Submitted:
22 September 2025
Posted:
25 September 2025
You are already at the latest version
Abstract
The human microbiome is a critical factor in health and disease, including breast pathology. While its association with breast cancer (BC) is increasingly studied, the specificity of the microbiome and its role in benign breast diseases (BBD) remain poorly understood. This review synthesizes current evidence on the origins of the mammary gland (MG) microbiota, which is distinct from the skin and formed via exogenous (e.g., cutaneous, retrograde via ducts) and endogenous (e.g., enteromammary, hematogenous) pathways. We detail the mechanisms of host-microbiota interaction, such as regulation of estrogen metabolism, immunomodulation, and epigenetic modifications, which can influence disease pathogenesis. The analysis reveals that while taxonomic profiles of tissue and gut microbiota share similarities between BBD and BC, key differences exist in the abundance of specific taxa (e.g., Pseudomonadota, Bacillota) and associated metabolic pathways. The review summarizes microbiota alterations associated with specific BBDs, including fibroadenomas, cysts, lactational and non-lactational mastitis (e.g., linked to Corynebacterium kroppenstedtii), and purulent-septic complications. Major limitations in the field are identified, such as the low microbial biomass of breast tissue, a lack of data on the virome and mycobiome, and the inability of current studies to establish causality. We conclude that microbial dysbiosis is implicated in BBD. However, further research is essential to elucidate cause-effect relationships. Understanding the microbiome’s role holds significant promise for developing novel diagnostic, preventive, and personalized therapeutic strategies for benign breast conditions.
Keywords:
1. Introduction
2. Pathways of Mammary Gland Microbiota Formation and Mechanisms of Its Interaction with the Host Organism
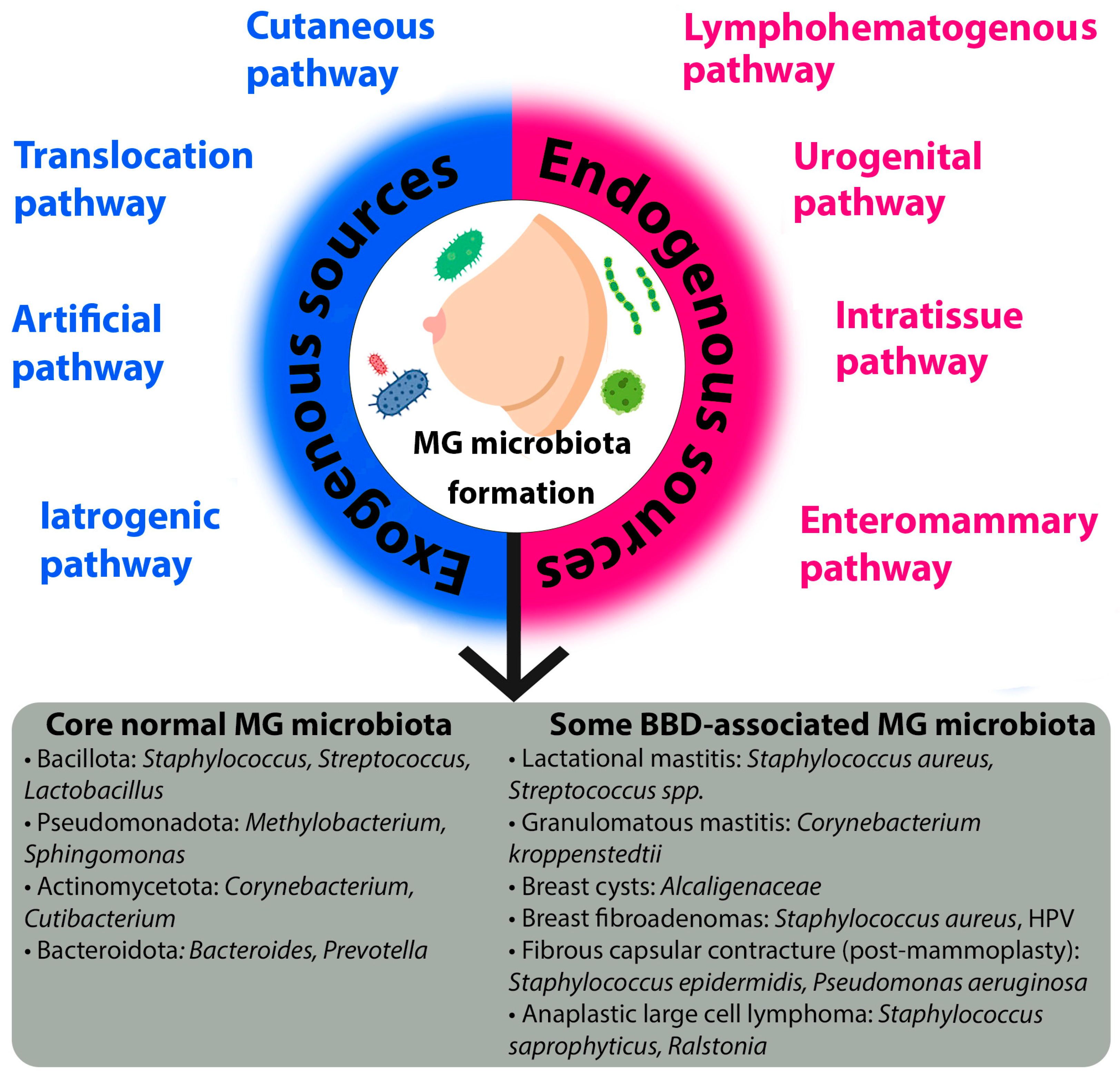
2.1. Origin of the Mammary Gland Microbiota
2.2. Microbiota of Unaltered Mammary Gland Tissues
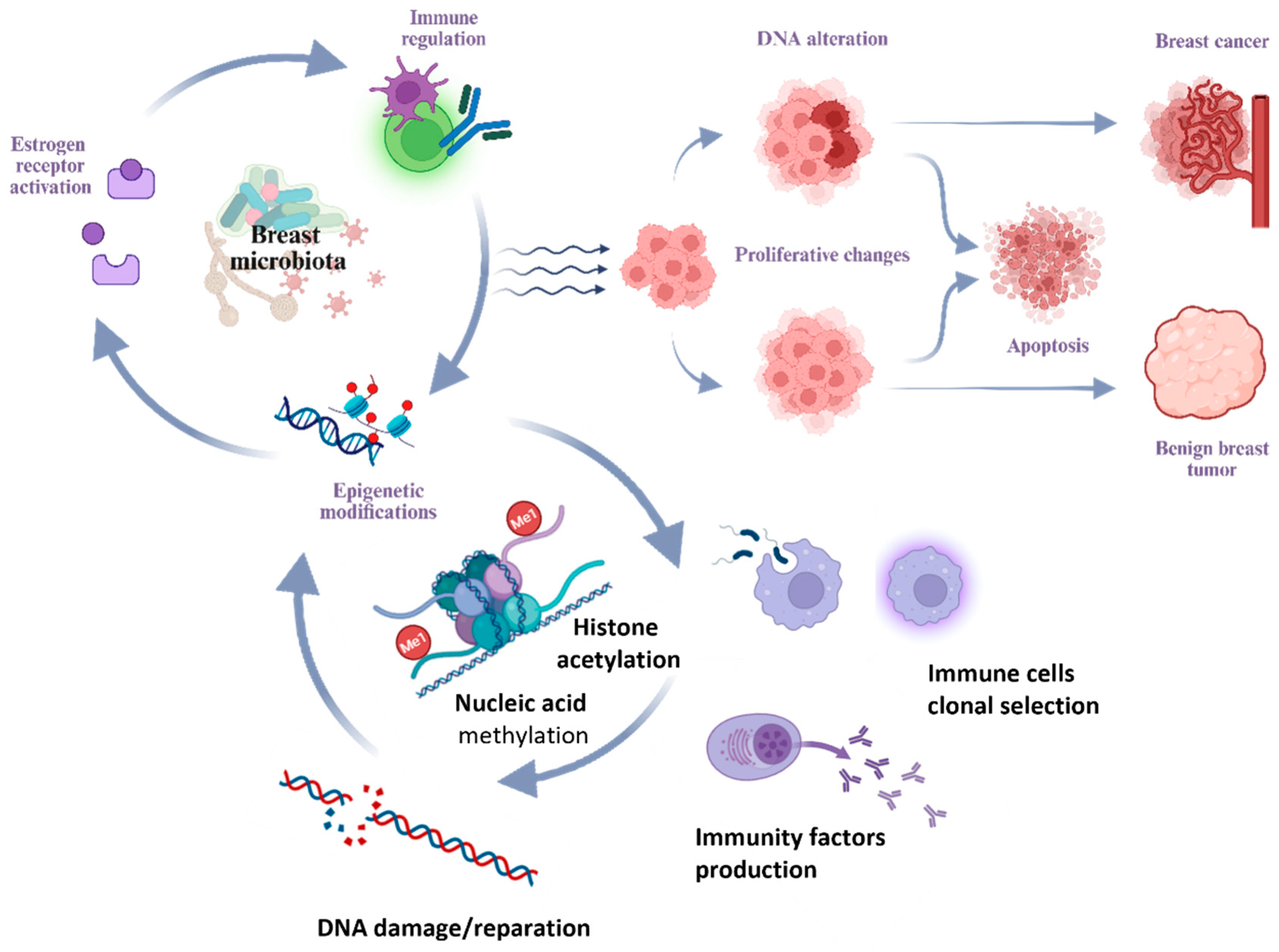
2.3. The Microbiota-Human Organism Interaction
2.4. Mechanisms of Microbiota-Induced Breast Pathogenesis
3. Microbiota Composition and Diversity in BBD
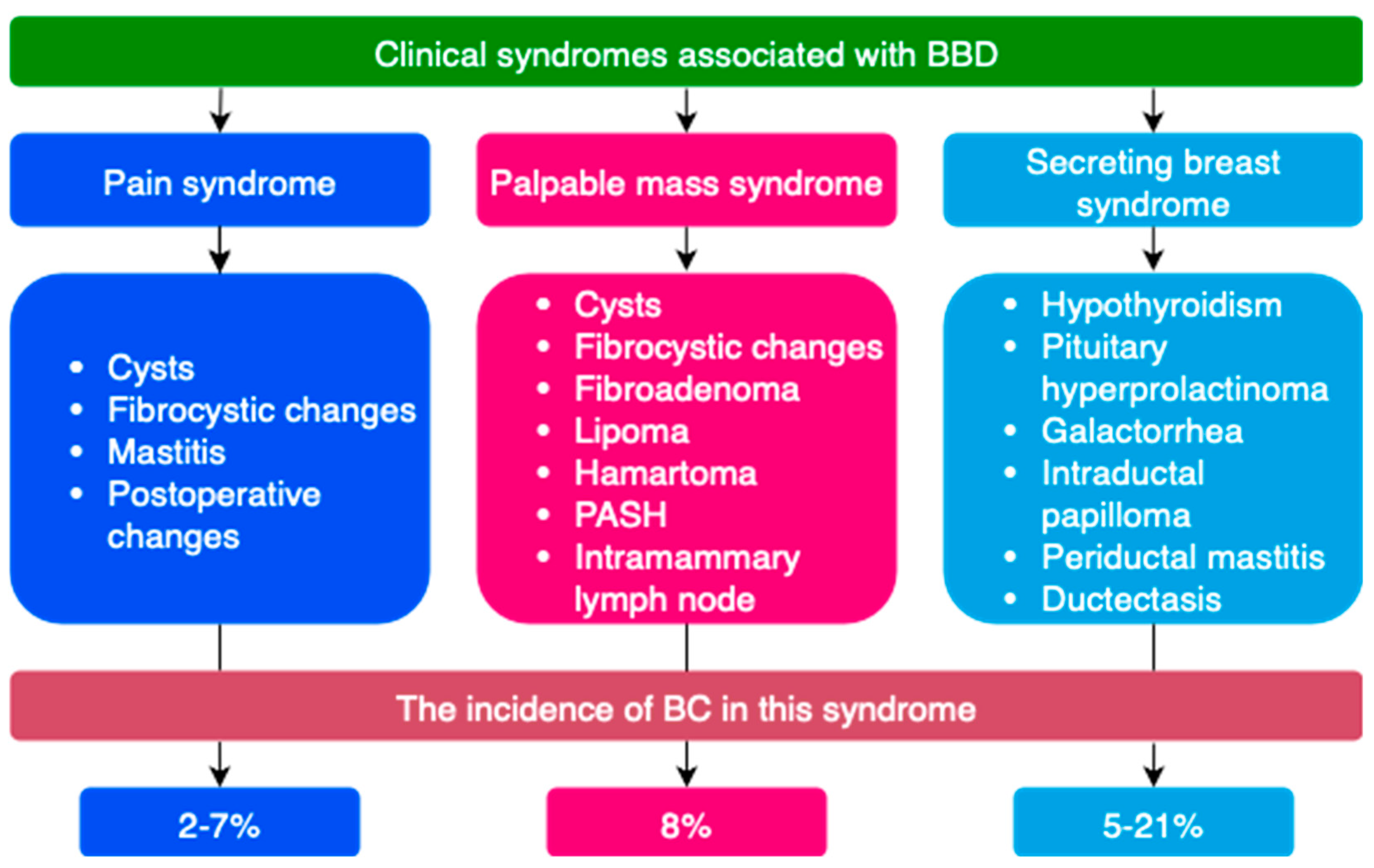
3.1. General Characteristics of BBD
3.2. General Characteristics of Microbiota in BBD
3.3. Features of Gut Microbiota in Breast Pathology
3.4. Mammary Gland Microbiota in Specific Types of BBD
3.4.1. Breast Cysts
3.4.2. Breast Fibroadenomas
3.4.3. Lactational Mastitis
3.4.4. Non-Lactational Mastitis
3.4.5. Granulomatous Mastitis
3.4.6. Ductal Changes
3.4.7. Purulent-Septic Changes of the Breast
3.4.8. Fibrous Capsular Contracture
3.4.9. Anaplastic Large Cell Lymphoma
4. Breast Microbiota in Men
- Developmental anomalies (amastia, polymastia, nipple inversion, athelia, polythelia, etc.)
- Inflammatory and reactive changes (mastitis, abscess, Mondor’s disease, etc.)
- Ductal changes (duct ectasia, intraductal papilloma, etc.)
- Systemic diseases/symptoms of systemic diseases (diabetic mastopathy, gynecomastia)
- Benign neoplasms (lipoma, angiolipoma, cavernous hemangioma, myofibroblastoma, epidermal cysts, pseudoangiomatous stromal hyperplasia (PASH), hamartoma, etc.)
- Traumatic and post-traumatic changes (hematoma, fat necrosis)
5. Limitations in Studying the Microbiota in BBD
- Insufficient study of the microbiota of breast tissue and other areas in the normal state.
- Low biomass of the MG microbiota in normal and pathological conditions.
- Lack of quality, representative data on the breast virome and micromycetes.
- Most research is focused on malignant breast neoplasms.
- Presence of intracellular forms of microorganisms complicates their detection [78].
- Inability to establish cause-and-effect relationships when studying the influence of the microbiome on breast pathology. Available data do not answer the question of whether microbial dysbiosis is a consequence or a cause of breast disease development [45].
- Individual variability of the microbiota and the multifactorial nature of BBD development.
- Presence of borderline states – ‘lesions of uncertain malignant potential (B3)’.
- Inaccessibility of tissue samples: unaltered tissues and tissues with benign changes are often unavailable for research, as biopsies are usually performed when malignancy is suspected. Using adjacent unaltered tissue from cancer cases as a reference can distort research data.
- Creation of diagnostic test systems for the differential diagnosis of benign and malignant neoplasms.
- Differential diagnosis of the secreting breast syndrome.
- Assessment of BC development risks.
- Investigation of etiopathogenetic aspects of BBD development.
- Treatment and prevention of BBD.
6. Materials and Methods
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Swidsinski A, Amann R, Guschin A, Swidsinski S, Loening-Baucke V, Mendling W, Sobel JD, Lamont RF, Vaneechoutte M, Baptista PV, Bradshaw CS, Kogan IY, Savicheva АM, Mitrokhin OV, Swidsinski NW, Sukhikh GT, Priputnevich TV, Apolikhina IA, Dörffel Y. Polymicrobial consortia in the pathogenesis of biofilm vaginosis visualized by FISH. Historic review outlining the basic principles of the polymicrobial infection theory. Microbes Infect. 2024 Nov-Dec;26(8):105403. Epub 2024 Aug 8. PMID: 39127090. [CrossRef]
- Cerca N, Vaneechoutte M, Guschin A, Swidsinski A. Polymicrobial infections and biofilms in women’s health: Gahro Expert Group Meeting Report. Res Microbiol. 2017 Nov-Dec;168(9-10):902-904. Epub 2017 Jul 14. PMID: 28716397. [CrossRef]
- NIH HMP Working Group et al., “The NIH Human Microbiome Project.,” Genome Res, vol. 19, no. 12, pp. 2317–23, Dec. 2009. [CrossRef]
- V. D’Afonseca, E. V. Muñoz, A. L. Leal, P. M. A. S. Soto, and C. Parra-Cid, “Implications of the microbiome and metabolic intermediaries produced by bacteria in breast cancer,” Genet Mol Biol, vol. 47, no. suppl 1, 2024. [CrossRef]
- El-Sayed, L. Aleya, and M. Kamel, “Microbiota’s role in health and diseases.,” Environ Sci Pollut Res Int, vol. 28, no. 28, pp. 36967–36983, Jul. 2021. [CrossRef]
- Y. Wang, Y. Wang, Y. Zhou, Y. Feng, T. Sun, and J. Xu, “Tumor-related fungi and crosstalk with gut fungi in the tumor microenvironment.,” Cancer Biol Med, vol. 21, no. 11, pp. 977–94, Nov. 2024. [CrossRef]
- J. Manos, “The human microbiome in disease and pathology.,” APMIS, vol. 130, no. 12, pp. 690–705, Dec. 2022. [CrossRef]
- J. Zhang et al., “Impact of intestinal dysbiosis on breast cancer metastasis and progression.,” Front Oncol, vol. 12, p. 1037831, 2022. [CrossRef]
- H. Wang et al., “Breast tissue, oral and urinary microbiomes in breast cancer,” Oncotarget, vol. 8, no. 50, pp. 88122–88138, Oct. 2017. [CrossRef]
- Samkari et al., “Body Microbiota and Its Relationship With Benign and Malignant Breast Tumors: A Systematic Review,” Cureus, May 2022. [CrossRef]
- P. Kovács et al., “Lithocholic Acid, a Metabolite of the Microbiome, Increases Oxidative Stress in Breast Cancer.,” Cancers (Basel), vol. 11, no. 9, Aug. 2019. [CrossRef]
- M. Levy et al., “Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling.,” Cell, vol. 163, no. 6, pp. 1428–43, Dec. 2015. [CrossRef]
- E. D. Sonnenburg, S. A. Smits, M. Tikhonov, S. K. Higginbottom, N. S. Wingreen, and J. L. Sonnenburg, “Diet-induced extinctions in the gut microbiota compound over generations.,” Nature, vol. 529, no. 7585, pp. 212–5, Jan. 2016. [CrossRef]
- T. S. Stappenbeck and H. W. Virgin, “Accounting for reciprocal host–microbiome interactions in experimental science,” Nature, vol. 534, no. 7606, pp. 191–199, Jun. 2016. [CrossRef]
- World Health Organization. Breast Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/breast-cancer. (accessed on 1st September 2025).
- E. J. Fernandez-Rodriguez et al., “Study on the additional financial burden of breast cancer disease on cancer patients and their families. Financial toxicity in cancer,” Front Public Health, vol. 12, Jul. 2024. [CrossRef]
- S. Chen et al., “Estimates and Projections of the Global Economic Cost of 29 Cancers in 204 Countries and Territories From 2020 to 2050,” JAMA Oncol, vol. 9, no. 4, p. 465, Apr. 2023. [CrossRef]
- E. M. van de Voort, G. M. Struik, S. P. van Streun, C. Verhoef, C. A. Uyl-de Groot, and T. M. Klem, “Hospital costs and cosmetic outcome of benign and high-risk breast lesions managed by vacuum-assisted excision versus surgical excision,” Br J Radiol, vol. 95, no. 1136, Aug. 2022. [CrossRef]
- L. Zhu et al., “Prevalence of breast fibroadenoma in healthy physical examination population in Guangdong province of China: a cross-sectional study,” BMJ Open, vol. 12, no. 6, p. e057080, Jun. 2022. [CrossRef]
- Stachs, J. Stubert, T. Reimer, and S. Hartmann, “Benign Breast Disease in Women.,” Dtsch Arztebl Int, vol. 116, no. 33–34, pp. 565–574, Aug. 2019. [CrossRef]
- Urbaniak et al., “Microbiota of Human Breast Tissue,” Appl Environ Microbiol, vol. 80, no. 10, pp. 3007–3014, May 2014. [CrossRef]
- X. Wang, H. Gao, Y. Zeng, and J. Chen, “Exploring the relationship between gut microbiota and breast diseases using Mendelian randomization analysis,” Front Med (Lausanne), vol. 11, Nov. 2024. [CrossRef]
- T. J. Hieken et al., “The Microbiome of Aseptically Collected Human Breast Tissue in Benign and Malignant Disease,” Sci Rep, vol. 6, no. 1, p. 30751, Aug. 2016. [CrossRef]
- S. Banerjee et al., “Distinct microbial signatures associated with different breast cancer types,” Front Microbiol, vol. 9, no. MAY, May 2018. [CrossRef]
- Xuan et al., “Microbial Dysbiosis Is Associated with Human Breast Cancer,” PLoS One, vol. 9, no. 1, p. e83744, Jan. 2014. [CrossRef]
- J. M. Rodríguez, “The Origin of Human Milk Bacteria: Is There a Bacterial Entero-Mammary Pathway during Late Pregnancy and Lactation?,” Advances in Nutrition, vol. 5, no. 6, pp. 779–784, Nov. 2014. [CrossRef]
- J. An et al., “Diagnostic Kit of breast cancer via urine microbiome,” European Journal of Surgical Oncology, vol. 46, no. 2, p. e33, Feb. 2020. [CrossRef]
- K. Wang, K. Nakano, N. Naderi, M. Bajaj-Elliott, and A. Mosahebi, “Is the skin microbiota a modifiable risk factor for breast disease?: A systematic review,” The Breast, vol. 59, pp. 279–285, Oct. 2021. [CrossRef]
- Boix-Amorós et al., “Mycobiome Profiles in Breast Milk from Healthy Women Depend on Mode of Delivery, Geographic Location, and Interaction with Bacteria.,” Appl Environ Microbiol, vol. 85, no. 9, May 2019. [CrossRef]
- Laborda-Illanes et al., “Breast and Gut Microbiota Action Mechanisms in Breast Cancer Pathogenesis and Treatment.,” Cancers (Basel), vol. 12, no. 9, Aug. 2020. [CrossRef]
- Gay M, Koleva P, Slupsky C, Toit E, Eggesbo M, Johnson C, Wegienka G, Shimojo N, Campbell D, Prescott S, Munblit D, Geddes D, Kozyrskyj A, InVIVO LactoActive Study Investigators. 2018. Worldwide variation in human milk metabolome: indicators of breast physiology and maternal lifestyle? Nutrients 10:1151. [CrossRef]
- S. Moossavi et al., “Composition and Variation of the Human Milk Microbiota Are Influenced by Maternal and Early-Life Factors.,” Cell Host Microbe, vol. 25, no. 2, pp. 324-335.e4, Feb. 2019. [CrossRef]
- Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. 2012. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr 96:544–551. [CrossRef]
- Owens and K. Stoessel, “Surgical site infections: epidemiology, microbiology and prevention,” Journal of Hospital Infection, vol. 70, pp. 3–10, Nov. 2008. [CrossRef]
- Z. Ye, L. Gao, Z. Guo, and Q. Wang, “Oral and intestinal flora translocation and tumor development.,” J Cancer Res Ther, vol. 21, no. 2, pp. 323–333, May 2025. [CrossRef]
- Z. Zhong et al., “Bifidobacterium animalis subsp. lactis Probio-M8 undergoes host adaptive evolution by glcU mutation and translocates to the infant’s gut via oral-/entero-mammary routes through lactation.,” Microbiome, vol. 10, no. 1, p. 197, Nov. 2022. [CrossRef]
- R. Soto-Pantoja et al., “Diet Alters Entero-Mammary Signaling to Regulate the Breast Microbiome and Tumorigenesis.,” Cancer Res, vol. 81, no. 14, pp. 3890–3904, Jul. 2021. [CrossRef]
- Angelopoulou, D. Field, C. A. Ryan, C. Stanton, C. Hill, and R. P. Ross, “The microbiology and treatment of human mastitis,” Med Microbiol Immunol, vol. 207, no. 2, pp. 83–94, Apr. 2018. [CrossRef]
- H. Wu, S. Ganguly, and T. O. Tollefsbol, “Modulating Microbiota as a New Strategy for Breast Cancer Prevention and Treatment,” Microorganisms, vol. 10, no. 9, p. 1727, Aug. 2022. [CrossRef]
- T. J. Hieken et al., “The breast tissue microbiome, stroma, immune cells and breast cancer.,” Neoplasia, vol. 27, p. 100786, May 2022. [CrossRef]
- J. Chen et al., “Associating microbiome composition with environmental covariates using generalized UniFrac distances.,” Bioinformatics, vol. 28, no. 16, pp. 2106–13, Aug. 2012. [CrossRef]
- Urbaniak, G. B. Gloor, M. Brackstone, L. Scott, M. Tangney, and G. Reid, “The Microbiota of Breast Tissue and Its Association with Breast Cancer,” Appl Environ Microbiol, vol. 82, no. 16, pp. 5039–5048, Aug. 2016. [CrossRef]
- Chan et al., “Characterization of the microbiome of nipple aspirate fluid of breast cancer survivors,” Sci Rep, vol. 6, no. 1, p. 28061, Jun. 2016. [CrossRef]
- L. Costantini et al., “Characterization of human breast tissue microbiota from core needle biopsies through the analysis of multi hypervariable 16S-rRNA gene regions,” Sci Rep, vol. 8, no. 1, p. 16893, Nov. 2018. [CrossRef]
- Niccolai et al., “Breast cancer: the first comparative evaluation of oncobiome composition between males and females,” Biol Sex Differ, vol. 14, no. 1, p. 37, Jun. 2023. [CrossRef]
- K. J. Thompson et al., “A comprehensive analysis of breast cancer microbiota and host gene expression,” PLoS One, vol. 12, no. 11, p. e0188873, Nov. 2017. [CrossRef]
- L. Narunsky-Haziza et al., “Pan-cancer analyses reveal cancer-type-specific fungal ecologies and bacteriome interactions.,” Cell, vol. 185, no. 20, pp. 3789-3806.e17, Sep. 2022. [CrossRef]
- Maldonado-Rodríguez et al., “Presence of Human Papillomavirus DNA in Malignant Neoplasia and Non-Malignant Breast Disease.,” Curr Issues Mol Biol, vol. 44, no. 8, pp. 3648–3665, Aug. 2022. [CrossRef]
- J. S. Lawson, W. H. Günzburg, and N. J. Whitaker, “Viruses and human breast cancer.,” Future Microbiol, vol. 1, no. 1, pp. 33–51, Jun. 2006. [CrossRef]
- S. Guglietta, X. Li, and D. Saxena, “Role of Fungi in Tumorigenesis: Promises and Challenges.,” Annu Rev Pathol, vol. 20, no. 1, pp. 459–482, Jan. 2025. [CrossRef]
- M. Levy, C. A. Thaiss, and E. Elinav, “Metagenomic cross-talk: the regulatory interplay between immunogenomics and the microbiome.,” Genome Med, vol. 7, p. 120, Nov. 2015. [CrossRef]
- T. Kovács, E. Mikó, G. Ujlaki, Z. Sári, and P. Bai, “The Microbiome as a Component of the Tumor Microenvironment,” 2020, pp. 137–153. [CrossRef]
- M. Luu and A. Visekruna, “Microbial metabolites: novel therapeutic tools for boosting cancer therapies,” Trends Cell Biol, vol. 31, no. 11, pp. 873–875, Nov. 2021. [CrossRef]
- T. Kovács et al., “Cadaverine, a metabolite of the microbiome, reduces breast cancer aggressiveness through trace amino acid receptors.,” Sci Rep, vol. 9, no. 1, p. 1300, Feb. 2019. [CrossRef]
- T. Kovács et al., “The involvement of oncobiosis and bacterial metabolite signaling in metastasis formation in breast cancer,” Cancer and Metastasis Reviews, vol. 40, no. 4, pp. 1223–1249, Dec. 2021. [CrossRef]
- Z. Ma, M. Qu, and X. Wang, “Analysis of Gut Microbiota in Patients with Breast Cancer and Benign Breast Lesions,” Pol J Microbiol, vol. 71, no. 2, pp. 217–226, May 2022. [CrossRef]
- J. Zhu et al., “Breast cancer in postmenopausal women is associated with an altered gut metagenome,” Microbiome, vol. 6, no. 1, p. 136, Dec. 2018. [CrossRef]
- J. J. Goedert et al., “Investigation of the Association Between the Fecal Microbiota and Breast Cancer in Postmenopausal Women: a Population-Based Case-Control Pilot Study,” JNCI: Journal of the National Cancer Institute, vol. 107, no. 8, Aug. 2015. [CrossRef]
- P. Yang, Z. Wang, Q. Peng, W. Lian, and D. Chen, “Comparison of the Gut Microbiota in Patients with Benign and Malignant Breast Tumors: A Pilot Study,” Evolutionary Bioinformatics, vol. 17, Jan. 2021. [CrossRef]
- S. Parida and D. Sharma, “The Microbiome–Estrogen Connection and Breast Cancer Risk,” Cells, vol. 8, no. 12, p. 1642, Dec. 2019. [CrossRef]
- M. F. Fernández, I. Reina-Pérez, J. M. Astorga, A. Rodríguez-Carrillo, J. Plaza-Díaz, and L. Fontana, “Breast Cancer and Its Relationship with the Microbiota,” Int J Environ Res Public Health, vol. 15, no. 8, p. 1747, Aug. 2018. [CrossRef]
- R. F. Schwabe and C. Jobin, “The microbiome and cancer,” Nat Rev Cancer, vol. 13, no. 11, pp. 800–812, Nov. 2013. [CrossRef]
- K. Al-Shami et al., “Estrogens and the risk of breast cancer: A narrative review of literature,” Heliyon, vol. 9, no. 9, p. e20224, Sep. 2023. [CrossRef]
- S. F. Doisneau-Sixou, C. M. Sergio, J. S. Carroll, R. Hui, E. A. Musgrove, and R. L. Sutherland, “Estrogen and antiestrogen regulation of cell cycle progression in breast cancer cells.,” Endocr Relat Cancer, pp. 179–186, Jun. 2003. [CrossRef]
- Mikó et al., “Microbiome—Microbial Metabolome—Cancer Cell Interactions in Breast Cancer—Familiar, but Unexplored,” Cells, vol. 8, no. 4, p. 293, Mar. 2019. [CrossRef]
- X. Deng et al., “Bibliometric analysis of global research trends between gut microbiota and breast cancer: from 2013 to 2023,” Front Microbiol, vol. 15, Jul. 2024. [CrossRef]
- R. Flores et al., “Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: a cross-sectional study,” J Transl Med, vol. 10, no. 1, p. 253, Dec. 2012. [CrossRef]
- B. J. Fuhrman et al., “Associations of the fecal microbiome with urinary estrogens and estrogen metabolites in postmenopausal women.,” J Clin Endocrinol Metab, vol. 99, no. 12, pp. 4632–40, Dec. 2014. [CrossRef]
- Peterson, N. P. McNulty, J. L. Guruge, and J. I. Gordon, “IgA Response to Symbiotic Bacteria as a Mediator of Gut Homeostasis,” Cell Host Microbe, vol. 2, no. 5, pp. 328–339, Nov. 2007. [CrossRef]
- S. Haque et al., “Microbial dysbiosis and epigenetics modulation in cancer development – A chemopreventive approach,” Semin Cancer Biol, vol. 86, pp. 666–681, Nov. 2022. [CrossRef]
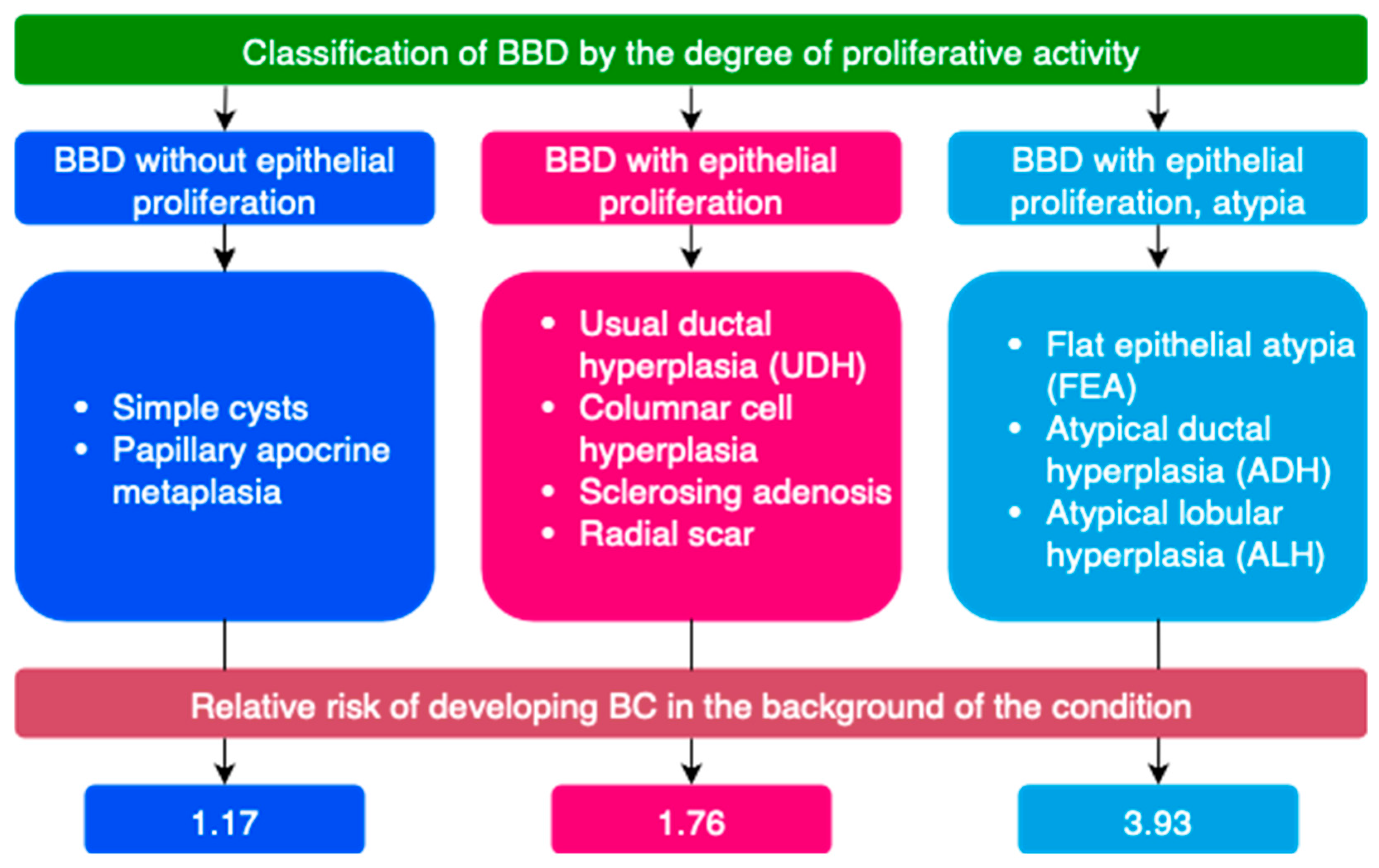
- S. W. Dyrstad, Y. Yan, A. M. Fowler, and G. A. Colditz, “Breast cancer risk associated with benign breast disease: systematic review and meta-analysis,” Breast Cancer Res Treat, vol. 149, no. 3, pp. 569–575, Feb. 2015. [CrossRef]
- M. Onstad and A. Stuckey, “Benign Breast Disorders,” Obstet Gynecol Clin North Am, vol. 40, no. 3, pp. 459–473, Sep. 2013. [CrossRef]
- J. Iddon and J. M. Dixon, “Mastalgia,” BMJ, vol. 347, no. dec12 2, p. bmj.f3288-bmj.f3288, Dec. 2013. [CrossRef]
- M. B. Barton, J. G. Elmore, and S. W. Fletcher, “Breast Symptoms among Women Enrolled in a Health Maintenance Organization: Frequency, Evaluation, and Outcome,” Ann Intern Med, vol. 130, no. 8, pp. 651–657, Apr. 1999. [CrossRef]
- S.-J. Lee et al., “ACR Appropriateness Criteria ® Evaluation of Nipple Discharge,” Journal of the American College of Radiology, vol. 14, no. 5, pp. S138–S153, May 2017. [CrossRef]
- S. C. Dupont, J. C. Boughey, R. E. Jimenez, T. L. Hoskin, and T. J. Hieken, “Frequency of diagnosis of cancer or high-risk lesion at operation for pathologic nipple discharge,” Surgery, vol. 158, no. 4, pp. 988–995, Oct. 2015. [CrossRef]
- Ellis et al., “Best Practice No 179. Guidelines for breast needle core biopsy handling and reporting in breast screening assessment.,” J Clin Pathol, vol. 57, no. 9, pp. 897–902, Sep. 2004. [CrossRef]
- M. J. Worsham et al., “Risk factors for breast cancer from benign breast disease in a diverse population,” Breast Cancer Res Treat, vol. 118, no. 1, pp. 1–7, Nov. 2009. [CrossRef]
- Johansson et al., “Characterization of Benign Breast Diseases and Association With Age, Hormonal Factors, and Family History of Breast Cancer Among Women in Sweden,” JAMA Netw Open, vol. 4, no. 6, p. e2114716, Jun. 2021. [CrossRef]
- S. Banerjee et al., “Prognostic correlations with the microbiome of breast cancer subtypes,” Cell Death Dis, vol. 12, no. 9, p. 831, Sep. 2021. [CrossRef]
- D. Nejman et al., “The human tumor microbiome is composed of tumor type-specific intracellular bacteria.,” Science, vol. 368, no. 6494, pp. 973–980, May 2020. [CrossRef]
- N. Swamy, M. Rohilla, S. Raichandani, and G. Bryant-Smith, “Epidemiology of male breast diseases: A 10-year institutional review.,” Clin Imaging, vol. 72, pp. 142–150, Apr. 2021. [CrossRef]
- Bullerdiek and B. Rommel, “Factors targeting MED12 to drive tumorigenesis?,” F1000Res, vol. 7, p. 359, 2018. [CrossRef]
- Zhu et al., “Interactions between the breast tissue microbiota and host gene regulation in nonpuerperal mastitis,” Microbes Infect, vol. 24, no. 3, p. 104904, Apr. 2022. [CrossRef]
- S. M. Park et al., “Breast abscess caused by Staphylococcus aureus in 2 adolescent girls with atopic dermatitis,” Korean J Pediatr, vol. 61, no. 6, p. 200, 2018. [CrossRef]
- J. Wang et al., “Pathogens in patients with granulomatous lobular mastitis,” International Journal of Infectious Diseases, vol. 81, pp. 123–127, Apr. 2019. [CrossRef]
- Yu et al., “Clinical metagenomic analysis of bacterial communities in breast abscesses of granulomatous mastitis,” International Journal of Infectious Diseases, vol. 53, pp. 30–33, Dec. 2016. [CrossRef]
- Boakes, A. Woods, N. Johnson, and N. Kadoglou, “Breast Infection: A Review of Diagnosis and Management Practices,” Eur J Breast Health, Jun. 2018. [CrossRef]
- Liu et al., “Periductal Mastitis: An Inflammatory Disease Related to Bacterial Infection and Consequent Immune Responses?,” Mediators Inflamm, vol. 2017, pp. 1–9, 2017. [CrossRef]
- Moazzez, “Breast Abscess Bacteriologic Features in the Era of Community-Acquired Methicillin-Resistant Staphylococcus aureus Epidemics,” Archives of Surgery, vol. 142, no. 9, p. 881, Sep. 2007. [CrossRef]
- D. C. Wu, W. W. Chan, A. I. Metelitsa, L. Fiorillo, and A. N. Lin, “Pseudomonas Skin Infection,” Am J Clin Dermatol, vol. 12, no. 3, pp. 157–169, Jun. 2011. [CrossRef]
- S. M. Rudresh, “Non Diphtheritic Corynebacteria : An Emerging Nosocomial Pathogen in Skin and Soft Tissue Infection,” JOURNAL OF CLINICAL AND DIAGNOSTIC RESEARCH, 2015. [CrossRef]
- Torrens, R. Marí, A. Alier, L. Puig, F. Santana, and S. Corvec, “Cutibacterium acnes in primary reverse shoulder arthroplasty: from skin to deep layers,” J Shoulder Elbow Surg, vol. 28, no. 5, pp. 839–846, May 2019. [CrossRef]
- Fazli et al., “Nonrandom Distribution of Pseudomonas aeruginosa and Staphylococcus aureus in Chronic Wounds,” J Clin Microbiol, vol. 47, no. 12, pp. 4084–4089, Dec. 2009. [CrossRef]
- Stachon et al., “Intraoperative Assessment of Endogenous Microbiota in the Breast.,” Rev Bras Ginecol Obstet, vol. 43, no. 10, pp. 759–764, Oct. 2021. [CrossRef]
- Cook, C. J. Holmes, R. Wixtrom, M. I. Newman, and J. N. Pozner, “Characterizing the Microbiome of the Contracted Breast Capsule Using Next Generation Sequencing,” Aesthet Surg J, vol. 41, no. 4, pp. 440–447, Mar. 2021. [CrossRef]
- Carvajal, M. Carvajal, and G. Hernández, “Back to Basics: Could the Preoperative Skin Antiseptic Agent Help Prevent Biofilm-Related Capsular Contracture?,” Aesthet Surg J, vol. 39, no. 8, pp. 848–859, Jul. 2019. [CrossRef]
- Y. Bachour et al., “PCR Characterization of Microbiota on Contracted and Non-Contracted Breast Capsules,” Aesthetic Plast Surg, vol. 43, no. 4, pp. 918–926, Aug. 2019. [CrossRef]
- Hu et al., “Bacterial Biofilm Infection Detected in Breast Implant–Associated Anaplastic Large-Cell Lymphoma,” Plast Reconstr Surg, vol. 137, no. 6, pp. 1659–1669, Jun. 2016. [CrossRef]
- N. Walker et al., “Insights into the Microbiome of Breast Implants and Periprosthetic Tissue in Breast Implant-Associated Anaplastic Large Cell Lymphoma,” Sci Rep, vol. 9, no. 1, p. 10393, Jul. 2019. [CrossRef]
- Charlot et al., “Pathologies of the male breast.,” Diagn Interv Imaging, vol. 94, no. 1, pp. 26–37, Jan. 2013. [CrossRef]
- Firmin-Lefebvre, D., & Misery, L. (2013). Pathologie du sein de l’homme [Male breast diseases]. Annales de dermatologie et de venereologie, 140(6-7), 436–443. [CrossRef]
- J. Chadha, D. Nandi, Y. Atri, and A. Nag, “Significance of human microbiome in breast cancer: Tale of an invisible and an invincible,” Semin Cancer Biol, vol. 70, pp. 112–127, May 2021. [CrossRef]
| Phylum | Family | Genus | Species | References |
|---|---|---|---|---|
| Pseudomonadota (Proteobacteria) | Sphingomonadaceae | - | - | [43] |
| Methylobacteriaceae | Methylobacterium | - | [4,9] | |
| Burkholderiaceae | Ralstonia | - | [4,43,44] | |
| Sphingomonadaceae | Sphingomonas | yanoikuyae | [4] | |
| - | [21,23,43] | |||
| Pseudomonadaceae | Pseudomonas | - | [21,43,45] | |
| Comamonadaceae | - | - | [21] | |
| Enterobacteriaceae | - | - | [21] | |
| Moraxellaceae | Acinetobacter | - | [21] | |
| Pasteurellaceae | Haemophilus | - | [45] | |
| Neisseriaceae | Neisseria | - | [45] | |
| - | - | - | [4,9,21,25,44] | |
| Bacillota (Firmicutes) | Veillonellaceae | Veillonella | - | [45] |
| Staphylococcaceae | Staphylococcus | - | [21,43] | |
| Streptococcaceae | Lactococcus | - | [43] | |
| Streptococcus | - | [43] | ||
| Clostridiaceae | Clostridium | - | [43] | |
| Lactobacillaceae | Lactobacillus | - | [43] | |
| Listeriaceae | Listeria | welshimeri | [21] | |
| - | - | - | [4,9,21,25,44,46] | |
| Actinomycetota (Actinobacteria) | Corynebacteriaceae | Corynebacterium | - | [43] |
| Micrococcaceae | Micrococcus | - | [4] | |
| Propionibacteriaceae | Propionibacterium | - | [21,43] | |
| - | - | - | [4,25] | |
| Bacteroidota (Bacteroidetes) | Bacteroidaceae | Bacteroides | - | [9] |
| Prevotellaceae | Prevotella | - | [21,43] | |
| - | - | - | [9,25,43] |
| Bacterial metabolite | Metabolic effect | References |
|---|---|---|
| Сytolethal distending toxin (CDT) and colibactin | Promotes DNA double strand breaks (DSB) | [35] |
| Rho GTPase family proteins | Reorganizing actin cytoskeleton | [35] |
| Cadaverine | Endothelial to mesenchymal transition modulation | [4] |
| Lithocholic acid (LCA) | Increases oxidative stress. Regulates KEAP1, NRF2, TGR5, GPX3 expression | [4] |
| Lipopolysaccharides (LPS) | Associated with S100A7 expression – regulates mammary cell proliferation | [4] |
| Trimethylamine N-oxide (TMAO) | Effects cell proliferation by α-casein | [4] |
| β-glucuronidase and/or β-glucosidase | Promote recirculation of estrogen and estrogen-like metabolites | [30] |
| Short chain fatty acids, folates, biotin | Activate epigenetically silenced genes in cells such as p21, BAK etc. | [12,30] |
| Disease/Condition | Microorganisms/Taxa (Associated) | Microorganisms/Taxa (Protective/Risk-Reducing) | Location/Notes | References |
|---|---|---|---|---|
| Breast cysts | Family Alcaligenaceae (gut) | Genus Eubacterium ruminantium, Lactococcus (gut) | Association is based on analysis of gut microbiota. HPV is detected in 40% of cases. | [22,56,82] |
| Breast fibroadenomas | Staphylococcus aureus, HPV (DNA detected in 38.9% of cases) | Not specified | S. aureus is considered a factor contributing to MED12 gene mutation. | [48,56,83] |
| Lactational mastitis | Staphylococcus aureus (main pathogen), coagulase-negative staphylococci, Streptococcus, Pseudomonas aeruginosa, Escherichia coli. Genera Anaerofilum, Anaerotruncus (gut) | Genus Butyricimonas, orders Coriobacteriales, Pasteurellales, Verrucomicrobiales (gut) | Lactational mastitis accounts for 33% of all breast diseases. | [20,22,56,84] |
| Non-lactational mastitis (including duct ectasia, periductal mastitis) | Family Prevotellaceae (gut). Genera Ruminococcus, Coprococcus, Clostridium (breast tissue). | Not specified | The breast tissue microbiota composition differs from that of healthy patients. May be associated with autoimmune reactions. | [22,28,84,85,86] |
| Granulomatous mastitis | Corynebacterium kroppenstedtii (key pathogen), genera Pseudomonas, Brevundimonas, Stenotrophomonas, Acinetobacter, fungi of the genus Aspergillus. | Not specified | Rare disease. Etiology is unknown; possible roles include dyshormonal changes and autoimmune reactions. | [20,28,84,86,87] |
| Periductal mastitis (duct changes) | Genera Enterococcus, Streptococcus, Bacteroides. | Not specified | Bacterial flora is detected in 50-62% of cases, often against the background of duct ectasia. | [84,88,89] |
| Purulent-septic changes (abscesses) | Coagulase-negative staphylococci, Peptostreptococci, Staphylococcus aureus (including MRSA), Corynebacterium, Pseudomonas aeruginosa. | Not specified | Often occur as a complication of non-lactational mastitis. May be associated with impaired skin barrier function (e.g., dermatitis). | [20,28,84,90,91,92] |
| Fibrous capsular contracture (post-mammoplasty) | Staphylococcus epidermidis (most common), Escherichia coli, Diaphorobacter nitroreducens, Cutibacterium acnes, Staphylococcus aureus, Staphylococcus spp., Pseudomonas aeruginosa, Sphingomonas paucimobilis. | Not specified | Associated with bacterial colonization of the implant and biofilm formation. | [27,34,93,94,95,96,97,98] |
| Anaplastic large cell lymphoma (BIA-ALCL) | Staphylococcus saprophyticus, representatives of the genus Ralstonia. | Not specified | Associated with bacterial colonization and biofilms on breast implants. | [96,99,100] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





