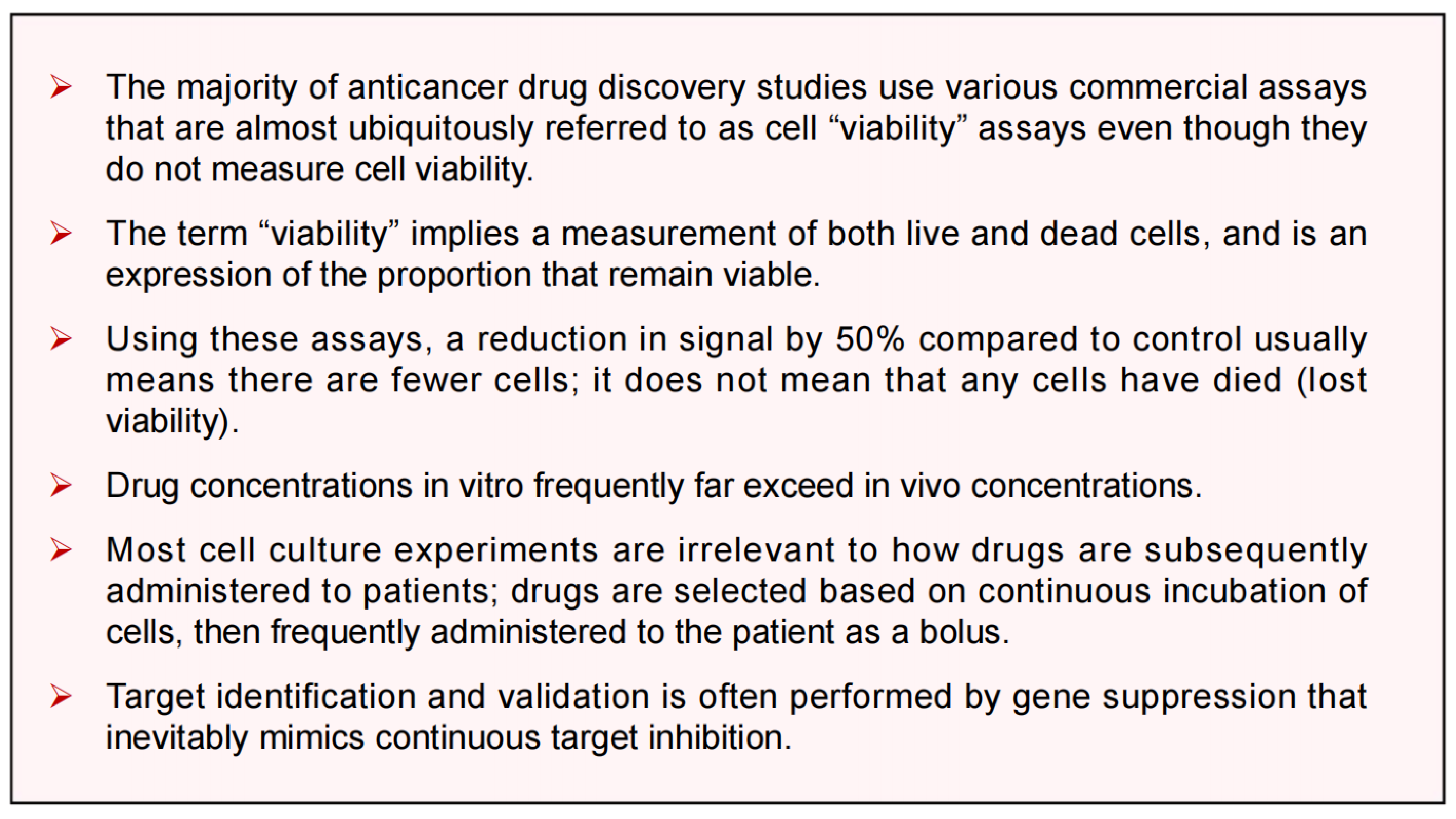
5. What Are the Reasons for Repeated Failures in Treating Solid Tumor Malignancies?
5.1. Most Preclinical Anticancer Studies Generate Clinically Irrelevant Information
5.2. The Consequences of Dishonesty in Data Reporting
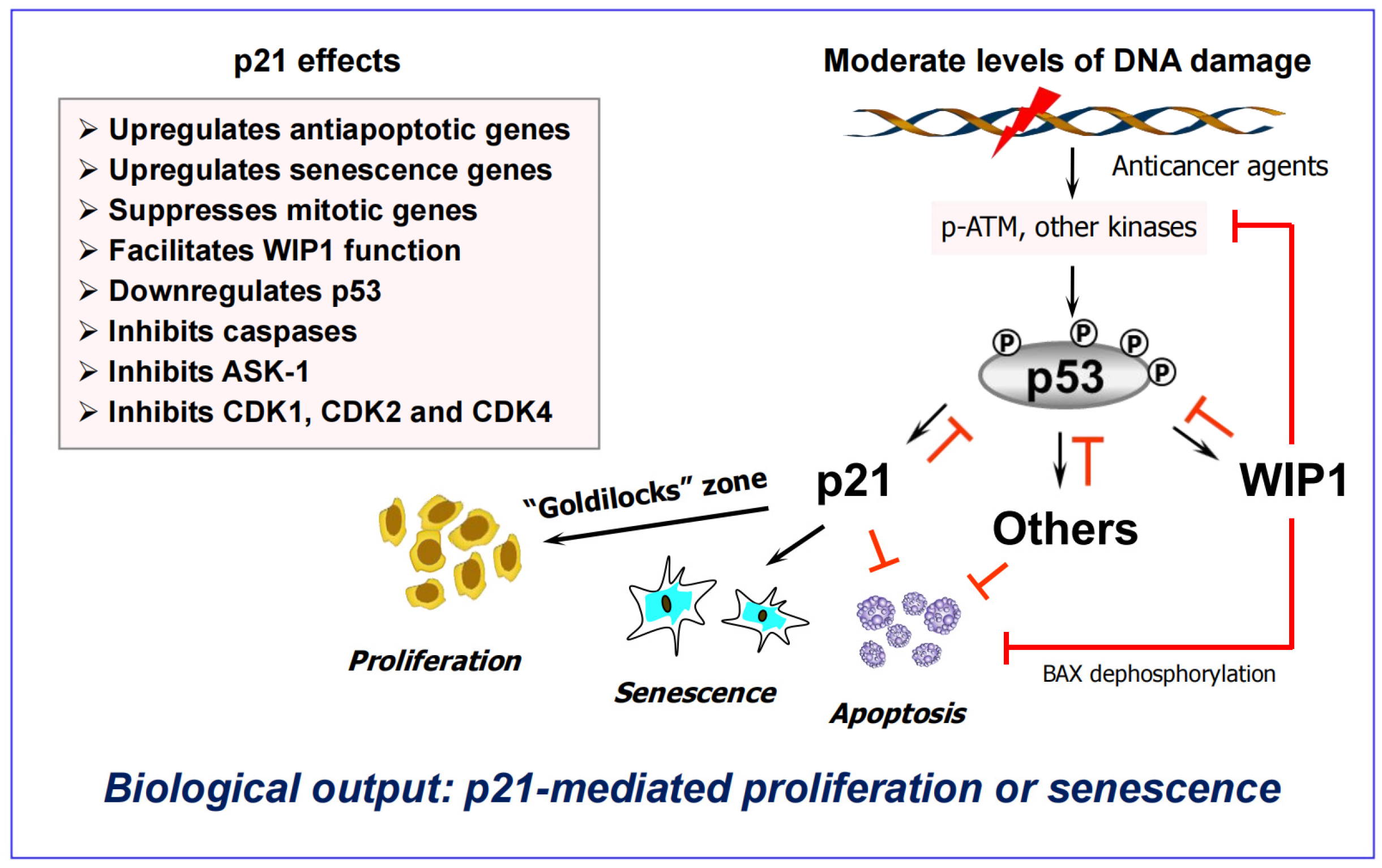
6. Activation of Wild-Type p53 Signaling Following Clinically Relevant Anticancer Treatment Serves to Suppress (“Treacherous”) Apoptosis
6.1. Impact of p53 on Apoptosis Under Non-Physiological Versus Clinically-Relevant Conditions
- ➢
- ➢
- Strong p53 activation (above the apoptotic threshold) is observed under non-physiological conditions, such as cell exposure to very high doses of genotoxic agents (cisplatin; UVC) that induce bulky (transcription-blocking) DNA lesions [115].
- ➢
- Under these conditions, bulky lesions prevent transcriptional activation of MDM2 and other p53 negative regulators (e.g., p21, WIP1), resulting in a strong accumulation of p53 protein that triggers apoptosis presumably via its proline-rich region [115].
- ➢
- ➢
6.2. The “Goldilocks Zone” for Cancer Cell Proliferation Following Clinically Relevant Chemotherapy Exposure
7. Conclusions
7.1. Who Would Disregard the Treacherous Side of Apoptosis in Treating Solid Tumors?
7.2. Call For Contrarian Logic In Cancer Research
- ➢
- What is apoptosis? Is it an irreversible mode of cell death based on cell “viability” and other misleading preclinical assays? Or engaging apoptosis in solid tumors represents a treacherous, tumor repopulating outcome? (I think it is the latter.)
- ➢
- Is “evading apoptosis” a hallmark of cancer, contributing to tumor progression and therapy resistance, as hypothesized by Hanahan and Weinberg over two decades ago [125]? Or, like normal cells, cancer cells simply employ the homeostatic process of anastasis to survive after engaging regulated cell death? (I think it is the latter.)
- ➢
- Is deregulated anastasis a hallmark of cancer? The availability of anastasis markers such as cell surface CD24 expression will hopefully lead to addressing this and other outstanding questions in cancer progression and therapy.
Funding
Conflicts of Interest
Appendix A
References
- Pabla, S. Contrarian Thinking in Bioinformatics: Unlocking Breakthroughs by Challenging Assumptions. 2025. Available online: https://www.linkedin.com/pulse/contrarian-thinking-bioinformatics-unlocking-sarabjot-pabla-y693e (accessed on 5 October 2025).
- Mirzayans, R.; Murray, D. What are the reasons for continuing failures in cancer therapy? Are misleading/inappropriate preclinical assays to be blamed? Might some modern therapies cause more harm than benefit? Int. J. Mol. Sci. 2022, 23, 13217. [Google Scholar] [CrossRef] [PubMed]
- Mirzayans, R. Changing the landscape of solid tumor therapy from apoptosis-promoting to apoptosis-inhibiting strategies. Curr. Issues Mol. Biol. 2024, 46, 5379–5396. [Google Scholar] [CrossRef]
- Mirzayans, R. Anastasis and other apoptosis-related prosurvival pathways call for a paradigm shift in oncology: Significance of deintensification in treating solid tumors. Int. J. Mol. Sci. 2025, 26, 1881. [Google Scholar] [CrossRef] [PubMed]
- Prasad, V. Perspective: The precision-oncology illusion. Nature 2016, 537, S63. [Google Scholar] [CrossRef]
- Maeda, H.; Khatami, M. Analyses of repeated failures in cancer therapy for solid tumors: Poor tumor-selective drug delivery, low therapeutic efficacy and unsustainable costs. Clin. Transl. Med. 2018, 7, 11. [Google Scholar] [CrossRef]
- Joyner, M.J.; Paneth, N. Promises, promises, and precision medicine. J. Clin. Invest. 2019, 129, 946–948. [Google Scholar] [CrossRef]
- Marine, J.C.; Dawson, S.J.; Dawson, M.A. Non-genetic mechanisms of therapeutic resistance in cancer. Nat. Rev. Cancer 2020, 20, 743–756. [Google Scholar] [CrossRef]
- Pich, O.; Bailey, C.; Watkins, T.B.K.; Zaccaria, S.; Jamal-Hanjani, M.; Swanton, C. The translational challenges of precision oncology. Cancer Cell 2022, 40, 458–478. [Google Scholar] [CrossRef]
- Heng, J.; Heng, H.H. Genome chaos, information creation, and cancer emergence: Searching for new frameworks on the 50th anniversary of the “war on cancer”. Genes 2022, 13, 101. [Google Scholar] [CrossRef]
- Lohse, S. Mapping uncertainty in precision medicine: A systematic scoping review. J. Eval. Clin. Pract. 2023, 29, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Fojo, T. Journeys to failure that litter the path to developing new cancer therapeutics. JAMA Netw. Open 2023, 6, e2324949. [Google Scholar] [CrossRef] [PubMed]
- Suehnholz, S.P.; Nissan, M.H.; Zhang, H.; Kundra, R.; Nandakumar, S.; Lu, C.; Carrero, S.; Dhaneshwar, A.; Fernandez, N.; Xu, B.W.; et al. Quantifying the expanding landscape of clinical actionability for patients with cancer. Cancer Discov. 2024, 14, 49–65. [Google Scholar] [CrossRef]
- Kailen, W.G. Preclinical cancer target validation: How not to be wrong. NIH Wednesday Afternoon Lectures (WELS) series, January 24, 2018 [https://videocast.nih.gov/watch=27066] (accessed on 5 October 2025).
- Belluz, J. Most cancer drugs fail in testing. This might be a big reason why. Science—VOX blog (2019) https://www.vox.com/2019/9/16/20864066/cancer-studies-fail (acceaassed on 5 October 2025).
- Horgan, J. The cancer industry: hype vs. reality. Cancer medicine generates enormous revenues but marginal benefits for patients (2020) https://www.scientificamerican.com/blog/cross-check/the-cancer-industry-hype-vs-reality/ (accessed on 5 October 2025).
- Horgan, J. The cancer industry: Hype versus reality (2023) https://johnhorgan.org/cross-check/the-cancer-industry-hype-versus-reality (accessed on 5 October 2025).
- Azra, R. The First Cell: And the Human Costs of Pursuing Cancer to the Last. New York, Basic Books, 2019.
- Conley, B.A.; Staudt, L.; Takebe, N.; Wheeler, D.A.; Wang, L.; Cardenas, M.F.; Korchina, V.; Zenklusen, J.C.; McShane, L.M.; Tricoli, J.V.; et al. The Exceptional Responders initiative: Feasibility of a National Cancer Institute pilot study. J. Natl. Cancer Inst. 2021, 113, 27–37. [Google Scholar] [CrossRef]
- Ichim, G.; Tait, S.W.G. A fate worse than death: Apoptosis as an oncogenic process. Nat. Rev. Cancer 2016, 16, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Shekhar, M.P.V. The dark side of apoptosis. In Molecular Mechanisms of Tumor Cell Resistance to Chemotherapy, Resistance to Targeted Anti-Cancer Therapeutics 1; Bonavida, B., Ed.; Springer: New York, NY, USA, 2013; pp. 245–258.
- Dhanasekaran, R. Treacherous apoptosis—Cancer cells sacrifice themselves at the altar of heterogeneity. Hepatology 2022, 76, 549–550. [Google Scholar] [CrossRef] [PubMed]
- Zaitceva, V.; Kopeina, G.S.; Zhivotovsky, B. Anastasis: Return journey from cell death. Cancers 2021, 13, 3671. [Google Scholar] [CrossRef]
- Tang, H.M.; Tang, H.L. Anastasis: Recovery from the brink of cell death. R. Soc. Open Sci. 2018, 5, 180442. [Google Scholar] [CrossRef]
- Castillo Ferrer, C.; Berthenet, K.; Ichim, G. Apoptosis—Fueling the oncogenic fire. FEBS J. 2021, 288, 4445–4463. [Google Scholar] [CrossRef]
- Jinesh, G.G. Exposing the deadly dark side of apoptotic cancer stem cells. Oncoscience 2017, 4, 124–125. [Google Scholar] [CrossRef]
- Tang, H.L.; Tang, H.M.; Mak, K.H.; Hu, S.; Wang, S.S.; Wong, K.M.; Wong, C.S.T.; Wu, H.Y.; Law, H.T.; Liu, K.; et al. Cell survival, DNA damage, and oncogenic transformation after a transient and reversible apoptotic response. Mol. Biol. Cell 2012, 23, 2240–2252. [Google Scholar] [CrossRef]
- Kalkavan, H.; Rühl, S.; Shaw, J.J.P.; Green, D.R. Non-lethal outcomes of engaging regulated cell death pathways in cancer. Nat. Cancer 2023, 4, 795–806. [Google Scholar] [CrossRef]
- Nano, M.; Montell, D.J. Apoptotic signaling: Beyond cell death. Semin Cell Dev. Biol. 2024, 156, 22–34. [Google Scholar] [CrossRef]
- Green, D.R. Cell death: Revisiting the roads to ruin. Dev. Cell 2024, 59, 2523–2531. [Google Scholar] [CrossRef] [PubMed]
- Green, D.R. Not dead yet: Cell death and survival in cancer and normal physiology. Front. Cell Death 2024, 3, 147734. [Google Scholar] [CrossRef]
- Tang, H.M.; Tang. H.L. Unravelling the pathological roles of anastasis in cancer recurrence. Open Biol. 2025, 15, 240270.
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.; Kin, T.; Beck, W.T. Impact of complex apoptotic signaling pathways on cancer cell sensitivity to therapy. Cancers 2024, 16, 984. [Google Scholar] [CrossRef] [PubMed]
- Vitale, I.; Pietrocola, F.; Guilbaud, E.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostini, M.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; et al. Apoptotic cell death in disease-Current understanding of the NCCD 2023. Cell Death Differ. 2023, 30, 1097–1154. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, R.; Liu, X.; Liu, M.; Sun, L.; Pan, X.; Hu, H.; Jiang, B.; Zou, Y.; Liu, Q.; et al. Chemotherapy-induced executioner caspase activation increases breast cancer malignancy through epigenetic de-repression of CDH12. Oncogenesis 2023, 12, 34. [Google Scholar] [CrossRef]
- Wang, R.; Wang, Y.; Liu, X.; Liu, M.; Sun, L.; Pan, X.; Hu, H; Jiang, B.; Zou, Y.; Liu, et al. Anastasis enhances metastasis and chemoresistance of colorectal cancer cells through upregulating cIAP2/NFκB signaling. Cell Death Dis. 2023, 14, 388.
- Sun, L.; Yao, C.; Li, X.; Wang, Y.; Wang, R.; Wang, M.; Liu, Q.; Montell, D.J.; Shao, C.; Gong, Y.; Sun, G. Anastasis confers ovarian cancer cells increased malignancy through elevated p38 MAPK activation. Cell Death Differ. 2023, 30, 809–824. [Google Scholar] [CrossRef]
- Vasileva, M.H.; Bennemann, A.; Zachmann, K.; Schön, M.P.; Frank, J.; Ulaganathan, V.K. CD24 flags anastasis in melanoma cells. Apoptosis 2025, 30, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ulaganathan, V.K. CD24: A Cell Surface Marker for Anastasis in Melanoma. 2024. Available online: https://communities.springernature.com/posts/cd24-a-cell-surface-marker-for-anastasis-in-melanoma (accessed on 5 October 2025).
- Corsi, F.; Capradossi, F.; Pelliccia, A.; Briganti, S.; Bruni, E.; Traversa, E.; Torino, F.; Reichle, A.; Ghibelli, L. Apoptosis as driver of therapy-induced cancer repopulation and acquired cell-resistance (CRAC): A simple in vitro model of Phoenix Rising in prostate cancer. Int. J. Mol. Sci. 2022, 23, 1152. [Google Scholar] [CrossRef] [PubMed]
- Eskandari, E.; Eaves, C.J. Paradoxical roles of caspase-3 in regulating cell survival, proliferation, and tumorigenesis. J. Cell Biol. 2022, 221, e202201159. [Google Scholar] [CrossRef] [PubMed]
- Park, W.Y.; Gray, J.M.; Holewinski, R.J.; Andresson, T.; So, J.S.; Carmona-Rivera, C.; Hollander, M.C.; Yang, H.H.; Lee, M.; Kaplan, M.J.; et al. Apoptosis-induced nuclear expulsion in tumor cells drives S100a4-mediated metastatic outgrowth through the RAGE pathway. Nat. Cancer 2023, 4, 419–435. [Google Scholar] [CrossRef]
- Yang, L.; Fang, J.; Chen, J. Tumor cell senescence response produces aggressive variants. Cell Death Discov. 2017, 3, 17049. [Google Scholar] [CrossRef]
- Jinesh, G.G.; Brohl, A.S. Classical epithelial-mesenchymal transition (EMT) and alternative cell death process-driven blebbishield metastatic-witch (BMW) pathways to cancer metastasis. Signal Transduct. Target. Ther. 2022, 7, 296. [Google Scholar] [CrossRef]
- Ergün, S.; Aslan, S.; Demir, D.; Kayaoğlu, S.; Saydam, M.; Keleş, Y.; Kolcuoğlu, D.; Hekim, N.T.; Güneş, S. Beyond death: Unmasking the intricacies of apoptosis escape. Mol. Diagn. Ther. 2024, 28, 403–423. [Google Scholar] [CrossRef]
- Tatebe, S.; Ishida, M.; Kasagi, N.; Tsujitani, S.; Kaibara, N.; Ito, H. Apoptosis occurs more frequently in metastatic foci than in primary lesions of human colorectal carcinomas: Analysis by terminal-deoxynucleotidyl-transferase-mediated dUTP-biotin nick end labeling. Int. J. Cancer 1996, 65, 173–177. [Google Scholar] [CrossRef]
- Huang, Q.; Li, S.; Cheng, I.; Deng, M.; He, X.; Wang, Z.; Yang, C.H.; Zhao, X.Y.; Huang, J. High expression of anti-apoptotic protein Bcl-2 is a good prognostic factor in colorectal cancer: Result of a meta-analysis. World J. Gastroenterol. 2017, 23, 5018–5033. [Google Scholar] [CrossRef]
- Flanagan, L.; Meyer, M.; Fay, J.; Curry, S.; Bacon, O.; Duessmann, H.; John, K.; Boland, K.C.; McNamara, D.A.; Kay, E.W.; et al. Low levels of Caspase-3 predict favourable response to 5FU-based chemotherapy in advanced colorectal cancer: Caspase-3 inhibition as a therapeutic approach. Cell Death Dis. 2016, 7, e2087. [Google Scholar] [CrossRef]
- Alcaide, J.; Funez, R.; Rueda, A.; Perez-Ruiz, E.; Pereda, T.; Rodrigo, I.; Covenas, R.; Munoz, M.; Redondo, M. The role and prognostic value of apoptosis in colorectal carcinoma. BMC Clin. Pathol. 2013, 13, 24. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.; Morrison, I.; Heriot, A.G.; Bartlett, J.B.; Finlayson, C.; Dalgleish, A.G.; Kumar, D. The correlation between colorectal cancer rates of proliferation and apoptosis and systemic cytokine levels; plus their influence upon survival. Br. J. Cancer 2006, 94, 1412–1419. [Google Scholar] [CrossRef]
- Hilska, M.; Collan, Y.U.; Laine, V.J.O.; Kössi, J.; Hirsimäki, P.; Laato, M.; Roberts, P.J. The significance of tumor markers for proliferation and apoptosis in predicting survival in colorectal cancer. Dis. Colon. Rectum. 2005, 48, 2197–2208. [Google Scholar] [CrossRef] [PubMed]
- Bendardaf, R.; Ristamaki, R.; Kujari, H.; Laine, J.; Lamlum, H.; Collan, Y.; Pyrhonen, S. Apoptotic index and bcl-2 expression as prognostic factors in colorectal carcinoma. Oncology 2003, 64, 435–442. [Google Scholar] [CrossRef]
- Yang, X.; Zhong, D.N.; Qin, H.; Wu, P.R.; Wei, K.L.; Chen, G.; He, R.Q.; Zhong, J.C. Caspase-3 over-expression is associated with poor overall survival and clinicopathological parameters in breast cancer: A meta-analysis of 3091 cases. Oncotarget 2018, 9, 8629–8641. [Google Scholar] [CrossRef]
- Lindner, A.U.; Lucantoni, F.; Varešlija, D.; Resler, A.; Murphy, B.M.; Gallagher, W.M.; Hill, A.D.K.; Young, L.S.; Prehn, J.H.M. Low cleaved caspase-7 levels indicate unfavourable outcome across all breast cancers. Mol. Med. 2018, 96, 1025–1037. [Google Scholar] [CrossRef]
- Pu, X.; Storr, S.J.; Zhang, Y.; Rakha, E.A.; Green, A.R.; Ellis, I.O.; Martin, S.G. Caspase-3 and caspase-8 expression in breast cancer: Caspase-3 is associated with survival. Apoptosis 2017, 22, 357–368. [Google Scholar] [CrossRef]
- Koshida, Y.; Saegusa, M.; Okayasu, I. Apoptosis, cell proliferation and expression of Bcl-2 and Bax in gastric carcinomas: Immunohistochemical and clinicopathological study. Br. J. Cancer 1997, 75, 367–373. [Google Scholar] [CrossRef]
- Beer, T.W.; Carr, N.J.; Whittaker, M.A.; Pullinger, N. Mitotic and in situ end-labeling apoptotic indices as prognostic markers in malignant mesothelioma. Ann. Diagn. Pathol. 2000, 4, 143–148. [Google Scholar] [CrossRef]
- Kahlos, K.; Soini, Y.; Paakko, P.; Saily, M.; Linnainmaa, K.; Kinnula, V.L. Proliferation, apoptosis, and manganese superoxide dismutase in malignant mesothelioma. Int. J. Cancer 2000, 88, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Mangili, F.; Cigala, C.; Arrigoni, G.; Rovere, E.; Gattuso, C.; Santambrogio, G.; Garancini, P. Cell loss and proliferation in non-small cell lung carcinoma: Correlation with histological subtype. Eur. J. Histochem. 1998, 42, 287–295. [Google Scholar]
- Tormanen, U.; Eerola, A.K.; Rainio, P.; Vahakangas, K.; Soini, Y.; Sormunen, R.; Bloigu, R.; Lehto, V.P.; Paakko, P. Enhanced apoptosis predicts shortened survival in non-small cell lung carcinoma. Cancer Res. 1995, 55, 5595–5602. [Google Scholar]
- Meggiato, T.; Calabrese, F.; Valente, M.; Favaretto, E.; Baliello, E.; Del Favero, G. Spontaneous apoptosis and proliferation in human pancreatic cancer. Pancreas 2000, 20, 117–122. [Google Scholar] [CrossRef]
- Magistrelli, P.; Coppola, R.; Tonini, G.; Vincenzi, B.; Santini, D.; Borzomati, D.; Vecchio, F.; Valeri, S.; Castri, F.; Antinori, A. Apoptotic index or a combination of Bax/Bcl-2 expression correlate with survival after resection of pancreatic adenocarcinoma. J. Cell Biochem. 2006, 97, 98–108. [Google Scholar] [CrossRef]
- Naresh, K.N.; Lakshminarayanan, K.; Pai, S.A.; Borges, A.M. Apoptosis index is a predictor of metastatic phenotype in patients with early stage squamous carcinoma of the tongue: A hypothesis to support this paradoxical association. Cancer 2001, 91, 578–584. [Google Scholar] [CrossRef]
- Montell, D.J. Cellular Survival by Anastasis. 2024. Available online: https://denisemontell.mcdb.ucsb.edu/research/cellular-survival-anastasis (accessed on 5 October 2025).
- Khatib, S.A.; Ma, L.; Dang, H.; Forgues, M.; Chung, J.-Y.; Ylaya, K.; Hewitt, S.M.; Chaisaingmongkol, J.; Rucchirawat, M.; Wang, X.W. Single-cell biology uncovers apoptotic cell death and its spatial organization as a potential modifier of tumor diversity in HCC. Hepatology 2022, 76, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.C.; Liang, S.B.; Luo, M.; Wang, X.P. Intratumoral heterogeneity and drug resistance in cancer. Cancer Cell Int. 2025, 25, 103. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hemann, M.T. A dynamic view of chemotherapy effectiveness. Nature 2019, 572, 321–322. [Google Scholar] [CrossRef]
- Mirzayans, R.; Murray, D. Intratumor heterogeneity and treatment resistance of solid tumors with a focus on polyploid/senescent giant cancer cells (PGCCs). Int. J. Mol. Sci. 2023, 24, 11534. [Google Scholar] [CrossRef] [PubMed]
- Min, H.Y.; Lee, H.Y. Cellular dormancy in cancer: mechanisms and potential targeting strategies. Cancer Res. Treat. 2023, 55, 720–736. [Google Scholar] [CrossRef]
- Sun, X.X.; Yu, Q. Intra-tumor heterogeneity of cancer cells and its implications for cancer treatment. Acta Pharmacol. Sin. 2015, 36, 1219–1227. [Google Scholar] [CrossRef]
- Rouault, C.D.; Charafe-Jauffret, E.; C.; Ginestier. The interplay of DNA damage, epigenetics and tumour heterogeneity in driving cancer cell fitness. Nat. Commun. 2025, 16, 8733. [CrossRef]
- Liu, Y.; Su, Z.; Tavana, O.; Gu, W. Understanding the complexity of p53 in a new era of tumor suppression. Cancer Cell. 2024, 42, 946–967. [Google Scholar] [CrossRef]
- MacDonald, W.J.; Purcell, C.; Pinho-Schwermann, M.; Stubbs, N.M.; Srinivasan, P.R.; El-Deiry, W.S. Heterogeneity in cancer. Cancers 2025, 17, 441. [Google Scholar] [CrossRef]
- Maleki, E.H.; Bahrami, A.R.; Matin, M.M. Cancer cell cycle heterogeneity as a critical determinant of therapeutic resistance. Genes Dis. 2023, 11, 189–204. [Google Scholar] [CrossRef]
- Zhu, H.; Tian, Y.; Chen, H.; Y.; Qian, J. Li.; Niu, D.; Zhao, W.; Wu, Y.; Zhang, X.; Tang, T.; et al. Targeting DNA damage response pathways in tumor drug resistance: Mechanisms, clinical implications, and future directions. Drug Resist. Updat. 2025, 83, 101287. [CrossRef] [PubMed]
- Oh, M.S.; Abascal, J.; Rennels, A.K.; Salehi-Rad, R.; Dubinett, S.M.; Liu, B. Tumor heterogeneity and the immune response in non-small cell lung cancer: Emerging insights and implications for immunotherapy. Cancers 2025, 17, 1027. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Miao, K.; Sun, H.; Deng, C.X. Tumor heterogeneity reshapes the tumor microenvironment to influence drug resistance. Int. J. Biol. Sci. 2022, 18, 3019–3033. [Google Scholar] [CrossRef] [PubMed]
- Proietto, M.; Crippa, M.; Damiani, C.; Pasquale, V.; Sacco, E.; Vanoni, M.; Gilardi, M. Tumor heterogeneity: preclinical models, emerging technologies, and future applications. Front. Oncol. 2023, 13, 1164535. [Google Scholar] [CrossRef]
- Tomasik B, Garbicz F, Braun M, Bieńkowski M and Jassem J. Heterogeneity in precision oncology. Camb. Prism. Precis. Med. 2024, 2, e2.
- Tonello, S.; Rolla, R.; Tillio, P.A.; Sainaghi, P.P.; Colangelo, D. Microenvironment and tumor heterogeneity as pharmacological targets in precision oncology. Pharmaceuticals 2025, 18, 915. [Google Scholar] [CrossRef]
- Clinton, T.N.; Chen, Z.; Wise, H.; Lenis, A.T.; Chavan, S.; Donoghue, M.T.A.; Almassi, N.; Chu, C.E.; Dason, S. Genomic heterogeneity as a barrier to precision oncology in urothelial cancer. Cell Rep. 2022, 41, 111859. [Google Scholar] [CrossRef]
- Mirzayans, R.; Murray, D. Pharmacological modulation of p53 function in cancer therapy. Curr. Signal. Transduct. Ther. 2008, 3, 183–194. [Google Scholar] [CrossRef]
- Erenpreisa, J.; Cragg, M.S. Mitotic death: a mechanism of survival? A review. Cancer Cell Int. 2001, 1, 1–7. [Google Scholar] [CrossRef]
- Chen, J.; Niu, N.; Zhang, J.; Qi, L.; Shen, W.; Donkena, K.V.; Feng, Z.; Liu, J. Polyploid giant cancer cells (PGCCs): The evil roots of cancer. Curr. Cancer Drug Targets 2019, 19, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Erenpreisa, J.; Sikora, E. Polyploid giant cancer cells: An emerging new field of cancer biology. Semin. Cancer Biol. 2022, 81, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Heng, J.; Heng, H.H. Genome chaos: Creating new genomic information essential for cancer macroevolution. Semin. Cancer Biol. 2022, 81, 160–175. [Google Scholar] [CrossRef] [PubMed]
- Trabzonlu, L.; Pienta, K.J.; Trock, B.J.; De Marzo, A.M.; Amend. S.R. Presence of cells in the polyaneuploid cancer cell (PACC) state predicts the risk of recurrence in prostate cancer. Prostate 2023, 83, 277–285. [CrossRef]
- Murray, D.; Mirzayans, R. Cellular responses to platinum-based anticancer drugs and UVC: Role of p53 and implications for cancer therapy. Int. J. Mol. Sci. 2020, 21, 5766. [Google Scholar] [CrossRef]
- Mirzayans, R. When Therapy-induced cancer cell apoptosis fuels tumor relapse. Onco. 2024, 4, 37–45. [Google Scholar] [CrossRef]
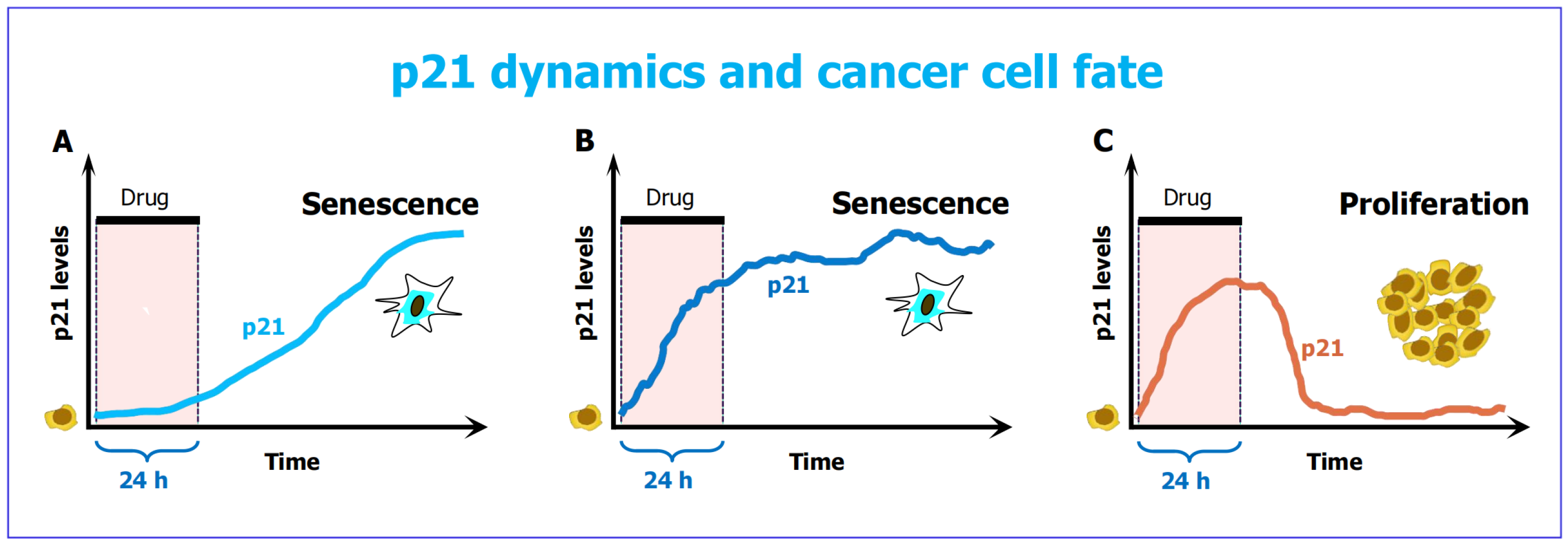
- Hsu, C.H.; Altschuler, S.J.; Wu, L.F. Patterns of early p21 dynamics determine proliferation-senescence cell fate after chemotherapy. Cell 2019, 178, 361–373. [Google Scholar] [CrossRef]
- Soon, J.A.; Franchini, F.; IJzermanm, N.J.; McArthur, G.A. Leveraging the potential for deintensification in cancer care. Nat. Cancer 2024, 5, 1597–1599. [Google Scholar] [CrossRef]
- Belluz, J. Most cancer drugs fail in testing. This might be a big reason why. Science—VOX blog (2019) https://www.vox.com/2019/9/16/20864066/cancer-studies-fail (accessed on 5 October 2025).
- Mirzayans, R. The cellular response to DNA damage: From DNA repair to polyploidy and beyond. Int. J. Mol. Sci. 2023, 24, 6852. [Google Scholar] [CrossRef]
- Mirzayans, R.; Murray, D. Intratumor heterogeneity and therapy resistance: Contributions of dormancy, apoptosis reversal (anastasis) and cell fusion to disease recurrence. Int. J. Mol. Sci. 2020, 21, 1308. [Google Scholar] [CrossRef] [PubMed]
- Eastman, A. Improving anticancer drug development begins with cell culture: Misinformation perpetrated by the misuse of cytotoxicity assays. Oncotarget 2017, 8, 8854–8866. [Google Scholar] [CrossRef] [PubMed]
- Forgie, B.N.; Prakash, R.; Goyeneche, A.A.; Telleria, C.M. Vitality, viability, long-term clonogenic survival, cytotoxicity, cytostasis and lethality: What do they mean when testing new investigational oncology drugs? Discov. Oncol. 2024, 15, 5. [Google Scholar] [CrossRef] [PubMed]
- Nicoletto, R.E.; Ofner, C.M. Cytotoxic mechanisms of doxorubicin at clinically relevant concentrations in breast cancer cells. Cancer Chemother. Pharmacol. 2022, 89, 285–311. [Google Scholar] [CrossRef]
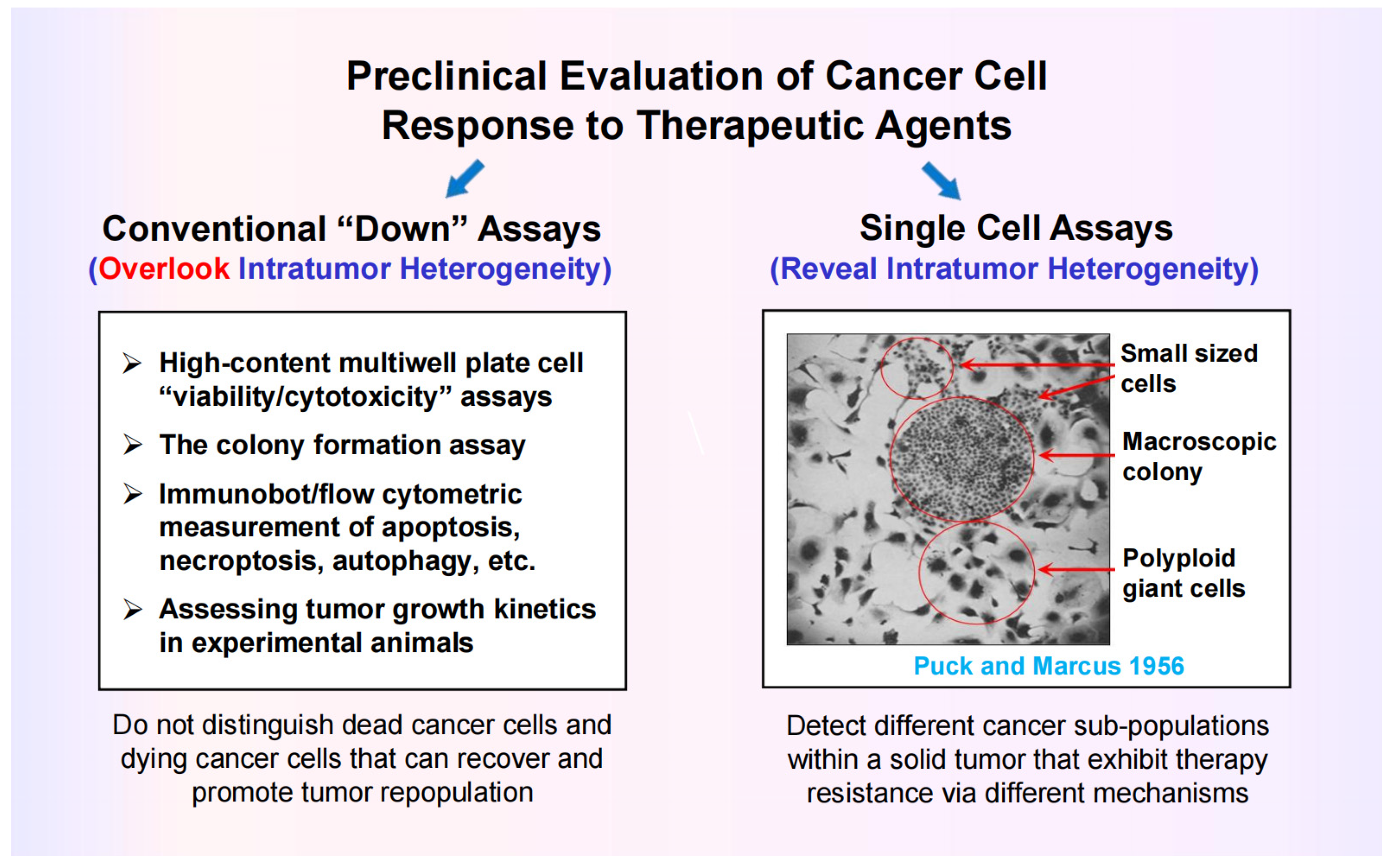
- Puck, T.T.; Marcus, P.I. Action of X-rays on mammalian cells. J. Exp. Med. 1956, 103, 653–666. [Google Scholar] [CrossRef]
- Retraction Watch: Tracking retractions as a window into the scientific process. Available online: https://retractionwatch.com/2015/07/06/cancer-research-retraction-is-fifth-for-robert-weinberg-fourth-for-his-former-student/ (accessed on 5 October 2025).
- Szabo, L. Cancer treatment hype gives false hope to many patients. Kaiser Health News (2017) https://www.usatoday.com/story/news/2017/04/27/cancer-treatment-hype-gives-false-hope-many-patients/100972794/ (last accessed on 5 October 2025).
- Hernández Borrero, L.J.; El-Deiry, WS. Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188556. [Google Scholar] [CrossRef]
- Lees, A.; Sessler, T.; McDade, S. Dying to survive—The p53 paradox. Cancers 2021, 13, 3257. [Google Scholar] [CrossRef]
- Zhang, S.; Carlsen, L.; Hernandez Borrero, L.; Seyhan, A.A.; Tian, X.; El-Deiry, W.S. Advanced strategies for therapeutic targeting of wild-type and mutant p53 in cancer. Biomolecules 2022, 12, 548. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, J.; Long, Y.; Maimaitijiang, A.; Su, Z.; Li, W.; Li, J. Unraveling the guardian: p53’s multifaceted role in the DNA damage response and tumor treatment strategies. Int. J. Mol. Sci. 2024, 25, 12928. [Google Scholar] [CrossRef] [PubMed]
- Chen, J. The cell-cycle arrest and apoptotic functions of p53 in tumor initiation and progression. Cold Spring Harb. Perspect. Med. 2016, 6, a026104. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.P. p53, guardian of the genome. Nature 1992, 358, 15–16. [Google Scholar] [CrossRef]
- Levine, A.J. Targeting the p53 protein for cancer therapies: The translational impact of p53 research. Cancer Res. 2022, 82, 362–364. [Google Scholar] [CrossRef]
- Oren, M.; Prives, C. p53: A tale of complexity and context. Cell 2024, 187, 1569–1573. [Google Scholar] [CrossRef]
- Kandouz, M. Cell death, by any other name. Cells 2024, 13, 325. [Google Scholar] [CrossRef]
- Murray, D.; Mirzayans, R.; McBride, W.H. Defenses against pro-oxidant forces—Maintenance of cellular and genomic integrity and longevity. Radiat. Res. 2018, 190, 331–349. [Google Scholar] [CrossRef] [PubMed]
- Barley, R.D.C.; Enns, L.; Paterson, M.C.; Mirzayans, R. Aberrant p21WAF1-dependent growth arrest as the possible mechanism of abnormal resistance to ultraviolet light cytotoxicity in Li-Fraumeni syndrome fibroblast strains heterozygous for TP53 mutations. Oncogene 1998, 17, 533–543. [Google Scholar] [CrossRef]
- Xia, Y.; Yang, Q.; Gong, X.; Yem, F.; Liou, Y.C. Dose-dependent mutual regulation between Wip1 and p53 following UVC irradiation. Int. J. Biochem. Cell Biol. 2011, 43, 535–544. [Google Scholar] [CrossRef]
- Kracikova, M.; G. ; Akiri, George, A.; Sachidanandam, R.; Aaronson, S.A. A threshold mechanism mediates p53 cell fate decision between growth arrest and apoptosis. Cell Death and Differ. 2013, 20, 576–588. [Google Scholar] [CrossRef]
- Mirzayans, R.; Andrais, B.; Scott, A.; Wang, Y.W.; Murray, D. Ionizing radiation-induced responses in human cells with differing TP53 status. Int. J. Mol. Sci. 2013, 14, 22409–22435. [Google Scholar] [CrossRef] [PubMed]
- Mirzayans, R.; Andrais, B.; Kumar, P.; Murray, D. Significance of wild-type p53 signaling in suppressing apoptosis in response to chemical genotoxic agents: Impact on chemotherapy outcome. Int. J. Mol. Sci. 2017, 18, 928. [Google Scholar] [CrossRef]
- Jänicke, R.U.; Sohn, D.; Schulze-Osthoff, K. The dark side of a tumor suppressor: Anti-apoptotic p53. Cell Death Differ. 2008, 15, 959–976. [Google Scholar] [CrossRef]
- Ashraf, H.M.; Moser, J.; Spencer, S.L. Senescence Evasion in Chemotherapy: A Sweet Spot for p21. Cell 2019, 178, 267–269. [Google Scholar] [CrossRef]
- Liu, Y.; Hemann, M.T. A dynamic view of chemotherapy effectiveness. Nature 2019, 572, 321–322. [Google Scholar] [CrossRef] [PubMed]
- Chang, B.D.; Broude, E.V.; Dokmanovic, M.; Zhu, H.; Ruth, A.; Xuan, Y.; Kandel, E.S.; Lausch. E.; Christov, K.; Roninson, I.B. A senescence-like phenotype distinguishes tumor cells that undergo terminal proliferation arrest after exposure to anticancer agents. Cancer Res. 1999, 59, 3761–3767.
- Chang, B.D.; Xuan, Y.; Broude, E.V.; Zhu, H.; Schott, B.; Fang, J.; Roninson, I.B. Role of p53 and p21waf1/cip1 in senescence-like terminal proliferation arrest induced in human tumor cells by chemotherapeutic drugs. Oncogene 1999, 18, 4808–4818. [Google Scholar] [CrossRef] [PubMed]
- Chang, B.D.; Broude, E.V.; J. Fang.; T.V. Kalinichenko.; R. Abdryashitov.; J.C. Poole.; I.B. Roninson. p21Waf1/Cip1/Sdi1-induced growth arrest is associated with depletion of mitosis-control proteins and leads to abnormal mitosis and endoreduplication in recovering cells. Oncogene 2000, 19, 2165–2170. [Google Scholar] [CrossRef]
- Yang, L.; Fang, J.; Chen, J. Tumor cell senescence response produces aggressive variants. Cell Death Discov. 2017, 3, 17049. [Google Scholar] [CrossRef]
- Mirzayans, R.; Andrais, B.; Murray, D. Viability assessment following anticancer treatment requires single-cell visualization. Cancers 2018, 10, 255. [Google Scholar] [CrossRef]
- Hanahan, D. , Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Mirzayans, R.; Murray, D. Amitotic cell division, malignancy, and resistance to anticancer agents: A tribute to Drs. Walen and Rajaraman. Cancers 2024, 16, 3106. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



