Submitted:
03 October 2025
Posted:
07 October 2025
You are already at the latest version
Abstract
Keywords:
Introduction
Material and Method
Unix Skill Levels in Bioinformatics
Tutorial Implementation
Tutorial Deployment
Results and Discussion
A Unix Competency Matrix for Learning Bioinformatics
Online Tutorials for Practical, Self-Paced and Autonomous Learning of Unix
- Pre-class activities in autonomy for professional bioinformatics training: “Introduction to the processing of genomic data obtained by high-throughput sequencing” at the French Bioinformatics Institute (EB3I 2023 and EB3I 2024, with 39 trainees in each session), a one-week intensive training session for life scientists.
- Pre-class activities in autonomy for a professional diploma at Université Paris Cité: “Production, analysis, and valorization of biological omics data” (DU omiques 2023 and DU omiques 2025, with 14 trainees in each session).
- Pre-class activities in autonomy for biologists and interns (14 participants) at the Institute for Integrative Biology of the Cell (I2BC) at Université Paris-Saclay
- Self-assessment of Linux competencies by 30 students of the IMALIS life science master’s program at the École Normale Supérieure (Paris).
- Self-assessment of Linux competencies by 40 students of the Polytech graduate school of engineering (specialty in Biological Engineering) at Aix-Marseille University.
References
- Attwood TK, Blackford S, Brazas MD, Davies A, Schneider MV. A global perspective on evolving bioinformatics and data science training needs. Brief Bioinform [Internet]. 2017 Aug 29 [cited 2025 Aug 20];20(2):398–404. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6433731/.
- Wilson Sayres MA, Hauser C, Sierk M, Robic S, Rosenwald AG, Smith TM, et al. Bioinformatics core competencies for undergraduate life sciences education. PLoS ONE [Internet]. 2018 June 5 [cited 2025 Aug 20];13(6):e0196878. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5988330/.
- Mulder N, Schwartz R, Brazas MD, Brooksbank C, Gaeta B, Morgan SL, et al. The development and application of bioinformatics core competencies to improve bioinformatics training and education. PLoS Comput Biol [Internet]. 2018 Feb 1 [cited 2025 Aug 20];14(2):e1005772. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5794068/.
- Perkel JM. Five reasons why researchers should learn to love the command line. Nature [Internet]. 2021 Feb 4 [cited 2025 Sept 9];590(7844):173–4. Available from: https://www.nature.com/articles/d41586-021-00263-0.
- Işık EB, Brazas MD, Schwartz R, Gaeta B, Palagi PM, Van Gelder CWG, et al. Grand challenges in bioinformatics education and training. Nat Biotechnol [Internet]. 2023 Aug [cited 2025 Aug 20];41(8):1171–4. Available from: https://www.nature.com/articles/s41587-023-01891-9.
- Schneider MV, Watson J, Attwood T, Rother K, Budd A, McDowall J, et al. Bioinformatics training: a review of challenges, actions and support requirements. Brief Bioinform [Internet]. 2010 Nov 1 [cited 2025 Aug 20];11(6):544–51. Available from: https://academic.oup.com/bib/article-lookup/doi/10.1093/bib/bbq021.
- Welch L, Brooksbank C, Schwartz R, Morgan SL, Gaeta B, Kilpatrick AM, et al. Applying, Evaluating and Refining Bioinformatics Core Competencies (An Update from the Curriculum Task Force of ISCB’s Education Committee). PLOS Comput Biol [Internet]. 2016 May 13 [cited 2025 Aug 20];12(5):e1004943. Available from: https://dx.plos.org/10.1371/journal.pcbi.1004943.
- Welch L, Lewitter F, Schwartz R, Brooksbank C, Radivojac P, Gaeta B, et al. Bioinformatics Curriculum Guidelines: Toward a Definition of Core Competencies. PLoS Comput Biol [Internet]. 2014 Mar 6 [cited 2025 Aug 20];10(3):e1003496. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3945096/.
- Wilson G. Software Carpentry: Getting Scientists to Write Better Code by Making Them More Productive. Comput Sci Eng [Internet]. 2006 Nov [cited 2025 Aug 20];8(6):66–9. Available from: http://ieeexplore.ieee.org/document/1717319/.
- Lui RWC, Dik MNY, Lee PTY. Evaluation of Online Learning Platforms for Interactive Self-paced Learning of Container-based Software Development. In: 2022 IEEE International Conference on Teaching, Assessment and Learning for Engineering (TALE) [Internet]. Hung Hom, Hong Kong: IEEE; 2022 [cited 2025 Aug 20]. p. 54–8. Available from: https://ieeexplore.ieee.org/document/10148308/.
- Baldwin L. Leveraging Katacoda technology exclusively within O’Reilly And the decision to shutter katacoda.com [Internet]. 2022. Available from: https://www.oreilly.com/online-learning/leveraging-katacoda-technology.html.
- Batut B, Hiltemann S, Bagnacani A, Baker D, Bhardwaj V, Blank C, et al. Community-Driven Data Analysis Training for Biology. Cell Syst [Internet]. 2018 June [cited 2025 Aug 20];6(6):752-758.e1. Available from: https://linkinghub.elsevier.com/retrieve/pii/S2405471218302308.
- Hiltemann S, Rasche H, Gladman S, Hotz HR, Larivière D, Blankenberg D, et al. Galaxy Training: A powerful framework for teaching! Ouellette F, editor. PLOS Comput Biol [Internet]. 2023 Jan 9 [cited 2025 Aug 20];19(1):e1010752. Available from: https://dx.plos.org/10.1371/journal.pcbi.1010752.
- Anderson LW, Krathwohl DR. A taxonomy for learning, teaching, and assessing: A revision of Bloom’s taxonomy of educational objectives. 2001. (New York: Longman).
- Chiapello H, Denecker T, Khamvongsa Charbonnier L, Poulain P, Puthier D, Sand O, et al. Matrice de compétence Unix pour l’apprentissage de la Bioinformatique [Internet]. Zenodo; 2024 [cited 2025 Aug 21]. Available from: https://zenodo.org/doi/10.5281/zenodo.12104694.
- Chiapello H, Denecker T, Khamvongsa Charbonnier L, Poulain P, Puthier D, Sand O, et al. Unix Competency Framework for Learning Bioinformatics [Internet]. Zenodo; 2025 [cited 2025 Sept 9]. Available from: https://zenodo.org/doi/10.5281/zenodo.17084249.
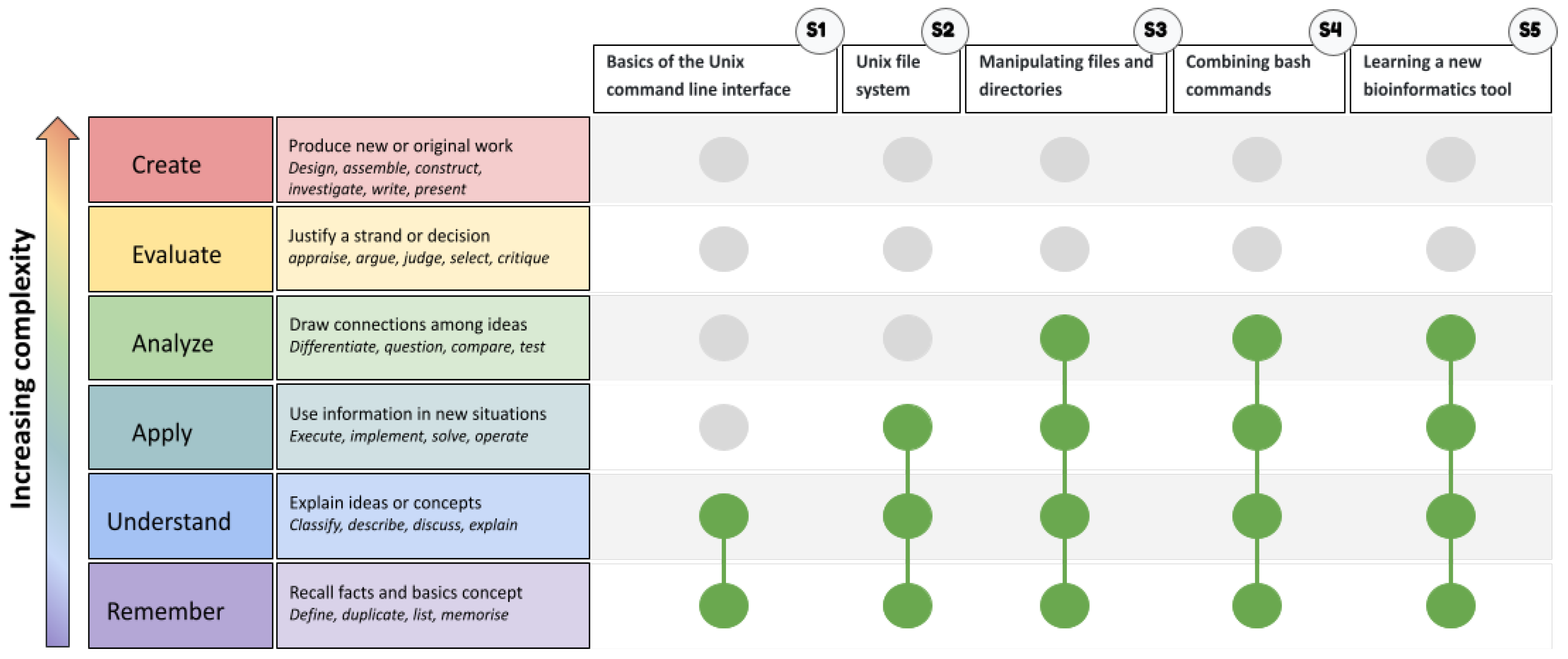

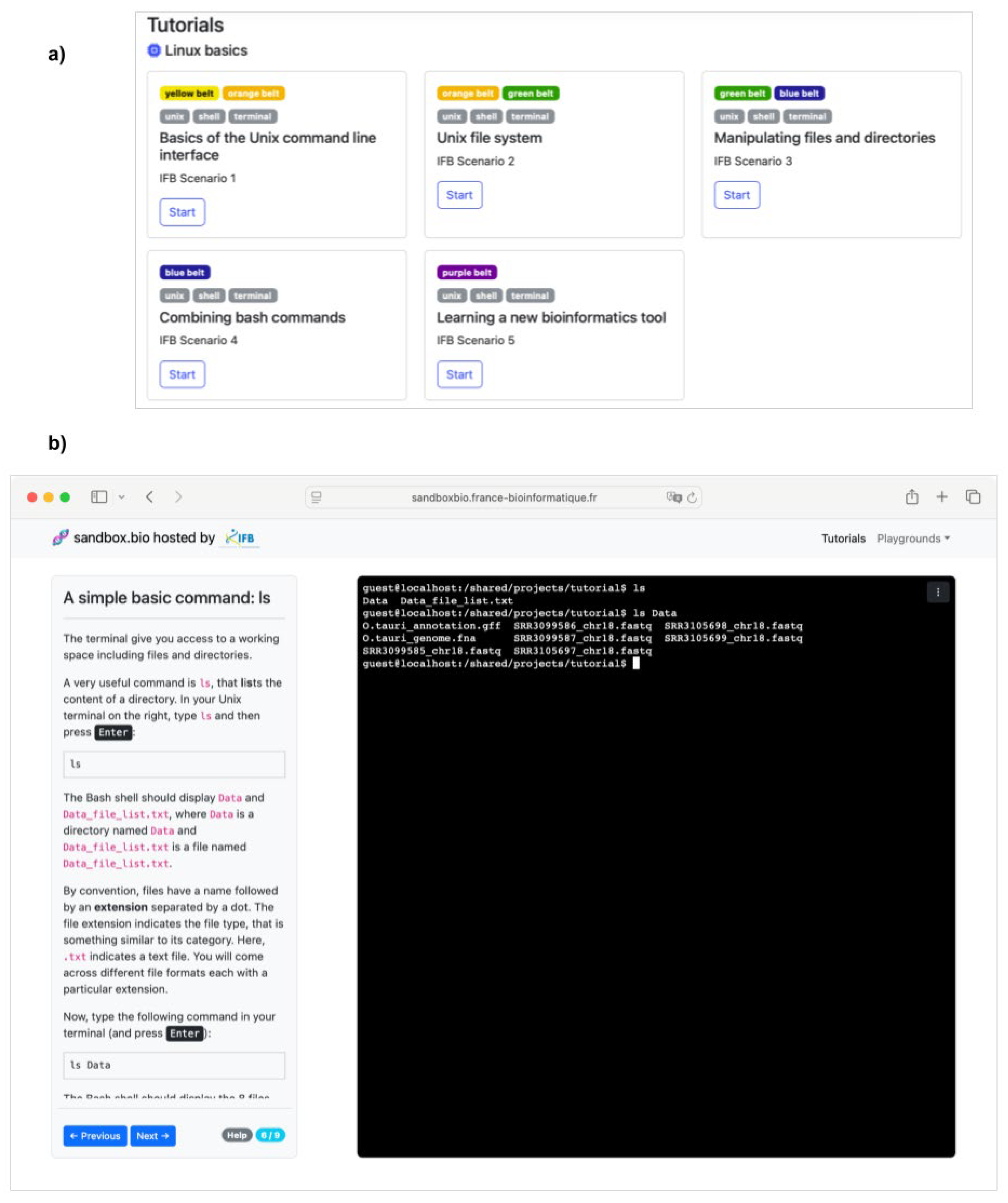
| Unix competency framework | Interactive online tutorials | |
|---|---|---|
| Description | A themed, judo-belt-inspired tool to guide the progressive learning of core Unix skills in bioinformatics | Open-access Unix tutorials focused on common bioinformatics applications, flexible and accessible without installation |
| Format | Textual framework / PDF | Web-based interactive |
| Target audience | Trainers, instructional designers and learners | Learners (beginners to intermediate) |
| Use cases | Curriculum design, learner self-assessment, certification | Self-training, classroom support, flipped classroom formats |
| Access | Downloadable / Shareable via Zenodo | Online (sandbox.bio) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





