1. Introduction
Poultry farming and production have contributed immensely to providing human-required animal protein worldwide, including in Nigeria (Fagbamila et al., 2017; Bettridge et al., 2014). The industry is rapidly expanding, and it increased from 150,700 million chickens (in 2005), to 192,313 million chickens in 2010 in Nigeria (Fagbamila et al. 2017). Despite this progress, the industry faces various challenges, e.g., from security threats to disease outbreaks (Anosike, 2017). Salmonellosis commonly affects poultry production in Nigeria, a zoonotic disease strongly associated with human foodborne pathogens (Chai et al., 2017; Majowicz et al., 2010; Antunes, 2016).
Salmonellosis is an enteric infection caused by the various serovars of Salmonella enterica ssp. enterica. These serovars can infect multiple animal species and humans; the zoonotic disease is mainly associated with consuming contaminated food or water. The causative agents, Salmonella are Gram-negative rod-shaped bacteria of the family Enterobacteriaceae (Akinyemi et al., 2007), and the two known significant species of Salmonella are S. enterica (Hurley et al. 2014) and S. bangori (Ikhimuikor et al., 2022). While S. bangori is less associated with human infections, most S. enterica and its sub-species are zoonotic and responsible for animals' and humans' non-typhoidal and typhoidal enteric diseases.
The clinical manifestation of salmonellosis is usually not specific but may include diarrhea, vomiting, and life-threatening conditions in high-risk individuals or groups with immunodepression conditions (Raufu et al., 2013). Salmonella usually spreads through contaminated farm workers and the consumption of fecal contaminated water and food.
In Nigeria, the prevalence of salmonellosis has been extensively studied. For instance, Asogwa et al. (2017) reported the prevalence of salmonellosis in Enugu State using culturing and morphological identification from fecal samples collected from chickens, Jibril et al. (2020) reported the prevalence of salmonellosis in the North Western States of Nigeria using culturing, serotyping and gene sequencing of pooled samples from dust and shoes of poultry workers and together with questioner predicted the associated risk factors of salmonellosis. But in the South Western States of Nigeria, only the reports of Mshelbwala et al. (2017) and Terese et al. (2022) have shed light on the prevalence of Salmonella in bird carcasses and cloaca swabs submitted for post-mortem and culturing in the Veterinary Teaching Hospital, Federal University of Agriculture Abeokuta using culturing and PCR but no study has shed light on genetic diversity of Salmonella enterica strains in Nigeria. Hence, this study examined the samples collected from poultry farms and workers for the presence of Salmonella species and characterized the positive samples by sequence analyses of the invA gene.
2. Materials and Methods
2.1. Study Area
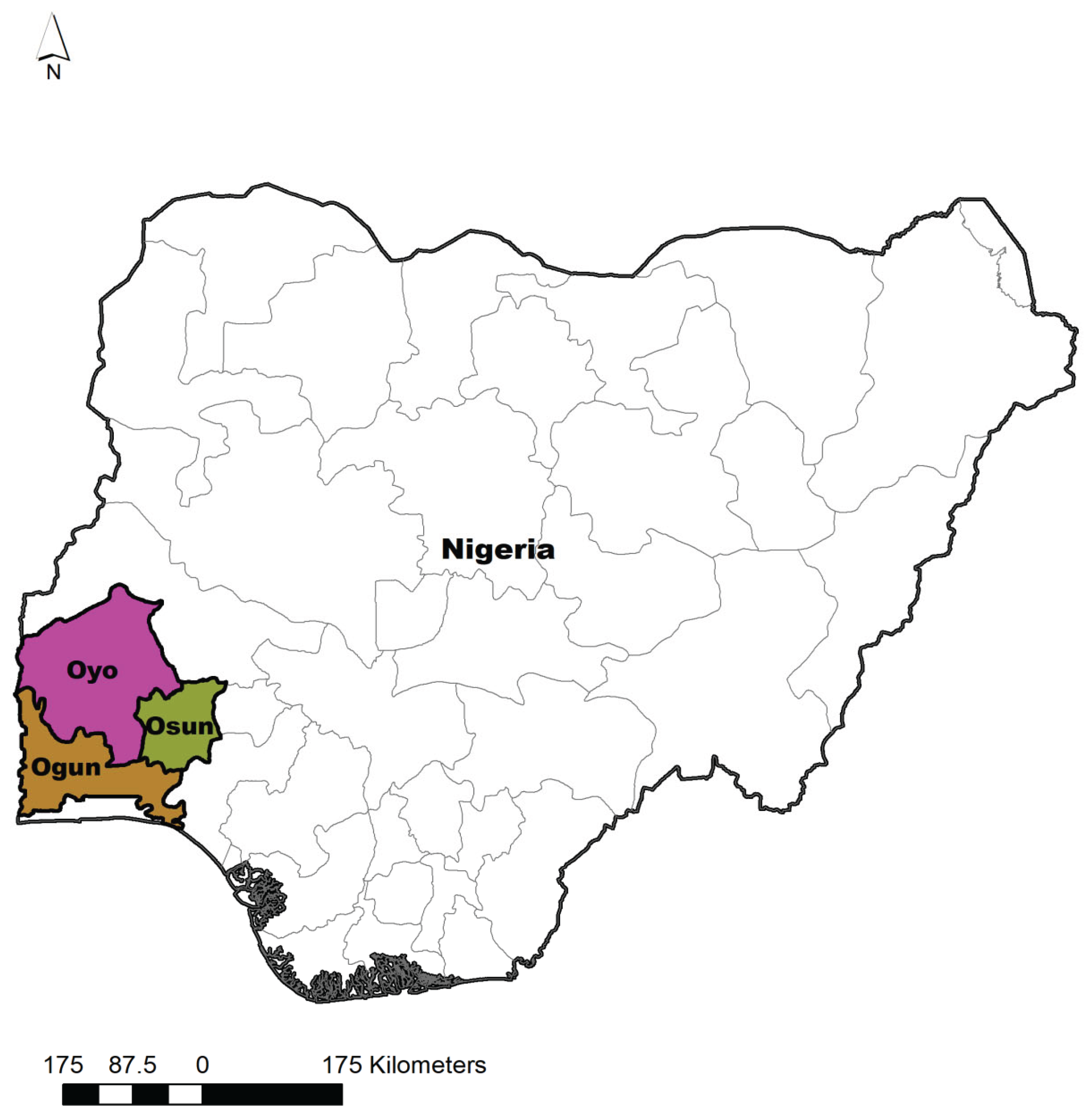
This study was carried out in the southwestern zone of Nigeria. This zone includes Ogun, Oyo, Osun, Ondo, Ekiti, and Lagos. Except for Lagos, they are all endowed with both thick forest and derived savannah (
Figure 1)
. A cross-sectional method using a multistage sampling method was used in this survey. In the first stage, the South Western zone was purposely selected because it forms the central zone where large-scale poultry farming is practiced. Three States (Ogun, Oyo, and Osun States) out of the six states in the South Western zone were selected by balloting without replacement in the second stage. All three chosen states have three senatorial districts. Hence, the third stage selection resulted in the choice of one local government for each of the three states by balloting. In the fourth stage, one village was randomly selected as a sampling site in each selected LGA. Hence, three villages were sampled per senatorial district to give nine villages in each State.
2.2. Sample Size Determination
Various studies have shown that the prevalence rate for salmonellosis in commercial poultry in Nigeria was 16%, which was used to determine the sample size. The sample size was calculated using the formula for cross-sectional studies (Thrusfield, 2007).
2.3. Sample Collection
The samples collected from each farm included poultry workers' feces, feed, dust, water, and palm swabs. A total of 314 samples were collected, which included 48 swabs from palms, 112 feces, 51 water, 52 dust, and 51 feeds. Sterile swabs were used to scrub the palms of the poultry workers, about 100 g per farm of dust was collected from different surfaces in each farm, commercially manufactured boot swabs (Technical Services Consultant, UK) were used to collect litter samples, fresh fecal dropping (about 100 g) collected from each farm, 100 -200 g feeds were collected from different point in the same farm and unmedicated water (100 ml) sample obtained from different poultry houses of the same farm. The samples were immediately transported in the mobile refrigerator (4oC) to the Microbiology Laboratory of the College of Veterinary Medicine, Federal University of Agriculture Abeokuta, for further analysis. The consent of the farmers was sought through the Ethical Committee of the College of Veterinary Medicine, Federal University of Agriculture Abeokuta.
2.4. Isolation of Salmonella
All the samples collected were emulsified in sterile water. Culture and isolation of salmonellae from the collected emulsified samples were carried out as described by Mshelbwala et al. (2017). Briefly, swabs, fecal, and environmental samples were pre-enriched in buffered peptone water and separately applied to the nutrient broth before incubation at 37oC for 24 hours. Two milliliters of the mixture were taken from the pre-enrichment medium and inoculated into 50 ml of Rappaport-Vasiliadis broth (Oxoid BasingStoke, UK), then onto tetrathionate glucose broth (Oxoid Basing Stoke, UK) for selective enrichment and incubated for 24 hours at 37oC. The typical colonies of Salmonella obtained from this culture were further sub-cultured on XLD agar and incubated for 24 hours at 37oC. The resulting Salmonella-suspected colonies were inoculated onto McConkey agar for purification. The cultures containing the growths or colonies were subjected to biochemical characterization for salmonella confirmation. These included lactose oxidation, glucose fermentation, gas, hydrogen sulfide (H2S) production, the lack of β-galactosidase (ONPG), and lysine decarboxylation.
2.5. DNA Extraction from Salmonella Isolates
Three colonies from each of the positive culture (sub-cultured thrice to prove the clonality) were picked with a sterile loop and dissolved in 200 µl distilled water to confirm the Salmonella was positive by biochemical characterization. DNA was extracted from the suspended culture using a Quick-DNA Fungal/Bacterial Miniprep Kit according to the manufacturer’s instructions (Zymo Research, USA). In short, 200 µl suspension containing the bacteria culture was added to the ZR Bashing Bead lysis tube containing 750 µl Bashing Bead buffer. The mixture was vortexed at the highest speed for 15 minutes and centrifuged at 10,000 x g for 1 minute. Approximately 400 µl of the supernatant was transferred to a Zymo-Spin III-F filter in a collection tube and centrifuged at 8,000 x g for 1 minute. 1,200 µl of genomic lysis buffer was added to the filtrate in a collection tube, and 800 µl of the mixture was transferred to a Zymo-Spin IICR column in a collection tube and centrifuged at 10,000 x g for 1 minute. The flow through in the collection tube was discarded, the spin column returned to the tube, and the centrifugation was repeated. 200 µl of DNA pre-wash buffer was added to the Zymo-Spin Column, inserted into a new collection tube, and centrifuged at 10,000 x g for 1 minute, after which 500 µl of g-DNA Wash buffer was added to the Zymo-spin column and centrifuged at 10,000 x g for 1 minute. The Zymo-spin IICR column was transferred to a clean, sterile 1.5 ml microcentrifuge tube, and 100 µl of DNA elution buffer was added directly to the Spin-column matrix and centrifuged at 10,000 x g for 30 seconds. The eluted DNA was kept at 20O C until use.
2.6. Amplification of the invA Gene of Salmonella
The invasive gene of Salmonella (invA gene) was targeted for amplification using a pair of primers described by Jibril et al. (2020); invA forward: GTGAAATTATCGCCACGTTCGGGCA and invA reverse: ATCGCACCGTCAAAGGAACC (synthesized by Bioneer Incorporation South Korea). The 25 μl final volume for the PCR reaction contained 12.5 μl of One Taq Quick-Load 2 x Master Mix (New England BIOLAB), one μl each of forward and reverse primers, 9.5 μl of nuclease-free water (New England BIOLAB) and 1 μl of template DNA. The reactions were placed in a Personal cycler series thermocycler (Biorad, Hercules, CA, USA). The reaction conditions were as follows: Initial denaturation at 940C for 4 min followed by 35 cycles of 940C for 1 min, 55 0C for 1 min, and 72 0C for 1 min; and final extension at 72 0C for 10 min. Ten microliters of the PCR products were subjected to electrophoresis through a 1% agarose gel in 1 x TAE buffer at 90 V for 60 min, along with 10 µl of GENEMate Quanti-Marker 100 bp DNA ladder (BioExpress, Kaysville, UT, USA). Gels were stained with ethidium bromide (Phenix Research Products, Candler, NC, USA) at 5 µl/100 ml of the agarose gel suspension. After electrophoresis, the gel was visualized on a UV transilluminator and was photographed using a handheld camera (Samsung, China). All positive samples were tested twice to confirm the PCR diagnosis, and positive DNA sample obtained from the pathogen laboratory of the Department Veterinary Microbiology, College of Veterinary Medicine, Federal University of Agriculture Abeokuta and negative (Nuclease free water) samples were used as controls in each run.
2.7. DNA Sequencing of the invA Gene of Salmonella Isolates
To confirm and validate our results, fourteen culture-positive samples of Salmonella were selected, and their PCR products were sequenced directly using the Big dye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) with the forward amplification PCR primers and AmpliTaq-FS DNA Polymerase. The sequences obtained were viewed and compared on Finch TV and Sequence Scanner (Applied Bioscience) before they were aligned with published sequences of various Salmonella species using the Molecular Evolutionary Genetic Analysis software (MEGA 5.05).
2.8. Sequence Alignment and Analysis
The invA gene sequences of the isolated Salmonella were used to do a BLAST search from the NCBI database. For comparison, invA gene sequences of Salmonella isolates from China, South Korea, South Africa, Egypt, Morocco, Taiwan, and the USA available in GenBank were selected. The alignment was done using the ClustalW method of Molecular Evolutionary Genetic Analysis (MEGA) software version 5.05 (Tamura et al., 2011). A phylogenetic tree was constructed using the Unweighted pair group method (UPGMA) algorithm of the phylogeny program of MEGA 5.05 (Tamura et al., 2011), which included two consensus sequences from this study and twelve sequences obtained from the GenBank with Citrobacter freundii (MZ202354) as the out-group to root the invA gene trees. The bootstrap confidence interval of the tree was determined based on l000 replicates.
2.9. Statistical Analysis
The data were summarized using descriptive statistics. The prevalence of salmonella in the studied area using cultural and molecular techniques was compared statistically using Student’s t-test (paired t-test).
3. Results
3.1. Prevalence of Salmonella in Samples Collected from Poultry Farms and Their Handlers
A total number of 314 samples including swabs from palms of attendants, feces, water and dust from 49 farms were collected from Abeokuta (18), Ibadan (20), and Oshogbo (11). The samples were selectively cultured for 24 hours to obtain typical
Salmonella colonies. The overall prevalence of
Salmonella in poultry farms sampled was 15/314 (4.78%), comprising 3 (6.25%) from 48 palm swabs from the poultry attendants, 4 (3.57%) from 112 fecal samples collected from the poultry houses, 5 (9.80%) from the 51 water samples, and 3 (5.88%) from the 51 feed sample collected (
Table 1).
In all 52 dust samples collected around the poultry houses Salmonella could not be detected.
Salmonella species were found in the three States sampled with a 2.75%, 6.0%, and 5.71% prevalence in Ogun, Osun, and Oyo States, respectively. This revealed the highest prevalence in Osun State (
Table 2).
3.2. Further Characterization of Salmonella Isolates by invA Gene Amplification and Sequencing
DNA was extracted from the Salmonella colonies, amplified (
invA gene) and the amplicons sequenced unidirectionally to further characterize and prove the genetic diversity of the isolates . Gel electrophoresis of the amplified PCR products derived from the DNA of the culture isolates reveals fourteen samples with visible bands of about 284 bp, the expected band size of
Salmonella enterica. The PCR product of negative control showed no band. Only two of the obtained sequences were usable and subjected to BLAST search for homology in the GeneBank because other sequences obtained were noisy. One revealed homology of about 99.59% with sequence with accession number LC320032, a Paratyphi serovar, and the other revealed homology of about 89.04% with sequences with accession number LC318423 an
Enteritidis serovar strain SYCH in GenBank. Analysis of the two sequences showed that sequence 01 has 248 bp length and sequence 02 has 225 bp length with 51.4% and 51.6 5 mean G-C contents, respectively. Alignment of the two sequences with each other revealed that Sequence 01 has nucleotide G inserted at point 81, CCCG inserted at points 101-104, GGTA inserted at points 127-130, TGA inserted at points 155-157, TTAT inserted at points 167-170 and GT inserted at 229-230. The sequence accession numbers used for constructing a phylogenetic tree are contained in
Table 3. The phylogenetic tree inferred from the InvA gene sequences of the
S. enterica separated the isolates from this study into two clades, with isolate 01 tightly clustered the sequence of
S. enterica serovar enteritidis from the USA while isolate 02 is separated into a lonely clade but next to
S. enterica serovar Typhimurium from Egypt (
Figure 2)
4. Discussion
Salmonella is a critical pathogen in food animals in Nigeria due to its substantial impact on public health and the agricultural sector (Lamichhane et al., 2024; Igbinosa et al., 2022). It is commonly found in livestock and poultry, where poor animal husbandry practices, such as inadequate sanitation and overcrowding, facilitate its spread. The contamination of meat, eggs, and dairy products with Salmonella poses a significant risk to human health, leading to foodborne illnesses that can range from mild gastroenteritis to severe systemic infections (Bintsis 2017; Ehuwa et A., 2021; Teklemariam et al., 2023). Additionally, the misuse of antibiotics in animal agriculture has led to the emergence of antibiotic-resistant strains, complicating treatment options and exacerbating the public health threat (Manyi-Loh et al., 2018; Muteeb et al., 2023). Addressing this issue requires comprehensive measures, including improved farm management practices, stringent food safety regulations, and robust surveillance systems to monitor and control the pathogen. Hence, to understand the prevalence and circulating serovars of Salmonella enterica ssp. enterica in and around commercial poultry farms in South West Nigeria, this study assessed the possible source of contaminants and characterized the detected Salmonella by sequencing and sequence analysis of invA gene.
The prevalence of Salmonella in Nigeria has been documented in several studies, showing varying rates. For instance, a prevalence of 21.4% was reported in poultry cloacal swab samples (Terese et al., 2022) collected from Oyo State, prevalence of 81.1% in intestinal organs submitted for post-mortem from Lagos, Ogun, and Oyo States to the Veterinary Teaching Hospital (Mshelbwala et al., 2023), prevalence of 54% in fecal droppings of chicken from various farming systems (Asogwa et al., 2022) from Imo State, 15.9% prevalence in North West (Kebbi, Sokoto and Zanfara States ) (Jibril et al., 2020), 14% prevalence from eggshell and its content from Ogun State (Agbaje et al., 2021) and 8.6% from poultry intestinal content sampled in Ilorin, Kwara State (Raji et al., 2021). These figures are higher than the 4.78% recorded in our study, indicating that the prevalence in Nigeria is relatively elevated. Several factors could be attributed to the results obtained from this study. Asogwa et al. (2022) and Terese et al. (2022) collected their samples, fecal droppings (120) and cloaca swabs (360), respectively, solely from a source that is believed to be richer in intestinal microbes, including Salmonella species. Against That, our study's samples (314) were obtained from various sources, including dust, palm swabs, water, fecal droppings, and feeds. This suggestion is further supported by the report of Ibrahim et al. (2020) from Nasarawa State, who reported more Salmonella typhimurium from diarrheic fecal samples than non-diarrheic samples. Also, the higher prevalence recorded by others might be connected with the higher number of samples collected and analyzed (Talukder et al., 2023). The report of another study that focused on the prevalence in commercial chicken eggs reported a rate of 6.5%, further highlighting the variability in prevalence depending on the specific sample and location within the region (Edward et al., 2023; Talukder et al., 2023). These variations highlight the importance of localized studies to accurately determine the prevalence of Salmonella and therefore, guide appropriate interventions.
Studies from other regions in African countries have shown even higher prevalence rates than our study. For instance, Edward et al. (2023), Ramtahal et al. (2022), and Waktole et al. (2024) reported a prevalence of 32.1%, 14.4%, and 5.5% in South Africa, Egypt, and Ethiopia, respectively, except the study of Bouchrif et al. (2009), which reported extremely lower prevalence (0.91%) in Morocco.
This study attempted to amplify and analyze the sequences of the invA gene that encodes a protein that is part of the Type III secretion system (T3SS), essential for injecting effector proteins into host cells correlating to the virulence of Salmonella. The invA gene is a highly conserved in Salmonella and is often targeted in PCR-based methods due to its specificity and reliability (Rahn, et al., 1992; Lampel et al., 2000; Murray and Lee 2000).
Molecular characterization of morphologically confirmed Salmonella isolates revealed a band size of 284 bp expected for the PCR product of the invA gene. This confirms the presence of Salmonella at the genus level in the study area. Our results agree with the findings of Naik et al. (2015), Kaushik et al. (2014), and Abhadionmhen et al. (2023). The InvA gene was amplified in fourteen of isolates amplified and sequenced in this study. This may suggest that the strain from which the InvA gene was not amplified may have undergone mutation (Yanestria et al., 2019) and not invasive in nature, but Naik et al. (2015) and Kadri et al. (2019) postulated that they may have another invasive mode of infecting their host. Though many literatures suggest that all Salmonella species have the InvA gene (Malorny et al., 2003; Lampel et al., 2000; Murray and Lee 2000), various studies in Nigeria could not have 100% amplification of this gene in all the Salmonella species analyzed in their studies, suggesting those not amplified were mutant Still, the studies of Mokgophi et al. (2024) in South Africa and Qiumei et al. (2012) in China reported 100% amplification of the same gene in their studies. In the reports of Abhadionmhen et al. (2023), Igbinosa et al. (2023), and Raufu et al. (2021) on amplification of InvA gene carried out in various States in Nigeria and Kadry et al. (2019) in Egypt, InvA gene was not amplified in all the Salmonella species subjected to PCR amplification. Hence, if the gene is indeed not present or has mutated in some of the Salmonella species, then it can be suggested that the InvA gene is not a good candidate for routine diagnosis of salmonellosis and their genetic diversity. Various literature that supports (Mokgophi et al., 2024) or does not support (Pavon and Rivera, 2021) the use of the InvA gene as a good candidate for Salmonella diagnosis and molecular characterization exist. Therefore, our study’s finding may also support the suggestion that the InvA gene may not be a good target for diagnosing salmonellosis in Nigeria, as many reports from the country revealed that not all the Salmonella isolates were amplified targeting the InvA gene amplification.
The phylogenetic analysis of the sequences obtained from this study and comparison with those obtained from GenBank revealed that those sequences are separated into two distinct clades, suggesting that they are different strains. This phylogenetic tree further supports the results of the search BLAST done on the isolates, which had homologies of 99.59% and 89.04% with Salmonella enterica serovar Paratyphi and Salmonella enterica serovar Enteritidis, respectively.
Salmonella enterica serovar Enteritidis is a significant pathogen responsible for causing foodborne illnesses in humans (Bintsis, 20017; Eng et al., 2015; Lamichhane et al., 2024). It commonly leads to gastroenteritis, characterized by symptoms such as diarrhea, fever, abdominal cramps, and vomiting, while Salmonella enterica serovar Paratyphi is a significant human pathogen responsible for paratyphoid fever, a severe systemic illness similar to typhoid fever and gastroenteritis in human (Smith et al., 2016). Detection of these two serovar in the study area may pose a severe public health challenge.
5. Conclusions
Salmonella enterica serovars Enteritidis and Typhimurium detected in and around poultry farms in Southwest Nigeria present a significant public health concern. These pathogens are major causes of foodborne illnesses that may lead to human gastrointestinal infections. These serovars indicate lapses in biosecurity measures and hygiene practices within the poultry industry. It underscores the need for stringent monitoring and control protocols to mitigate contamination risks. To address this issue, collaboration between poultry farmers, veterinarians, and regulatory agencies should be encouraged to implement adequate sanitation, vaccination, and surveillance strategies. Enhancing farmer education on biosecurity and proper handling practices is crucial to reducing the prevalence of these pathogens and ensuring the safety of poultry products.
References
- Bouchrif, B.; Paglietti, B.; Murgia, M.; Piana, A.; Cohen, N.; Ennaji, M.M.; Rubino, S.; Timinouni, M. Prevalence and antibiotic-resistance of Salmonella isolated from food in Morocco. J Infect Dev Ctries. 2009, 3, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Waktole, H.; Ayele, Y.; Ayalkibet, Y.; Teshome, T.; Muluneh, T.; Ayane, S.; Borena, B.M.; Abayneh, T.; Deresse, G.; Asefa, Z.; et al. Prevalence, Molecular Detection, and Antimicrobial Resistance of Salmonella Isolates from Poultry Farms across Central Ethiopia: A Cross-Sectional study in urban and peri-urban areas. Microorganisms 2024, 12, 767. [Google Scholar] [CrossRef]
- Ramtahal Melissa, A.; Somboro Anou, M.; Amoako Daniel, G.; Abia Akebe, L.K.; Perrett Keith Bester Linda, A.; Essack Sabiha, Y. Molecular Epidemiology of Salmonella enterica in Poultry in South Africa Using the Farm-to-Fork Approach. International Journal of Microbiology 2022, 5121273, 12. [Google Scholar] [CrossRef]
- Mokgophi, T.M.; Gcebe, N.; Fasina, F.; Adesiyun, A.A. Molecular characterization of virulence and resistance genes in Salmonella strains isolated from chickens sold at the informal chicken market in Gauteng Province, South Africa. Journal of Food Safety 2024, 44, e13110. [Google Scholar] [CrossRef]
- Malorny, B.; Bunge, C.; Helmuth, R. Discrimination of d-tartrate-fermenting and non-fermenting Salmonella enterica subsp. enterica isolates by genotypic and phenotypic methods. Journal of Clinical Microbiology 2003, 41, 4292–4297. [Google Scholar] [CrossRef] [PubMed]
- Raufu, I.A.; Ahmed, O.A.; Aremu, A.; Ameh, J.A.; Ambali, A. Molecular characterization of Salmonella enterica from poultry farms in Ilorin, north-central Nigeria. Sokoto Journal of Veterinary Sciences 2021, 19, 197–207. [Google Scholar] [CrossRef]
- Abhadionmhen AAnyiam, V.I.; Imarenezor, E.P.K.; Brown, S.T.C. Molecular Characterization of Salmonella species isolated from animal sources in southern Taraba State, North-East, Nigeria. International Journal of Pathogen Research 2023, 12, 33–40. [Google Scholar] [CrossRef]
- Naik, V.K.; Shakya, S.; Patyal, A.; Gade, N.E.; Bhoomika. Isolation and molecular characterization of Salmonella spp. from chevon and chicken meat collected from different districts of Chhattisgarh, India. Veterinary World 2015, 8, 702–706. [Google Scholar] [CrossRef]
- Kaushik, P.; Anjay Kumari, S.; Bharti, S.K.; Dayal, S. Isolation and prevalence of Salmonella from chicken meat and cattle milk collected from local markets of Patna, India. Veterinary World 2014, 7, 62–65. [Google Scholar] [CrossRef]
- Pavon, R.D.N.; Rivera, W.L. Molecular serotyping by phylogenetic analyses of a 1498 bp segment of InvA gene of Salmonella. ASM Science Journal 2021, 14, 2021. [Google Scholar]
- Eng, S.K.; Pusparajah, P.; Ab Mutalib, N.S.; Ser, H.L.; Chan, K.G.; Lee, L.H. Salmonella: A review on pathogenesis, epidemiology and antibiotic resistance. Frontiers in Life Science 2015, 8, 284–293. [Google Scholar] [CrossRef]
- Lamichhane, B.; Mawad, A.M.M.; Saleh, M.; Kelley, W.G.; Harrington, P.J.; Lovestad, C.W.; Amezcua, J.; Sarhan, M.M.; El Zowalaty, M.E.; Ramadan, H. Salmonellosis: An overview of epidemiology, pathogenesis, and innovative approaches to mitigate the antimicrobial resistant infections. Antibiotics 2024, 13, 76. [Google Scholar] [CrossRef]
- Smith, S.I.; Seriki, A.; Ajayi, A. Typhoidal and non-typhoidal Salmonella infections in Africa. European Journal of Clinical Microbiology and Infectious Diseases 2016, 35, 1913–1922. [Google Scholar] [CrossRef] [PubMed]
- Garba, B.; Saidu, B.; Mahmuda, A. Current Trend on the Economic and Public Health Significance of Salmonellosis in Iraq. Advances in Animal and Veterinary Sciences 2019, 7, 484–491. [Google Scholar] [CrossRef]
- Talukder, H.; Roky, S.A.; Debnath, K.; Sharma, B.; Ahmed, J.; Roy, S. Prevalence and Antimicrobial Resistance Profile of Salmonella Isolated from Human, Animal and Environment Samples in South Asia: A 10-Year Meta-analysis. Journal of Epidemiology and Global Health 2023, 13, 637–652. [Google Scholar] [CrossRef]
- Orum, T.G.; Ishola, O.O.; Adebowale, O.O. Occurrence and antimicrobial susceptibility patterns of Salmonella species from poultry farms in Ibadan, Nigeria. African Journal of Laboratory Medicine 2022, 11, 1606. [Google Scholar] [CrossRef] [PubMed]
- Asogwa, N.V.; Udeani, T.K.; Obeagu, E.I.; Asogwa, B.E.; Chukwueze, C.M. Prevalence of salmonella species infection in poultry farming systems in Enugu metropolis Nigeria. European Journal of Pharmaceutical and Medical Research 2022, 9, 87–91. [Google Scholar]
- Jibril, A.H.; Okeke, I.N.; Dalsgaard, A.; Kudirkiene, E.; Akinlabi, O.C.; Bello, M.B.; Olsen, J.E. Prevalence and risk factors of Salmonella in commercial poultry farms in Nigeria. PLoS ONE 2020, 15, 17–e0238190. [Google Scholar] [CrossRef]
- Agbaje, M.; Ayo-Ajayi, P.; Kehinde, O.; Omoshaba, E.; Dipeolu, M.; Fasina, F.O. Salmonella characterization in poultry eggs sold in farms and markets in relation to handling and biosecurity practices in Ogun State, Nigeria. Antibiotics 2021, 10, 773. [Google Scholar] [CrossRef]
- Raji, M.A.; Kazeem, H.M.; Magyigbe, K.A.; Ahmed, A.O.; Lawal, D.N.; Raufu, I.A. Salmonella Serovars, Antibiotic Resistance, and Virulence Factors Isolated from Intestinal Content of Slaughtered Chickens and Ready-to-Eat Chicken Gizzards in the Ilorin Metropolis, Kwara State, Nigeria. International Journal of Food Science 2021, 2021, 8872137, 11. [Google Scholar] [CrossRef]
- Edward, W.; Chisnall, T.; Tano-Debrah, K.; Card, R.M.; Duodu, S. Prevalence and genomic characterization of Salmonella isolates from commercial chicken eggs retailed in traditional markets in Ghana. Frontiers in Microbiology 2023, 14, 283835. [Google Scholar] [CrossRef] [PubMed]
- Bintsis, T. Foodborne pathogens. AIMS Microbiol. 2017, 3, 529–563. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Teklemariam, A.D.; Al-Hindi, R.R.; Albiheyri, R.S.; Alharbi, M.G.; Alghamdi, M.A.; Filimban, A.A.R.; Al Mutiri, A.S.; Al-Alyani, A.M.; Alseghayer, M.S.; Almaneea, A.M.; et al. Human Salmonellosis: A Continuous Global Threat in the Farm-to-Fork Food Safety Continuum. Foods 2023, 12, 1756. [Google Scholar] [CrossRef]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Muteeb, G.; Rehman, M.T.; Shahwan, M.; Aatif, M. Origin of Antibiotics and Antibiotic Resistance, and Their Impacts on Drug Development: A Narrative Review. Pharmaceuticals 2023, 16, 1615. [Google Scholar] [CrossRef]
- Ehuwa, O.; Jaiswal, A.K.; Jaiswal, S. Salmonella, Food Safety and Food Handling Practices. Foods 2021, 10, 907. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lamichhane, B.; Mawad, A.M.M.; Saleh, M.; Kelley, W.G.; Harrington, P.J., II.; Lovestad, C.W.; Amezcua, J.; Sarhan, M.M.; El Zowalaty, M.E.; Ramadan, H.; et al. Salmonellosis: An Overview of Epidemiology, Pathogenesis, and Innovative Approaches to Mitigate the Antimicrobial Resistant Infections. Antibiotics 2024, 13, 76. [Google Scholar] [CrossRef]
- Igbinosa, E.O.; Beshiru, A.; Igbinosa, I.H.; Okoh, A.I. Antimicrobial resistance and genetic characterization of Salmonella enterica from retail poultry meats in Benin City, Nigeria. LWT, Food Science and Technology 2022, 169, 2022–114049. [Google Scholar] [CrossRef]
- Bamiro, B.S.; Coker, A.O. Salmonellosis in Lagos, Nigeria: Incidence of Plasmodium falciparum-associated co-infection, patterns of antimicrobial resistance, and emergence of reduced susceptibility to fluoroquinolones. Journal of Health Population 2007, 25, 351–358. [Google Scholar]
- Hurley, D.; McCusker, M.P.; Fanning, S.; Martins, M. Salmonella–Host Interactions – Modulation of the Host Innate Immune System. Frontiers in Immunology 2014, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Fagbamila, I.O.; Barco, L.; Mancini, M.; Kwaga, J.; Ngulukun, S.S.; Zavagnin, P.; et al. Salmonella serovars and their distribution in Nigerian commercial chicken layer farms. PLoS ONE 2017, 12, e0173097. [Google Scholar] [CrossRef]
- Chai, S; Cole, D; Nisler, A; Mahon, B. Poultry: Themostcommonfoodinoutbreakswithknownpathogens, United States, 1998–2012. Epidemiol. Infect. 2017, 145, 316–325. [Google Scholar] [CrossRef] [PubMed]
- Majowicz, E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O’Brien, S.J.; Jones, T.F.; Fazil, A.; Hoekstra, R.M. The global burden of non-typhoidal salmonella gastroenteritis. Clin. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef]
- Antunes, P.; Mourão, J.; Campos, J.; Peixe, L. Salmonellosis: The role of poultry meat. Clinical Microbiology and Infection 2016, 22, 110–121. [Google Scholar] [CrossRef]
- Raufu, I.; Bortolaia, V.; Svendsen, C.A.; Ameh, J.A.; Ambali, A.G.; Aarestrup, F.M. The first attempt of an active integrated laboratory-based Salmonella surveillance program in the north-eastern region of Nigeria. Journal of Applied Microbiology 2013, 115, 1059–1067. [Google Scholar] [CrossRef]
- Asogwa, N.V.; Udeani, T.K.; Obeagu, E.I.; Asogwa, B.E.; Chukwueze, C.M. Prevalence of Salmonella species infection in poultry farming systems in Enugu metropolis Nigeria. European Journal of Pharmaceutical and medical research 2022, 9, 87–91. [Google Scholar]
- Jibril, A.H.; Okeke, I.N.; Dalsgaard, A.; Kudirkiene, E.; Akinlabi, O.C.; Bello, M.B.; et al. Prevalence and risk factors of Salmonella in commercial poultry farms in Nigeria. PLoS ONE 2020, 15, e0238190. [Google Scholar] [CrossRef]
- Mshelbwala, F.M.; Ibrahim, N.D.G.; Saidu, S.N.; Azeez, A.A.; Akinduti, P.A.; Kwanashie, C.N.; Fakilahyel-Kadiri, A.K.; Muhammed, M.; Fagbamila, I.O.; Luka, P.D. Motile Salmonella serotypes causing high mortality in poultry farms in three SouthWestern States of Nigeria. Veterinary Record Open 2017, 4, e000247. [Google Scholar] [CrossRef]
- Bettridge, J.M.; Lynch, S.E.; Brena, M.C.; Melese, K.; Dessie, T.; Terfa, Z.G. Infection-interactions in Ethiopian village chickens. Preventive Veterinary Medicine 2014, 117, 358–66. [Google Scholar] [CrossRef] [PubMed]
- Thrusfield, M. Veterinary epidemiology, 3rd ed.; Oxford, UK: Blackwell Science Ltd., A Blackwell Publishing Company, 2007. [Google Scholar]
- Lampel, K.A.; Orlandi, P.A.; Kornegay, L. Improved template preparation for PCR-based assay for detection of food-borne bacterial pathogens. Applied and Environmental Microbiology 2000, 66, 4539–4542. [Google Scholar] [CrossRef] [PubMed]
- Murray, R.A.; Lee, C.A. Invasion genes are not required for Salmonella enterica serovar Typhimurium to breach the intestinal epithelium: Evidence that Salmonella pathogenicity island 1 has alternative functions during infections. Journal of Infection and Immunology 2000, 68, 5050–5055. [Google Scholar] [CrossRef] [PubMed]
- Yanestria, S.M.; Rahmaniar, R.P.; Wibisono, F.J.; Effendi, M.H. Detection of invA gene of Salmonella from milkfish (Chanos chanos) at Sidoarjo wet fish market, Indonesia, using polymerase chain reaction technique. Vet World. 2019, 12, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Rahn, K.; De Grandis, S.A.; Clarke, R.C.; McEwen, S.A.; Galan, J.E.; Ginocchio, C.; Curtis III, R.; Gyles, C.L. Amplification of invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Probes 1992, 6, 271–279. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).