Submitted:
06 February 2025
Posted:
06 February 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Selection Criteria, Database Search and Study Design
2.2. Downloading and Pre-Processing of 16S rRNA Datasets
2.3. Alpha and Beta Diversity Analyses
2.4. Differential Abundance and Prevalence Between Different Breast Tissues
2.5. Reanalysis of Breast Tissue Microbiome Data from the Cancer Genome Atlas
2.6. Microbial Correlation Analysis with Tumor Phenotype and Clinical Survival Data
2.7. Statistical Analysis
3. Results
3.1. Study Selection
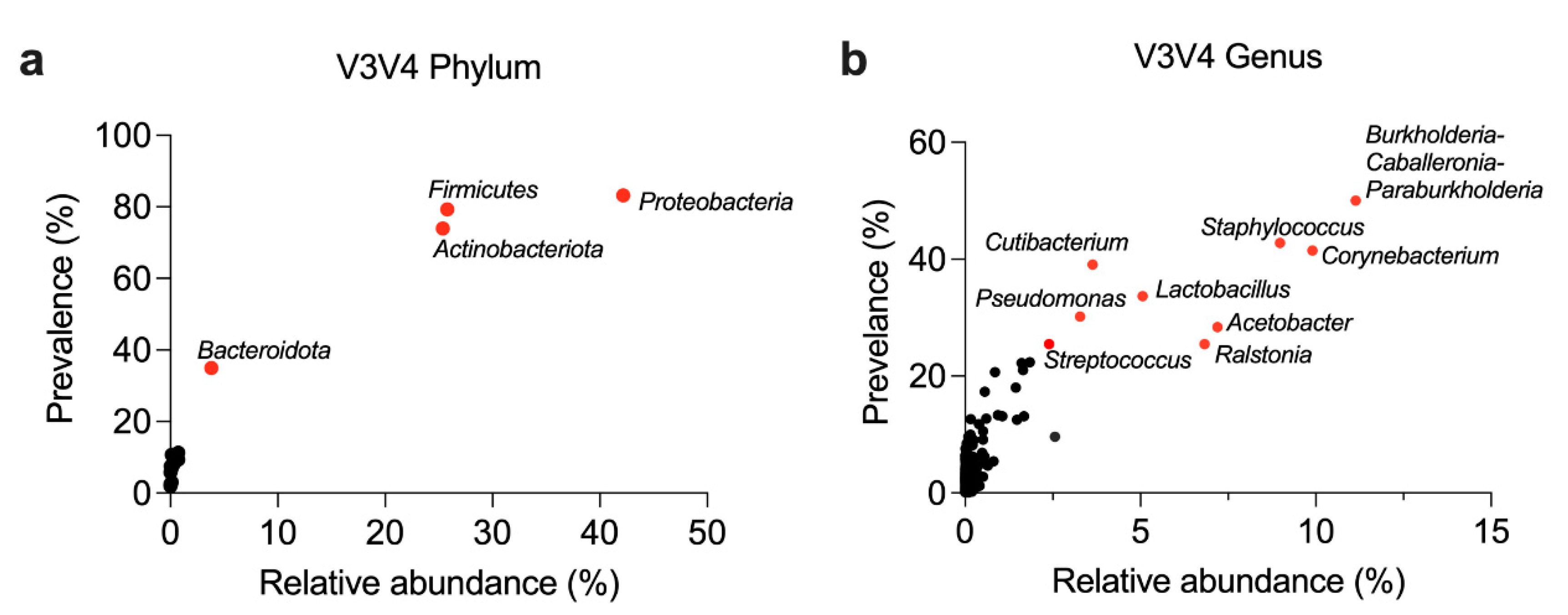
3.2. General Composition of the Breast Microbiome at the Phylum and Genus Level
| Phylum | Relative Abundance (Mean ± SD) | Prevalence (%) |
| Proteobacteria | 42.2 ± 33.1% | 83.2 % |
| Firmicutes | 25.8 ± 28.3 % | 79.3 % |
| Actinobacteriota | 25.4 ± 29.7 % | 73.9 % |
| Bacteroidota | 3.8 ± 12.7 % | 35.0 % |
| Genera | Relative Abundance (Mean ± SD) | Prevalence (%) |
| Burkholderia-Caballeronia-Paraburkholderia | 11.1 ± 20.6 % | 50.0 % |
| Corynebacterium | 9.9 ± 21.1 % | 41.5 % |
| Staphylococcus | 9.0 ± 20.3 % | 42.8 % |
| Acetobacter | 7.2 ± 16.8 % | 28.4 % |
| Ralstonia | 6.8 ± 18.5 % | 25.5 % |
| Lactobacillus | 5.1 ± 12.9 % | 33.7 % |
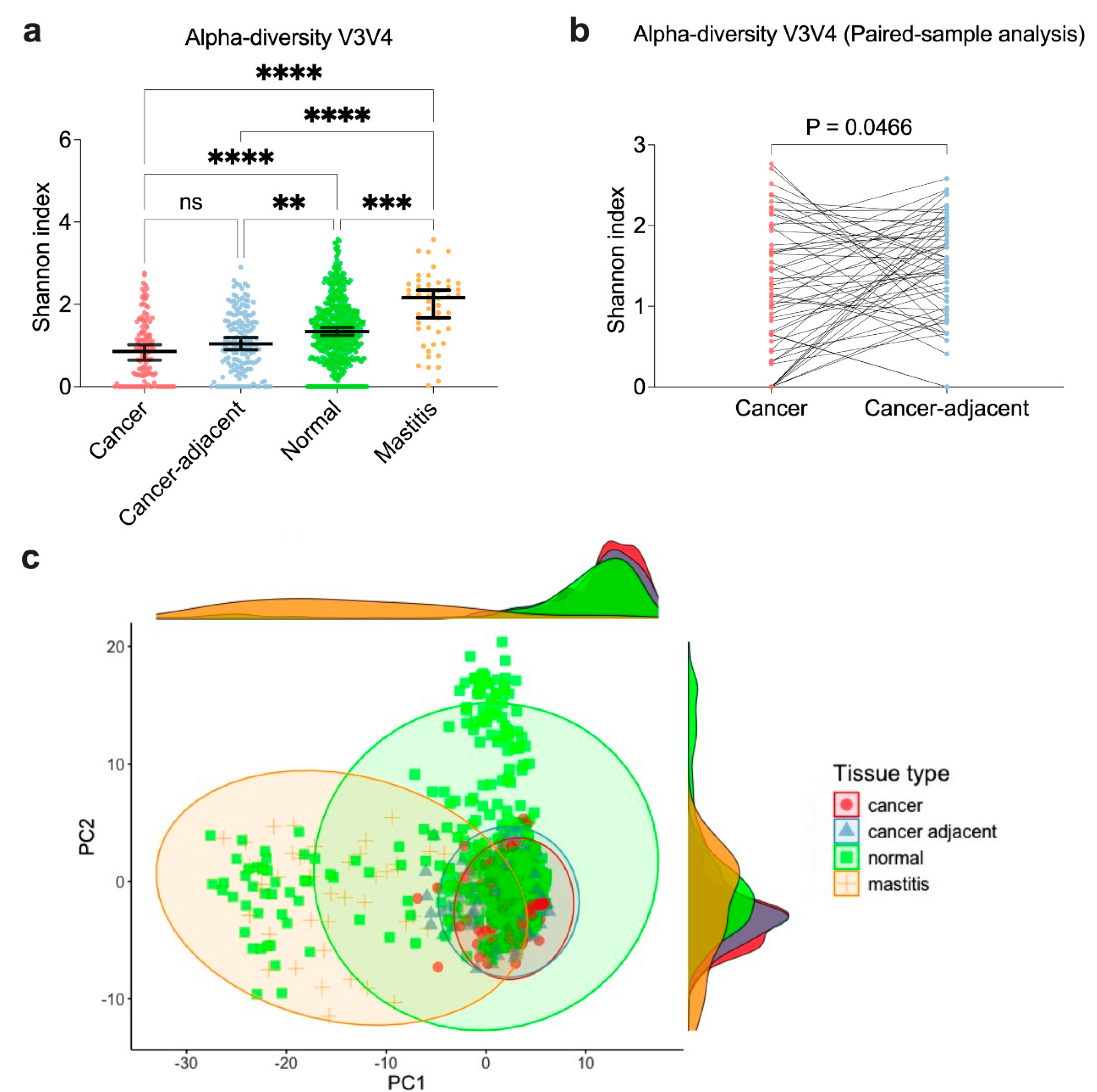
3.3. Similar Microbial Diversity Between Cancer and Cancer-Adjacent Breast Tissue, but Distinct from Mastitis and Normal Tissues
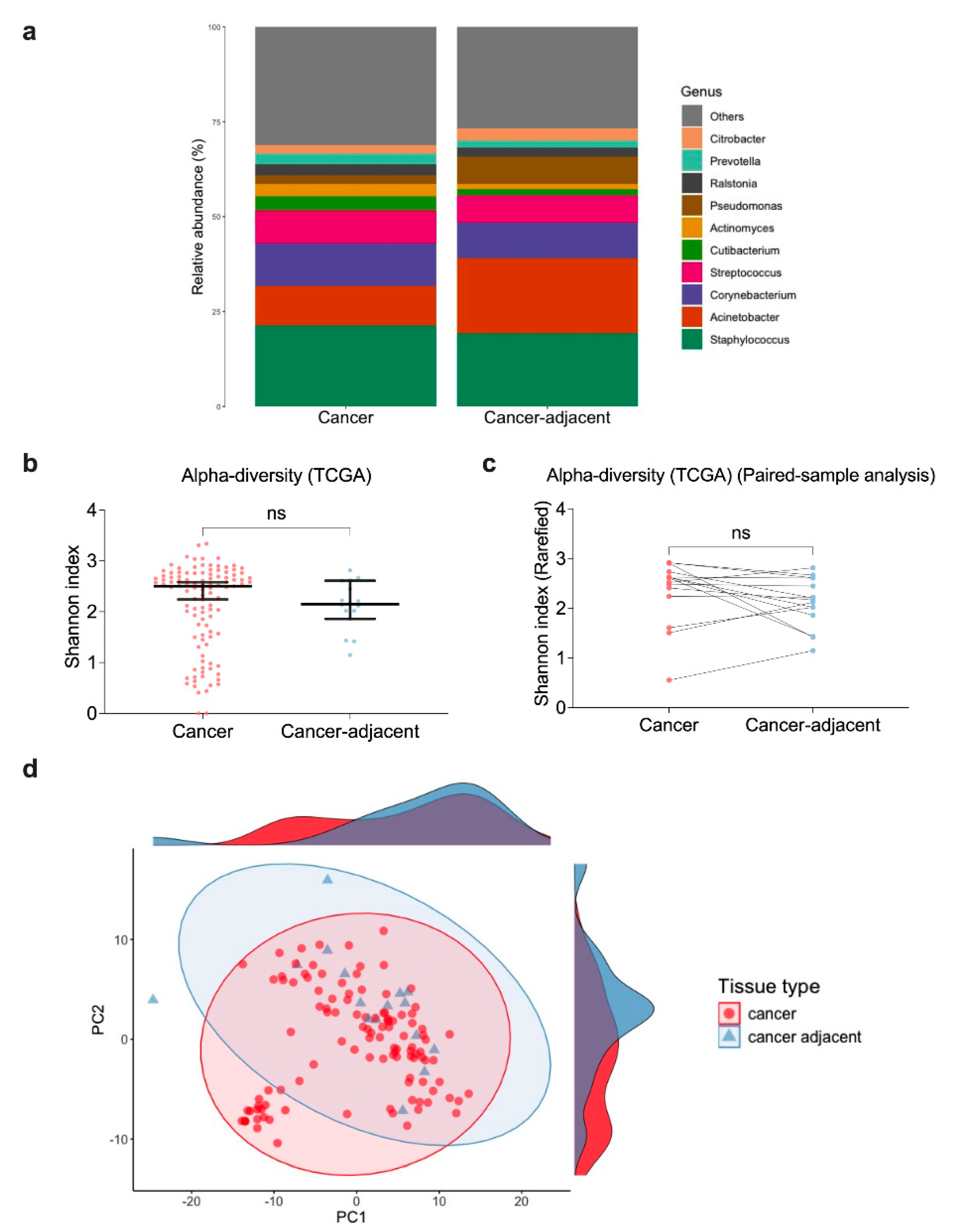
3.4. Similar Genus-Level Abundance Between Cancer and Cancer-Adjacent Breast Tissue Samples
3.5. Microbial Profiles of Cancer and Cancer-Adjacent Breast Tissues in TCGA-BRCA Are Consistent with 16S rRNA Sequencing
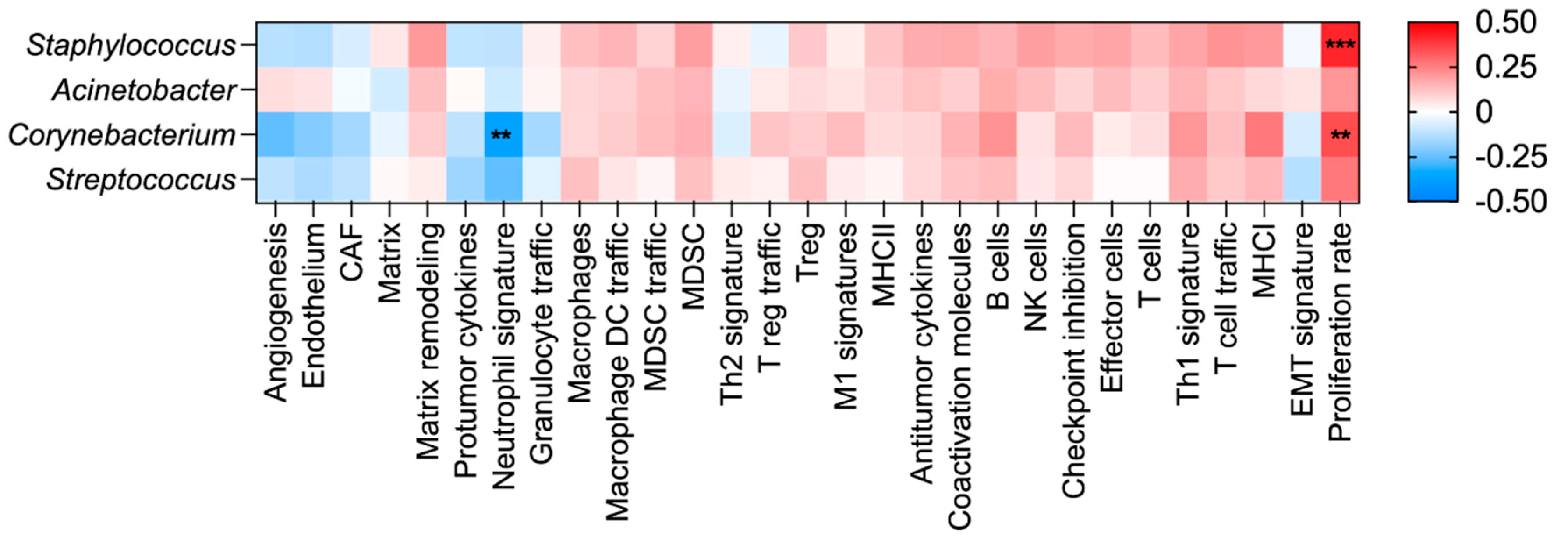
3.6. Evaluation of Microbial Abundance with Tumor Phenotype and Overall Survival
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| FGES | Functional gene expression signatures |
| FFPE | Formalin-fixed paraffin-embedded |
| TCGA | The Cancer Genome Atlas |
| TCGA-BRCA | The Cancer Genome Atlas Breast Cancer |
| DADA | Divisive Amplicon Denoising Algorithm |
| FDR | False discovery rate |
| ND | Not detected |
| CLR | Centered log ratio |
References
- Cao, Y.; Xia, H.; Tan, X.; Shi, C.; Ma, Y.; Meng, D.; Zhou, M.; Lv, Z.; Wang, S.; Jin, Y. Intratumoural microbiota: a new frontier in cancer development and therapy. Signal Transduction and Targeted Therapy 2024, 9, 15.
- Laborda-Illanes, A.; Sanchez-Alcoholado, L.; Dominguez-Recio, M.E.; Jimenez-Rodriguez, B.; Lavado, R.; Comino-Méndez, I.; Alba, E.; Queipo-Ortuño, M.I. Breast and gut microbiota action mechanisms in breast cancer pathogenesis and treatment. Cancers 2020, 12, 2465.
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast cancer. Nature reviews Disease primers 2019, 5, 66.
- Chen, J.; Douglass, J.; Prasath, V.; Neace, M.; Atrchian, S.; Manjili, M.H.; Shokouhi, S.; Habibi, M. The microbiome and breast cancer: a review. Breast cancer research and treatment 2019, 178, 493-496.
- Xavier, J.B.; Young, V.B.; Skufca, J.; Ginty, F.; Testerman, T.; Pearson, A.T.; Macklin, P.; Mitchell, A.; Shmulevich, I.; Xie, L. The cancer microbiome: distinguishing direct and indirect effects requires a systemic view. Trends in cancer 2020, 6, 192-204.
- Liu, C.-C.; Wolf, M.; Ortego, R.; Grencewicz, D.; Sadler, T.; Eng, C. Characterization of immunomodulating agents from Staphylococcus aureus for priming immunotherapy in triple-negative breast cancers. Scientific Reports 2024, 14, 756.
- Parhi, L.; Alon-Maimon, T.; Sol, A.; Nejman, D.; Shhadeh, A.; Fainsod-Levi, T.; Yajuk, O.; Isaacson, B.; Abed, J.; Maalouf, N. Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nature communications 2020, 11, 3259.
- Kim, H.-E.; Kim, J.; Maeng, S.; Oh, B.; Hwang, K.-T.; Kim, B.-S. Microbiota of breast tissue and its potential association with regional recurrence of breast cancer in Korean women. Journal of microbiology and biotechnology 2021, 31, 1643.
- German, R.; Marino, N.; Hemmerich, C.; Podicheti, R.; Rusch, D.B.; Stiemsma, L.T.; Gao, H.; Xuei, X.; Rockey, P.; Storniolo, A.M. Exploring breast tissue microbial composition and the association with breast cancer risk factors. Breast Cancer Research 2023, 25, 82.
- Hoskinson, C.; Zheng, K.; Gabel, J.; Kump, A.; German, R.; Podicheti, R.; Marino, N.; Stiemsma, L.T. Composition and functional potential of the human mammary microbiota prior to and following breast tumor diagnosis. Msystems 2022, 7, e01489-01421.
- Kartti, S.; Bendani, H.; Boumajdi, N.; Bouricha, E.M.; Zarrik, O.; El Agouri, H.; Fokar, M.; Aghlallou, Y.; El Jaoudi, R.; Belyamani, L. Metagenomics analysis of breast microbiome highlights the abundance of Rothia genus in tumor tissues. Journal of Personalized Medicine 2023, 13, 450.
- Thyagarajan, S.; Zhang, Y.; Thapa, S.; Allen, M.S.; Phillips, N.; Chaudhary, P.; Kashyap, M.V.; Vishwanatha, J.K. Comparative analysis of racial differences in breast tumor microbiome. Scientific reports 2020, 10, 14116.
- Zhu, J.; Wu, J.; Liang, Z.; Mo, C.; Qi, T.; Liang, S.; Lian, T.; Qiu, R.; Yu, X.; Tang, X. Interactions between the breast tissue microbiota and host gene regulation in nonpuerperal mastitis. Microbes and Infection 2022, 24, 104904.
- Hieken, T.J.; Chen, J.; Chen, B.; Johnson, S.; Hoskin, T.L.; Degnim, A.C.; Walther-Antonio, M.R.; Chia, N. The breast tissue microbiome, stroma, immune cells and breast cancer. Neoplasia 2022, 27, 100786.
- Esposito, M.V.; Fosso, B.; Nunziato, M.; Casaburi, G.; D’Argenio, V.; Calabrese, A.; D’Aiuto, M.; Botti, G.; Pesole, G.; Salvatore, F. Microbiome composition indicate dysbiosis and lower richness in tumor breast tissues compared to healthy adjacent paired tissue, within the same women. BMC cancer 2022, 22, 1-11.
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E. The human tumor microbiome is composed of tumor type–specific intracellular bacteria. Science 2020, 368, 973-980.
- Urbaniak, C.; Gloor, G.B.; Brackstone, M.; Scott, L.; Tangney, M.; Reid, G. The microbiota of breast tissue and its association with breast cancer. Applied and environmental microbiology 2016, 82, 5039-5048.
- Costantini, L.; Magno, S.; Albanese, D.; Donati, C.; Molinari, R.; Filippone, A.; Masetti, R.; Merendino, N. Characterization of human breast tissue microbiota from core needle biopsies through the analysis of multi hypervariable 16S-rRNA gene regions. Scientific reports 2018, 8, 16893.
- Li, X.; Sun, X.; Zhang, A.; Pang, J.; Li, Y.; Yan, M.; Xu, Z.; Yu, Y.; Yang, Z.; Chen, X. Breast microbiome associations with breast tumor characteristics and neoadjuvant chemotherapy: A case-control study. Frontiers in Oncology 2022, 12, 926920.
- Tzeng, A.; Sangwan, N.; Jia, M.; Liu, C.-C.; Keslar, K.S.; Downs-Kelly, E.; Fairchild, R.L.; Al-Hilli, Z.; Grobmyer, S.R.; Eng, C. Human breast microbiome correlates with prognostic features and immunological signatures in breast cancer. Genome Medicine 2021, 13, 1-17.
- Thompson, K.J.; Ingle, J.N.; Tang, X.; Chia, N.; Jeraldo, P.R.; Walther-Antonio, M.R.; Kandimalla, K.K.; Johnson, S.; Yao, J.Z.; Harrington, S.C. A comprehensive analysis of breast cancer microbiota and host gene expression. PloS one 2017, 12, e0188873.
- Niccolai, E.; Baldi, S.; Nannini, G.; Gensini, F.; Papi, L.; Vezzosi, V.; Bianchi, S.; Orzalesi, L.; Ramazzotti, M.; Amedei, A. Breast cancer: the first comparative evaluation of oncobiome composition between males and females. Biology of sex differences 2023, 14, 37.
- Chang, J.; Li, X.; Xia, Q.; Yang, S.; Zhang, H.; Yang, H. Potential values of formalin-fixed paraffin-embedded tissues for intratumoral microbiome analysis in breast cancer. Heliyon 2023, 9.
- Chiba, A.; Bawaneh, A.; Velazquez, C.; Clear, K.Y.; Wilson, A.S.; Howard-McNatt, M.; Levine, E.A.; Levi-Polyachenko, N.; Yates-Alston, S.A.; Diggle, S.P. Neoadjuvant chemotherapy shifts breast tumor microbiota populations to regulate drug responsiveness and the development of metastasis. Molecular Cancer Research 2020, 18, 130-139.
- Crowe, S.A.; Simister, R.L.; Spence, J.S.; Kenward, P.A.; Van Slyke, A.C.; Lennox, P.; Carr, N. Microbial community compositions in breast implant biofilms associated with contracted capsules. PLoS One 2021, 16, e0249261.
- Desalegn, Z.; Smith, A.; Yohannes, M.; Cao, X.; Anberber, E.; Bekuretsion, Y.; Assefa, M.; Bauer, M.; Vetter, M.; Kantelhardt, E.J. Human breast tissue microbiota reveals unique microbial signatures that correlate with prognostic features in adult Ethiopian women with breast cancer. Cancers 2023, 15, 4893.
- Hilmi, M.; Kamal, M.; Vacher, S.; Dupain, C.; Ibadioune, S.; Halladjian, M.; Sablin, M.P.; Marret, G.; Ajgal, Z.C.; Nijnikoff, M. Intratumoral microbiome is driven by metastatic site and associated with immune histopathological parameters: An ancillary study of the SHIVA clinical trial. European Journal of Cancer 2023, 183, 152-161.
- Hogan, G.; Eckenberger, J.; Narayanen, N.; Walker, S.P.; Claesson, M.J.; Corrigan, M.; O’Hanlon, D.; Tangney, M. Biopsy bacterial signature can predict patient tissue malignancy. Scientific Reports 2021, 11, 18535.
- Liu, E.; Zhang, F.; Xu, T.; Ye, L.; Ma, S.S.Q.; Ji, Z.-S. Relationship between tumor microbiota transcriptional activity and gene expression in breast cancer. BMC cancer 2023, 23, 252.
- Ma, J.; Sun, L.; Liu, Y.; Ren, H.; Shen, Y.; Bi, F.; Zhang, T.; Wang, X. Alter between gut bacteria and blood metabolites and the anti-tumor effects of Faecalibacterium prausnitzii in breast cancer. BMC microbiology 2020, 20, 1-19.
- Meng, S.; Chen, B.; Yang, J.; Wang, J.; Zhu, D.; Meng, Q.; Zhang, L. Study of microbiomes in aseptically collected samples of human breast tissue using needle biopsy and the potential role of in situ tissue microbiomes for promoting malignancy. Frontiers in oncology 2018, 8, 318.
- Ong, S.S.; Xu, J.; Sim, C.K.; Khng, A.J.; Ho, P.J.; Kwan, P.K.W.; Ravikrishnan, A.; Tan, K.-T.B.; Tan, Q.T.; Tan, E.Y. Profiling microbial communities in idiopathic granulomatous mastitis. International Journal of Molecular Sciences 2023, 24, 1042.
- Smith, A.; Pierre, J.F.; Makowski, L.; Tolley, E.; Lyn-Cook, B.; Lu, L.; Vidal, G.; Starlard-Davenport, A. Distinct microbial communities that differ by race, stage, or breast-tumor subtype in breast tissues of non-Hispanic Black and non-Hispanic White women. Scientific reports 2019, 9, 11940.
- Smith, A.; Cao, X.; Gu, Q.; Kubi Amos-Abanyie, E.; Tolley, E.A.; Vidal, G.; Lyn-Cook, B.; Starlard-Davenport, A. Characterization of the metabolome of breast tissues from non-Hispanic black and non-Hispanic white women reveals correlations between microbial Dysbiosis and enhanced lipid metabolism pathways in triple-negative breast tumors. Cancers 2022, 14, 4075.
- Walker, J.N.; Hanson, B.M.; Pinkner, C.L.; Simar, S.R.; Pinkner, J.S.; Parikh, R.; Clemens, M.W.; Hultgren, S.J.; Myckatyn, T.M. Insights into the microbiome of breast implants and periprosthetic tissue in breast implant-associated anaplastic large cell lymphoma. Scientific reports 2019, 9, 10393.
- Wang, H.; Altemus, J.; Niazi, F.; Green, H.; Calhoun, B.C.; Sturgis, C.; Grobmyer, S.R.; Eng, C. Breast tissue, oral and urinary microbiomes in breast cancer. Oncotarget 2017, 8, 88122.
- Luo, L.; Fu, A.; Shi, M.; Hu, J.; Kong, D.; Liu, T.; Yuan, J.; Sun, S.; Chen, C. Species-level characterization of the microbiome in breast tissues with different malignancy and hormone-receptor statuses using nanopore sequencing. Journal of personalized medicine 2023, 13, 174.
- Urbaniak, C.; Cummins, J.; Brackstone, M.; Macklaim, J.M.; Gloor, G.B.; Baban, C.K.; Scott, L.; O'Hanlon, D.M.; Burton, J.P.; Francis, K.P. Microbiota of human breast tissue. Applied and environmental microbiology 2014, 80, 3007-3014.
- Arnold, M.; Morgan, E.; Rumgay, H.; Mafra, A.; Singh, D.; Laversanne, M.; Vignat, J.; Gralow, J.R.; Cardoso, F.; Siesling, S. Current and future burden of breast cancer: Global statistics for 2020 and 2040. The Breast 2022, 66, 15-23.
- Banerjee, S.; Wei, Z.; Tian, T.; Bose, D.; Shih, N.N.; Feldman, M.D.; Khoury, T.; De Michele, A.; Robertson, E.S. Prognostic correlations with the microbiome of breast cancer subtypes. Cell death & disease 2021, 12, 831.
- Wang, N.; Yang, J.; Han, W.; Han, M.; Liu, X.; Jiang, L.; Cao, H.; Jing, M.; Sun, T.; Xu, J. Identifying distinctive tissue and fecal microbial signatures and the tumor-promoting effects of deoxycholic acid on breast cancer. Frontiers in Cellular and Infection Microbiology 2022, 12, 1029905.
- Thu, M.S.; Chotirosniramit, K.; Nopsopon, T.; Hirankarn, N.; Pongpirul, K. Human gut, breast, and oral microbiome in breast cancer: A systematic review and meta-analysis. Frontiers in Oncology 2023, 13, 1144021.
- Sohail, S.; Burns, M.B. Integrating current analyses of the breast cancer microbiome. PLoS One 2023, 18, e0291320.
- Yeo, K.; Wu, F.; Li, R.; Smith, E.; Wormald, P.-J.; Valentine, R.; Psaltis, A.J.; Vreugde, S.; Fenix, K. Is Short-Read 16S rRNA Sequencing of Oral Microbiome Sampling a Suitable Diagnostic Tool for Head and Neck Cancer? Pathogens 2024, 13, 826.
- Hieken, T.J.; Chen, J.; Hoskin, T.L.; Walther-Antonio, M.; Johnson, S.; Ramaker, S.; Xiao, J.; Radisky, D.C.; Knutson, K.L.; Kalari, K.R. The microbiome of aseptically collected human breast tissue in benign and malignant disease. Scientific reports 2016, 6, 30751.
- Oh, E.; Lee, H. Transcriptomic data in tumor-adjacent normal tissues harbor prognostic information on multiple cancer types. Cancer Medicine 2023, 12, 11960-11970.
- Sterne, J.A.; Savović, J.; Page, M.J.; Elbers, R.G.; Blencowe, N.S.; Boutron, I.; Cates, C.J.; Cheng, H.-Y.; Corbett, M.S.; Eldridge, S.M. RoB 2: a revised tool for assessing risk of bias in randomised trials. bmj 2019, 366.
- Estaki, M.; Jiang, L.; Bokulich, N.A.; McDonald, D.; González, A.; Kosciolek, T.; Martino, C.; Zhu, Q.; Birmingham, A.; Vázquez-Baeza, Y. QIIME 2 enables comprehensive end-to-end analysis of diverse microbiome data and comparative studies with publicly available data. Current protocols in bioinformatics 2020, 70, e100.
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nature methods 2016, 13, 581-583.
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal 2011, 17, 10-12.
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic acids research 2012, 41, D590-D596.
- Yeo, K.; Connell, J.; Bouras, G.; Smith, E.; Murphy, W.; Hodge, J.-C.; Krishnan, S.; Wormald, P.-J.; Valentine, R.; Psaltis, A.J. A comparison between full-length 16S rRNA Oxford nanopore sequencing and Illumina V3-V4 16S rRNA sequencing in head and neck cancer tissues. Archives of Microbiology 2024, 206, 248.
- Curry, K.D.; Wang, Q.; Nute, M.G.; Tyshaieva, A.; Reeves, E.; Soriano, S.; Wu, Q.; Graeber, E.; Finzer, P.; Mendling, W. Emu: species-level microbial community profiling of full-length 16S rRNA Oxford Nanopore sequencing data. Nature methods 2022, 19, 845-853.
- Wang, Y.; Lê Cao, K.-A. PLSDA-batch: a multivariate framework to correct for batch effects in microbiome data. Briefings in Bioinformatics 2023, 24, bbac622.
- Gloor, G.B.; Macklaim, J.M.; Pawlowsky-Glahn, V.; Egozcue, J.J. Microbiome datasets are compositional: and this is not optional. Frontiers in microbiology 2017, 8, 2224.
- Yeo, K.; Li, R.; Wu, F.; Bouras, G.; Mai, L.T.; Smith, E.; Wormald, P.-J.; Valentine, R.; Psaltis, A.J.; Vreugde, S. Identification of consensus head and neck cancer-associated microbiota signatures: a systematic review and meta-analysis of 16S rRNA and The Cancer Microbiome Atlas datasets. Journal of Medical Microbiology 2024, 73, 001799.
- Kodikara, S.; Ellul, S.; Lê Cao, K.-A. Statistical challenges in longitudinal microbiome data analysis. Briefings in Bioinformatics 2022, 23, bbac273.
- McMurdie, P.J.; Holmes, S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS one 2013, 8, e61217.
- Liu, C.; Cui, Y.; Li, X.; Yao, M. microeco: an R package for data mining in microbial community ecology. FEMS microbiology ecology 2021, 97, fiaa255.
- Anderson, M.J. Permutational multivariate analysis of variance (PERMANOVA). Wiley statsref: statistics reference online 2014, 1-15.
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.-A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS computational biology 2017, 13, e1005752.
- Sepich-Poore, G.D.; McDonald, D.; Kopylova, E.; Guccione, C.; Zhu, Q.; Austin, G.; Carpenter, C.; Fraraccio, S.; Wandro, S.; Kosciolek, T. Robustness of cancer microbiome signals over a broad range of methodological variation. Oncogene 2024, 43, 1127-1148.
- Feng, Y.; Ramnarine, V.R.; Bell, R.; Volik, S.; Davicioni, E.; Hayes, V.M.; Ren, S.; Collins, C.C. Metagenomic and metatranscriptomic analysis of human prostate microbiota from patients with prostate cancer. BMC genomics 2019, 20, 1-8.
- Bagaev, A.; Kotlov, N.; Nomie, K.; Svekolkin, V.; Gafurov, A.; Isaeva, O.; Osokin, N.; Kozlov, I.; Frenkel, F.; Gancharova, O. Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer cell 2021, 39, 845-865. e847.
- Banerjee, S.; Wei, Z.; Tan, F.; Peck, K.N.; Shih, N.; Feldman, M.; Rebbeck, T.R.; Alwine, J.C.; Robertson, E.S. Distinct microbiological signatures associated with triple negative breast cancer. Scientific reports 2015, 5, 15162.
- Banerjee, S.; Tian, T.; Wei, Z.; Shih, N.; Feldman, M.D.; Peck, K.N.; DeMichele, A.M.; Alwine, J.C.; Robertson, E.S. Distinct microbial signatures associated with different breast cancer types. Frontiers in microbiology 2018, 9, 951.
- Hadzega, D.; Minarik, G.; Karaba, M.; Kalavska, K.; Benca, J.; Ciernikova, S.; Sedlackova, T.; Nemcova, P.; Bohac, M.; Pindak, D. Uncovering microbial composition in human breast cancer primary tumour tissue using transcriptomic RNA-seq. International Journal of Molecular Sciences 2021, 22, 9058.
- Bachour, Y.; Poort, L.; Verweij, S.P.; van Selms, G.; Winters, H.A.; Ritt, M.J.; Niessen, F.B.; Budding, A.E. PCR characterization of microbiota on contracted and non-contracted breast capsules. Aesthetic plastic surgery 2019, 43, 918-926.
- Kim, D.; Yu, Y.; Jung, K.S.; Kim, Y.H.; Kim, J.-J. Tumor microenvironment can predict chemotherapy response of patients with triple-negative breast cancer receiving neoadjuvant chemotherapy. Cancer Research and Treatment: Official Journal of Korean Cancer Association 2023, 56, 162.
- Haddaway, N.R.; Page, M.J.; Pritchard, C.C.; McGuinness, L.A. PRISMA2020: An R package and Shiny app for producing PRISMA 2020-compliant flow diagrams, with interactivity for optimised digital transparency and Open Synthesis. Campbell Systematic Reviews 2022, 18, e1230 . [CrossRef]
- Juni, E. The genus Psychrobacter. In The Prokaryotes: A Handbook on the Biology of Bacteria: Ecophysiology, Isolation, Identification, Applications; Springer: 1992; pp. 3241-3246.
- Dohlman, A.B.; Mendoza, D.A.; Ding, S.; Gao, M.; Dressman, H.; Iliev, I.D.; Lipkin, S.M.; Shen, X. The cancer microbiome atlas: a pan-cancer comparative analysis to distinguish tissue-resident microbiota from contaminants. Cell host & microbe 2021, 29, 281-298. e285.
- Bars-Cortina, D.; Ramon, E.; Rius-Sansalvador, B.; Guinó, E.; Garcia-Serrano, A.; Mach, N.; Khannous-Lleiffe, O.; Saus, E.; Gabaldón, T.; Ibáñez-Sanz, G. Comparison between 16S rRNA and shotgun sequencing in colorectal cancer, advanced colorectal lesions, and healthy human gut microbiota. BMC genomics 2024, 25, 730.
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC biology 2014, 12, 1-12.
- Laurence, M.; Hatzis, C.; Brash, D.E. Common contaminants in next-generation sequencing that hinder discovery of low-abundance microbes. PloS one 2014, 9, e97876.
- Schierwagen, R.; Alvarez-Silva, C.; Servant, F.; Trebicka, J.; Lelouvier, B.; Arumugam, M. Trust is good, control is better: technical considerations in blood microbiome analysis. Gut 2020, 69, 1362-1363.
- Pannaraj, P.S.; Li, F.; Cerini, C.; Bender, J.M.; Yang, S.; Rollie, A.; Adisetiyo, H.; Zabih, S.; Lincez, P.J.; Bittinger, K. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA pediatrics 2017, 171, 647-654.
- Togo, A.; Dufour, J.-C.; Lagier, J.-C.; Dubourg, G.; Raoult, D.; Million, M. Repertoire of human breast and milk microbiota: a systematic review. Future microbiology 2019, 14, 623-641.
- Wang, K.; Nakano, K.; Naderi, N.; Bajaj-Elliott, M.; Mosahebi, A. Is the skin microbiota a modifiable risk factor for breast disease?: A systematic review. The Breast 2021, 59, 279-285.
- Hattar, K.; Reinert, C.P.; Sibelius, U.; Gökyildirim, M.Y.; Subtil, F.S.; Wilhelm, J.; Eul, B.; Dahlem, G.; Grimminger, F.; Seeger, W. Lipoteichoic acids from Staphylococcus aureus stimulate proliferation of human non-small-cell lung cancer cells in vitro. Cancer Immunology, Immunotherapy 2017, 66, 799-809.
- Kong, L.-X.; Wang, Z.; Shou, Y.-K.; Zhou, X.-D.; Zong, Y.-W.; Tong, T.; Liao, M.; Han, Q.; Li, Y.; Cheng, L. The FnBPA from methicillin-resistant Staphylococcus aureus promoted development of oral squamous cell carcinoma. Journal of Oral Microbiology 2022, 14, 2098644.
- Xie, W.; Huang, Y.; Xie, W.; Guo, A.; Wu, W. Bacteria peptidoglycan promoted breast cancer cell invasiveness and adhesiveness by targeting toll-like receptor 2 in the cancer cells. PLoS One 2010, 5, e10850.
- Wei, Y.; Sandhu, E.; Yang, X.; Yang, J.; Ren, Y.; Gao, X. Bidirectional functional effects of staphylococcus on carcinogenesis. Microorganisms 2022, 10, 2353.
- Stelzner, K.; Boyny, A.; Hertlein, T.; Sroka, A.; Moldovan, A.; Paprotka, K.; Kessie, D.; Mehling, H.; Potempa, J.; Ohlsen, K. Intracellular Staphylococcus aureus employs the cysteine protease staphopain A to induce host cell death in epithelial cells. PLoS Pathogens 2021, 17, e1009874.
- Akbari, A.; Farahnejad, Z.; Akhtari, J.; Abastabar, M.; Mobini, G.R.; Mehbod, A.S.A. Staphylococcus aureus enterotoxin B down-regulates the expression of transforming growth factor-beta (TGF-β) signaling transducers in human glioblastoma. Jundishapur journal of microbiology 2016, 9.
- Gong, Y.-T.; Zhang, L.-J.; Liu, Y.-C.; Tang, M.; Lin, J.-Y.; Chen, X.-Y.; Chen, Y.-X.; Yan, Y.; Zhang, W.-D.; Jin, J.-M. Neutrophils as potential therapeutic targets for breast cancer. Pharmacological Research 2023, 106996.
- Ma, S.; Shungin, D.; Mallick, H.; Schirmer, M.; Nguyen, L.H.; Kolde, R.; Franzosa, E.; Vlamakis, H.; Xavier, R.; Huttenhower, C. Population structure discovery in meta-analyzed microbial communities and inflammatory bowel disease using MMUPHin. Genome Biology 2022, 23, 208.
- Gibbons, S.M.; Duvallet, C.; Alm, E.J. Correcting for batch effects in case-control microbiome studies. PLoS computational biology 2018, 14, e1006102.
- Nearing, J.T.; Comeau, A.M.; Langille, M.G. Identifying biases and their potential solutions in human microbiome studies. Microbiome 2021, 9, 113.
- Weinroth, M.D.; Belk, A.D.; Dean, C.; Noyes, N.; Dittoe, D.K.; Rothrock Jr, M.J.; Ricke, S.C.; Myer, P.R.; Henniger, M.T.; Ramírez, G.A. Considerations and best practices in animal science 16S ribosomal RNA gene sequencing microbiome studies. Journal of animal science 2022, 100, skab346.
- Hong, B.-Y.; Driscoll, M.; Gratalo, D.; Jarvie, T.; Weinstock, G.M. Improved DNA extraction and amplification strategy for 16S rRNA gene amplicon-based microbiome studies. International Journal of Molecular Sciences 2024, 25, 2966.
- Lagier, J.-C.; Dubourg, G.; Million, M.; Cadoret, F.; Bilen, M.; Fenollar, F.; Levasseur, A.; Rolain, J.-M.; Fournier, P.-E.; Raoult, D. Culturing the human microbiota and culturomics. Nature Reviews Microbiology 2018, 16, 540-550.
- Errington, J.; Mickiewicz, K.; Kawai, Y.; Wu, L.J. L-form bacteria, chronic diseases and the origins of life. Philosophical Transactions of the Royal Society B: Biological Sciences 2016, 371, 20150494.
- Galeano Niño, J.L.; Wu, H.; LaCourse, K.D.; Srinivasan, H.; Fitzgibbon, M.; Minot, S.S.; Sather, C.; Johnston, C.D.; Bullman, S. INVADEseq to identify cell-adherent or invasive bacteria and the associated host transcriptome at single-cell-level resolution. Nature protocols 2023, 18, 3355-3389.
- Galeano Niño, J.L.; Wu, H.; LaCourse, K.D.; Kempchinsky, A.G.; Baryiames, A.; Barber, B.; Futran, N.; Houlton, J.; Sather, C.; Sicinska, E. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature 2022, 611, 810-817.
- Lötstedt, B.; Stražar, M.; Xavier, R.; Regev, A.; Vickovic, S. Spatial host–microbiome sequencing reveals niches in the mouse gut. Nature Biotechnology 2024, 42, 1394-1403.
- Buchta Rosean, C.; Bostic, R.R.; Ferey, J.C.; Feng, T.-Y.; Azar, F.N.; Tung, K.S.; Dozmorov, M.G.; Smirnova, E.; Bos, P.D.; Rutkowski, M.R. Preexisting commensal dysbiosis is a host-intrinsic regulator of tissue inflammation and tumor cell dissemination in hormone receptor–positive breast cancer. Cancer research 2019, 79, 3662-3675.
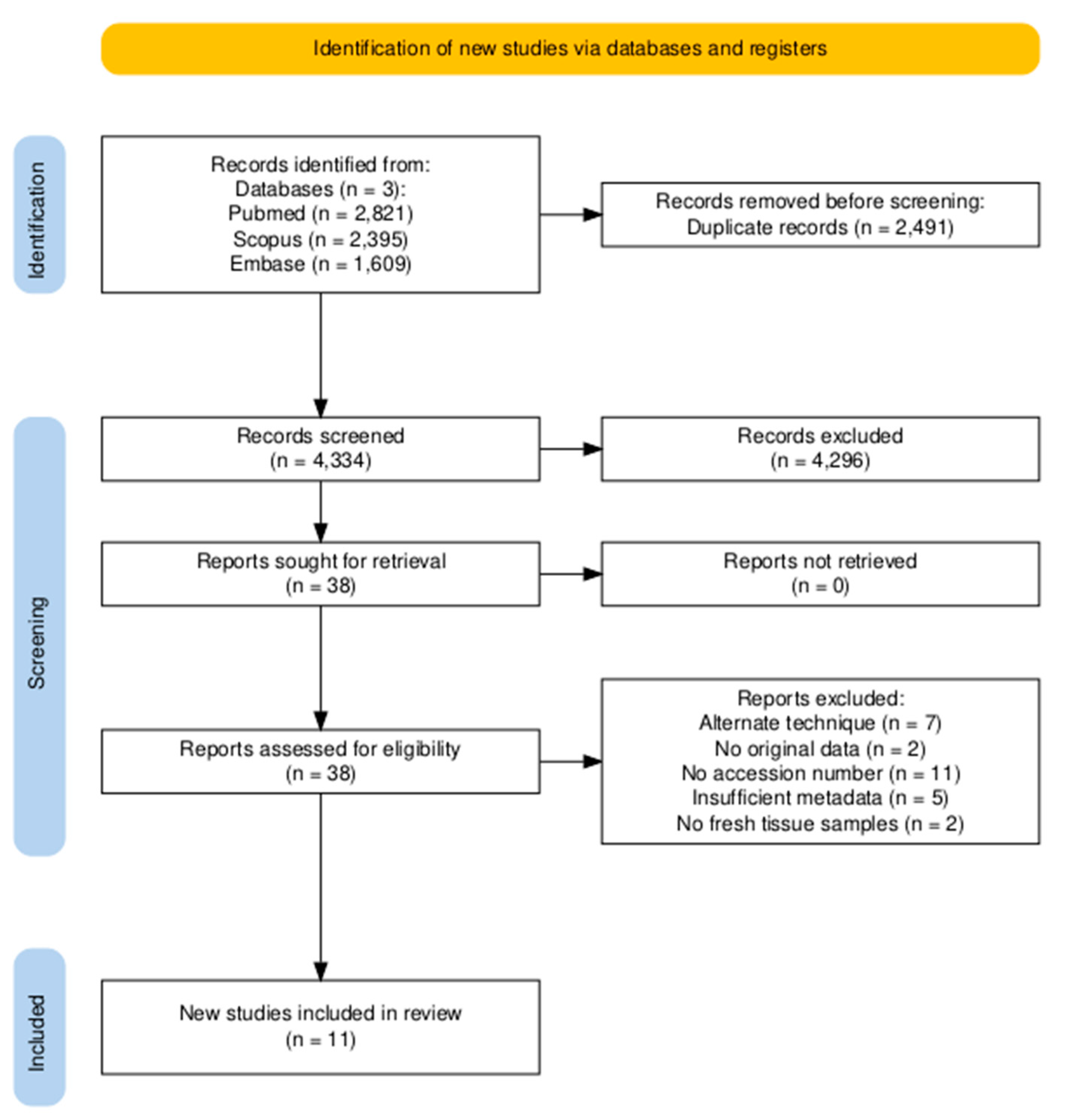
| Study | SRA Project Accession number | 16S rRNA hypervariable region | Cancer | Cancer-adjacent | Normal | Benign | Benign-adjacent | Mastitis |
| Kim (2021) [8] | PRJEB37724 | V1V3 | 37 | 38 | 0 | 0 | 0 | 0 |
| German (2023) [9] | PRJNA867176 | V3V4 | 31 | 61 | 398 | 0 | 0 | 0 |
| Hoskinson (2022) [10] | PRJNA723425 | V3V4 | 42 | 46 | 63 | 0 | 0 | 0 |
| Kartti (2023) [11] | PRJNA926328 | V3V4 | 17 | 14 | 0 | 0 | 0 | 0 |
| Thyagarajan (2020) [12] | PRJNA637875 | V3V4 | 31 | 33 | 0 | 0 | 0 | 0 |
| Zhu (2022) [13] | PRJNA667140 | V3V4 | 0 | 0 | 48 | 0 | 0 | 48 |
| Heiken (2016) [45] | PRJNA335375 | V3V5 | 0 | 16 | 0 | 12 | 0 | |
| Li (2022) [19] | PRJNA842933 | V4 | 0 | 79 | 0 | 15 | 0 | 0 |
| Esposito (2022) [15] | PRJNA759366 | V4V6 | 34 | 34 | 0 | 0 | 0 | 0 |
| Nejman (2020) [16] | PRJNA624822 | V6 | 77 | 18 | 0 | 0 | 0 | 0 |
| Urbaniak (2016) [17] | PRJNA323995 | V6 | 0 | 33 | 23 | 0 | 12 | 0 |
| Total | 269 | 372 | 532 | 15 | 24 | 48 |
| n | HR (95% CI) | p-value | ||
| Age (years) | < 65 ≥ 65 |
74 33 |
2.23 (0.85 – 5.84) |
0.103 |
| Staphylococcus | Low High Continuous |
33 32 107 |
4.06 (1.13 – 14.6) 1.40 (1.04 – 1.88) |
0.032* 0.027* |
| Acinetobacter | Low High Continuous |
33 31 107 |
0.90 (0.27 – 2.95) 1.01 (0.83 – 1.24) |
0.859 0.891 |
| Corynebacterium | Low High Continuous |
34 31 107 |
4.72 (1.02 – 21.9) 1.21 (0.93 – 1.56) |
0.048* 0.160 |
| Streptococcus | Low High Continuous |
32 31 107 |
2.06 (0.41 – 10.3) 1.20 (0.89 – 1.63) |
0.236 0.380 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





