Submitted:
21 September 2025
Posted:
22 September 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Synopsis of Neurotransmitters (NTs)
1.2. Knowledge Gap on Neurotransmitters and Their Estimation Using Biosensors
1.3. Challenges in Neurotransmitter Estimation by Electrochemical Techniques and Their Mitigation
1.4. Unresolved Issues in NT Estimation by Electrochemical Techniques and Potential AI-Driven Solutions
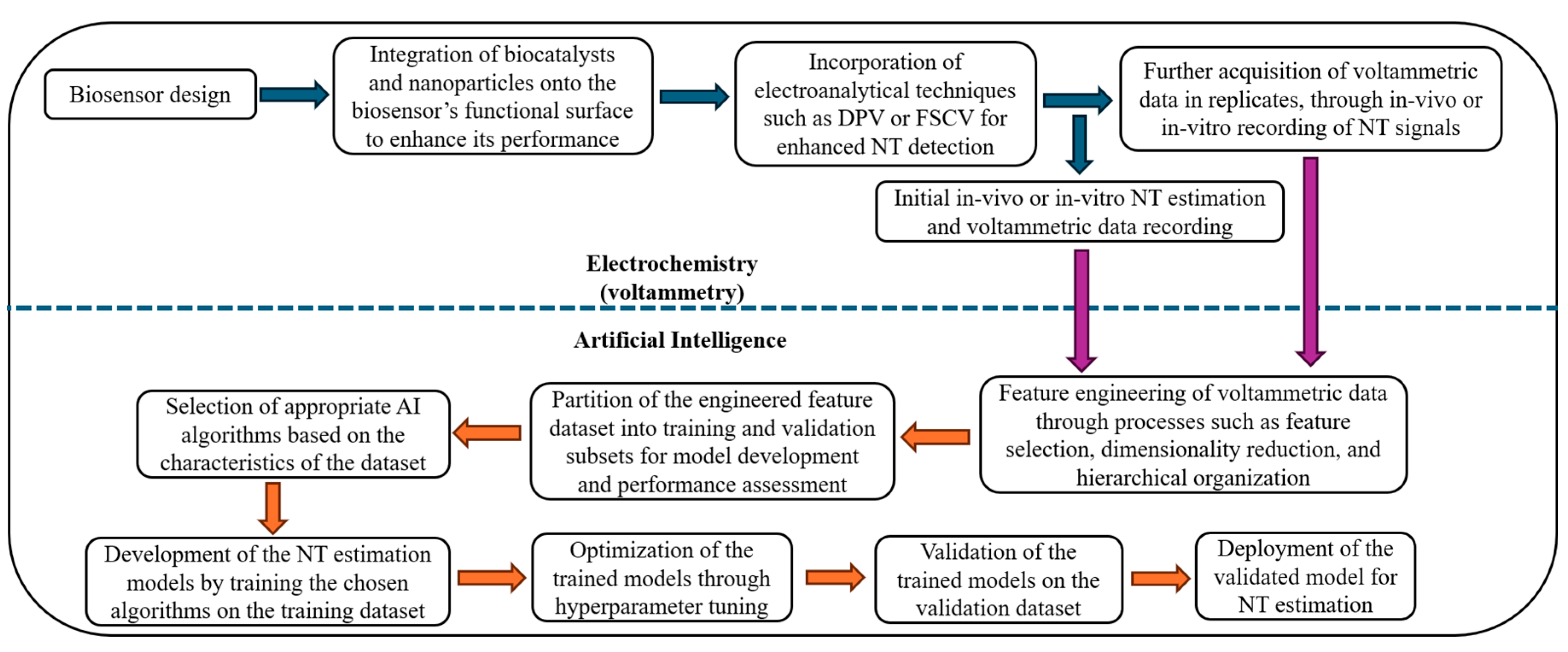
1.5. Related Reviews
1.6. Scope of this Review
1.7. Organization of the Review
2. Materials and Method
2.1. Review Methodology
2.2. Planning the Survey
2.2.1. Research Questions
- RQ1: Which NTs were studied? This question explores the various types of NTs, including their electroactivity, chemical structures, functions, central nervous system locations, and receptor interactions.
- RQ2: How were the multiplexed signal patterns of NTs recorded? This inquiry examines the different biosensors and neurochemical techniques used to capture multiplexed NT signals from complex biological fluids, as well as the experimental settings (in-vitro or in-vivo).
- RQ3: What were the characteristics of the datasets? We will review the datasets based on their size, features, and authenticity.
- RQ4: Which ML, PR or DL algorithms were employed for NT estimation? This question will assess the AI algorithms used, including whether they were supervised or unsupervised, classification or regression, parametric or non-parametric, linear or nonlinear. It will also cover the ML workflows involved, such as feature selection, normalization/regularization, dimensionality reduction, model selection, model training-validation-testing and the quality metrics used to evaluate the performance of the trained models.
2.2.2. Sources of Study
2.2.3. Search Strategy for the Review
| Criteria | Inclusion criteria | Exclusion criteria |
|---|---|---|
| Articles quality | Peer reviewed research Articles | Thesis and dissertations |
| Scope of the Articles | Articles with clear elaboration of the end-to-end AI algorithm used for the Pattern Recognition of NTs | Articles that do not meet this criterion |
| Publication Language | The articles must be published in English | Articles published in other languages other than English |
| Research Index Terms | A subset of the following words must be addressed by the articles: pattern recognition, simultaneous detection and prediction, automatic quantification + Neurotransmitters and Machine Learning, Deep Learning + Chemometrics | Any article not falling in these subsets |
| Target neurochemicals | Articles analyzing neurotransmitters | Articles analyzing neurochemicals other than neurotransmitters |
| Detection limits of the NTs | Articles focusing on in-vivo or in-vitro analysis of NTs having concentrations within the physiological ranges | Ranges Falling out the physiological ranges |
| Age of the publication | Articles published not earlier than the year 2009 | Articles published earlier than the year 2009 |
| Reference | Summary | Outcome | Publication Year |
|---|---|---|---|
| Sazonova et al [1] | Used two pattern recognition techniques: Principal Component Regression (PCR) and Partial Least Squared Regression (PLSR), with voltammetry to simultaneously estimate dopamine and serotonin, addressing signal overlap | Achieved estimation accuracies ranging from 81% to 91% for DA and 91% to 100% for SE | 2009 |
| Abbasi et al [2] | Developed a quantum/carbon dot tricolor fluorescent probe to enable rapid, pattern-recognition-based discrimination of catecholamine NTs from ascorbic acid (AA) in urine with Linear Discriminant Analysis (LDA). | Achieved 98% accuracy in NT discrimination using leave-many-out cross- validation | 2019 |
| Xiaotong et al [3] | Developed a metal nanoparticle-based nanozyme sensor array to enable pattern-recognition-driven discrimination of monoamine NTs in human serum using LDA and a Hierarchical Clustering Algorithm (HCA) | Successfully discriminated monoamine NTs at varying concentrations with 100% accuracy | 2022 |
| Jose et al [9] | Used TinyML embedded in portable biosensors to discriminate NTs from uric acid (UA) and ascorbic acid (AA) interference for real time applications. | Achieved NT-discrimination accuracies of 98.1% using a 32-bit floating-point unit and 96.01% after 8-bit quantization, | 2023 |
| Martens et al [21] | Predicted glutamate (GL) from whole-brain functional connectivity of the pregenual anterior cingulate cortex using elastic net (EN), PLSR and HCA | Achieved an R2 value (regression fit) of 0.143 and p-value (probability value) of less than 0.001 using EN for the prediction of GL | 2020 |
| Nchouwat et al [24] | Used nIRCat data to simultaneously detect and quantify age-dependent DA release in mice brain slices using CatBoost regressor, that was later distillated to kernelized ridge regressor (KRR) for improved performance. | Achieved a performance for the validation Mean Squared Error (MSE) of 0.001 and an R² value of 0.97 in estimating DA release. | 2025 |
| Salimian et al [33] | Used UV–vis spectrophotometry coupled with net analyte system and PCR to simultaneously detect levodopa (LD) and carbidopa (CD) in mixtures, drugs, and breast milk | Achieved mean recovery values of 96.86% for LD and 92.43% for CD using PCR, with corresponding mean squared prediction errors of 1.50 for LD and 7.14 for CD | 2022 |
| Dowek et al [34] | Developed a robust, pharmaceutical-grade method with PLSR to distinguish and quantify norepinephrine (NE) and epinephrine (EP) | Achieved R² values of 0.95 and 0.91 for the quantification of EP and NE respectively, with corresponding root mean square errors (RMSE) of 5.47 for EP and 7.27 for NE | 2022 |
| Jafarinejad et al [35] | Designed an optical sensor array with three fluorescent dyes and pattern recognition to detect DA, EP, and NE by tracking changes in their emission when gold ions are present using LDA, artificial neural networks (ANN) and multilinear regression (MLR) | Achieved an accuracy of 100% in discriminating the NTs and their mixtures using LDA | 2020 |
| Kallabis et al [36] | Applied MLR, KRR and Bayesian Linear regression (BLR) models to quantify dopamine concentrations amidst nonlinear variations induced by magnesium ion interactions. | Achieved a mean absolute percentage error of approximately 6–7% across all models, which is slightly above the experimental error observed in the absence of magnesium ions | 2024 |
| Jafarinejad et al [37] | Proposed a high-performance colorimetric sensor array and pattern recognition (PCA, LDA and HCA) to detect and distinguish catecholamines (DA, EP, NE and their mixtures) by their ability to reduce silver onto gold nanorods | Achieved a discrimination accuracy of 100% for the individual NTs and their mixtures using LDA | 2017 |
| Siamak et al [38] | Utilized nIRCat imaging combined with machine learning models: Support Vector Machine (SVM) and Random Forest (RF), to uncover distinct dopamine release patterns across different regions of mice brains | Achieved average detection accuracies of 55.5% and 83.2% using SVM and RF, respectively, in studies involving mice younger than 12 weeks. | 2023 |
| Komoto et al [40] |
Directly observed a single NT (DA, SE, NE or their mixtures) by Measuring tunneling current flowing through the single NT, using nanogap electrodes and XGBoost classifier. | Identified the spatial distribution patterns of NTs in the brain with high temporal resolution | 2020 |
| Hoseok et al [42] | Compared the performance of deep learning (DL) and principal component regression (PCR) in predicting NT concentrations focussing on DA, SE, EP and NE | Demonstrated that DL slightly outperformed PCR for NT detection, achieving an average accuracy of 96.23% compared to 95.39% with PCR | 2022 |
| Seongtak et al [43] | Used deep learning to simultaneously estimate tonic DA and SE with high temporal resolution in vitro. | Achieved statistically significant accuracy (p < 0.001) for the in vitro estimation of DA and SE | 2023 |
| Rantataro et al [44] | Selectively detected DA and SE at nanomolar concentration from complex in-vitro systems in real-time with electrochemical techniques | Achieved an average R² value of 0.99 for both DA and SE estimation using cyclic voltammetry (CV) and chronoamperometry | 2023 |
| Buchanan et al [45] | Used convolutional neural networks to evaluate SE neurochemistry in vivo | Achieved statistically significant accuracy (p < 0.0001) for the in vivo estimation of SE | 2024 |
| Simon et al [46] | Focused on linear and quadratic regression models to describe an FPGA-based system for measuring NT concentrations on a multi-sensor platform, utilizing a visible-light optical spectrometer | Achieved a mean training precision of 91.22% and a mean validation precision of 90.19% for NT estimation using quadratic regression | 2020 |
| Doyun Kim et al [47] | Automated cell detection method for TH-positive dopaminergic neurons in a mouse model of Parkinson’s disease using convolutional neural networks | Successfully detected TH-positive dopaminergic neurons with a recall of 78.07%, precision of 74.46%, and an F1 score of 76.51% | 2023 |
| Jian Lv et al [48] | Developed a nanopipette method coupled with an XGBoost classifier to detect DA in single exosomes | Achieved a classification accuracy of 99% for DA detection in single exosome | 2023 |
| Credico et al [49] | Applied ML algorithms (LDA, XGBoost and LightGBM) to identify phenotypic profile alterations of human dopaminergic neurons exposed to bisphenols and perfuoroalkyls | Achieved classification accuracies ranging from 88% to 96.5% across the three algorithms | 2023 |
| Arijit Pal Et al [50] | Detected DA using a machine-intelligent web app interface and paper sensor modified with MoS2 | Achieved classification accuracy of 99% | 2023 |
| Kammarchedu et al [52] | Electrochemical Screened NTs (DA, SE, EP and NE) using a customizable machine learning-based multimodal system based on K-nearest neighbors (KNNR) and decision tree regressors (DTR). | Successfully differentiated between the four NTs and selectively detected each when independently present in complex media | 2023 |
| Bang et al [53] | showed that ne tracks emotional modulation of attention in human amygdala and estimated NE, SE and DA in-vivo using deep learning | Achieved statistically significant accuracy (p < 0.001) for the in-vivo estimation of NE, DA and SE | 2023 |
| Sanjeet et al [54] | Simultaneously detected DA and SE in an optimized carbon thread-based miniaturized device using several ML algorithms | Achieved an R² value of 0.99 for both DA and SE estimation using k-nearest neighbors regressor and random forest regressor | 2024 |
| Goyal et al [55] | Applied voltammetry coupled with deep learning (DiscrimNet architecture) to estimate tonic concentrations of highly similar NTs (DA, SE and NE) and their mixtures | DiscrimNet accurately predicted changes in DA and SE levels, even in the presence of interfering substances like cocaine or oxycodone, demonstrating low RMSE across all NTs | 2024 |
| Unger et al [56] | Analyzed the directed evolution of a selective and sensitive SE sensor using ML (Random Forest and generalized linear model) | Used ML to demonstrate the detection of SE release in freely moving mice during fear conditioning, social interactions, and sleep–wake transitions | 2020 |
| Movassaghi1 et al [57] | Simultaneously monitored SE and DA across timescales by rapid pulse voltammetry (RPV) coupled with partial least squares regression (PLSR) | Demonstrated that RPV-PLSR outperforms FSCV-PCR in the simultaneous monitoring of DA and SE. | 2021 |
| Zhang et al [58] | Applied deep learning to automatically classify and predict NTs (GABA, acetylcholine and glutamate) synapses in electron microscopy | Successfully identified NT synapses from EM images to construct a complete neuronal connectivity map, achieving 98% validation accuracy | 2022 |
| Matsushita et al [59] | Automatically Identified phasic dopamine release using SVM | Accurately identified phasic DA using automatically extracted patches, achieving 89.18% accuracy and a best F-measure of 77.23% | 2018 |
| Matsushita et al [60] | Improved the automatic identification of phasic dopamine release from fast-scan cyclic voltammetry data using convolutional neural networks (CNN). | Achieved 97.66% accuracy in phasic DA detection using an end-to-end CNN object detection system based on YOLOv3 | 2019 |
| Xue et al [61] | Introduced a deep learning-voltammetry platform for the selective analysis of three neurochemicals (ascorbate, DA and sodium chloride) in live animal brains | Selectively and simultaneously estimated neurochemicals with high spatial and temporal resolution | 2021 |
| Nchouwat et al [111] | Used PCR and PLSR for the simultaneous estimation of NTs, reducing complexity for SE and DA | Simultaneously estimated DA and SE with 97.6% accuracy, while reducing the number of feature subsets required for the NT estimation | 2025 |
3. Conducting the Survey
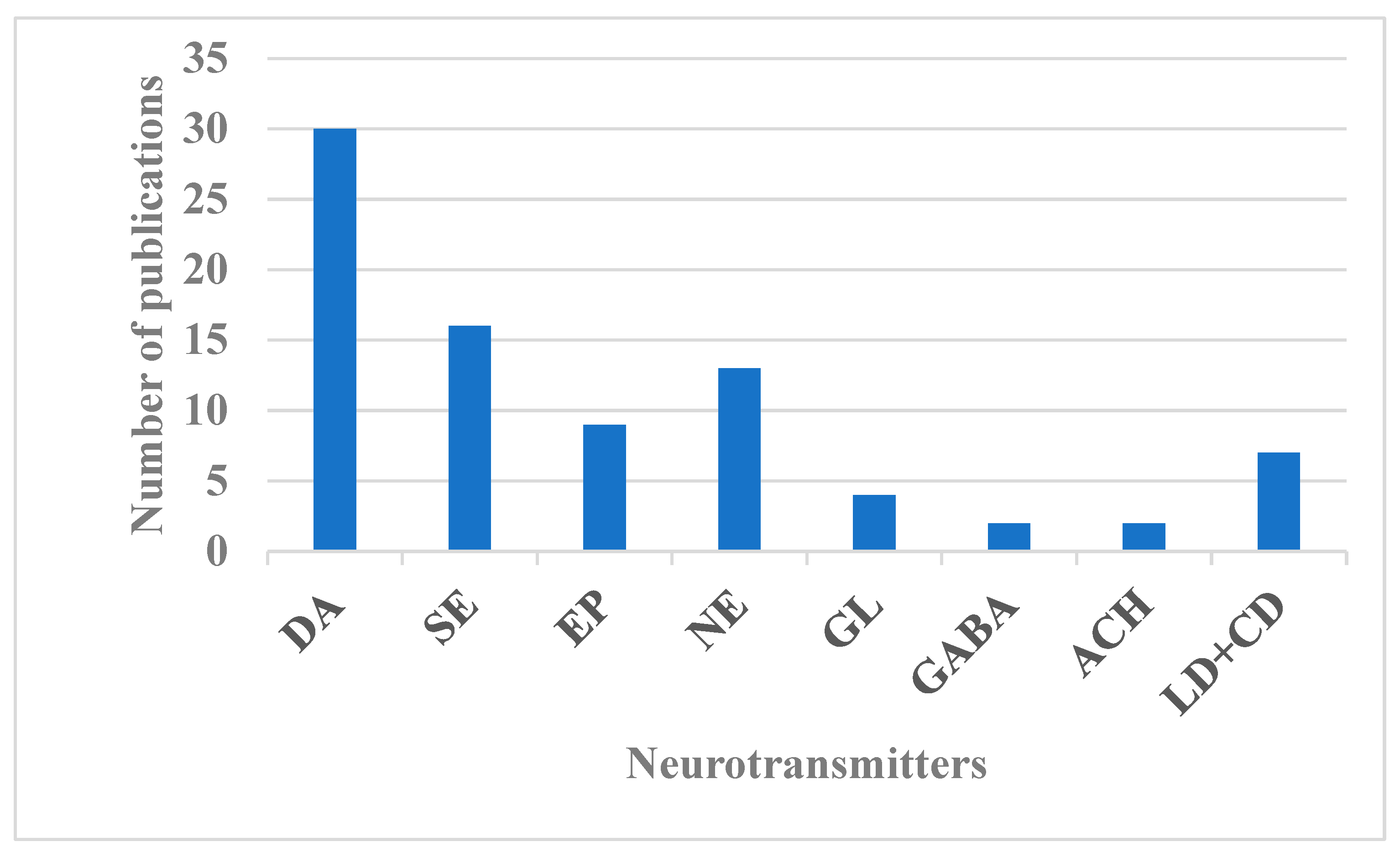
3.1. Neurotransmitters (NTs)
3.1.1. Dopamine (DA)
3.1.2. Serotonin (SE)
3.1.3. Glutamate (GL)
3.1.4. Acetylcholine (ACH)
3.1.5. Epinephrine (EP) and Norepinephrine (NEP)
3.1.6. Gamma-Aminobutyric Acid (GABA)
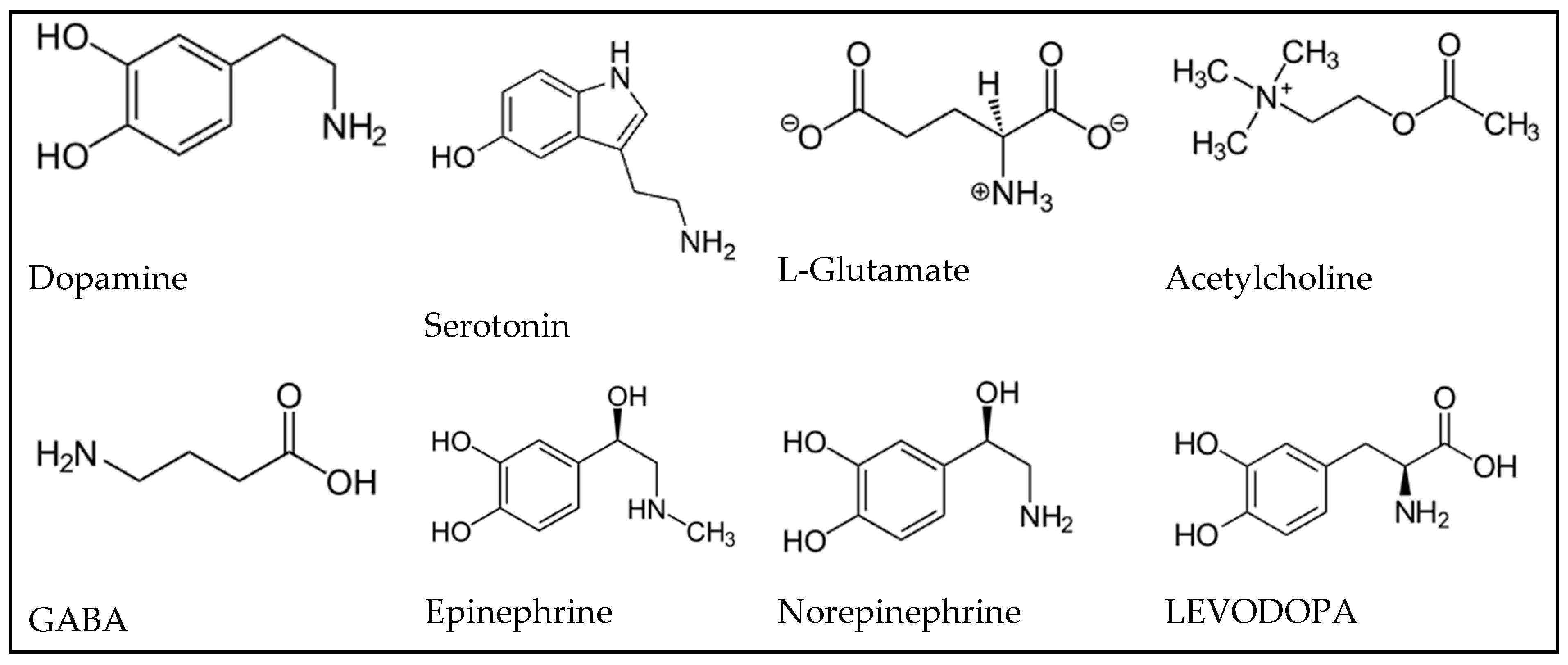
| Category | Neurotransmitter | Chemical formula | localization | Role | Pathology | Reference |
|---|---|---|---|---|---|---|
| Indoleamine | Serotonin (SE) | C₁₀H₁₂N₂O | midbrain, hypothalamus, limbic system, cerebellum, pineal gland, spinal cord. |
excitatory to other NTs, sleep, appetite, memory, cardiovascular regulation, temperature, walking | nausea, headaches, regulation of mood, schizophrenia, anxiety and depression |
[1,3,40], [41,42,43,44,45], [46,52,53,54,55,56,57,58] |
| Catecholamines | Dopamine (DA) | C₈H₁₁NO₂ | Hypothalamus, substantia nigra | excitatory to other NTs, pleasure, satisfaction, motivation, the regulation of emotional, social stress, learning, reward, addictive behavior, motion control |
Parkinson’s disease, Huntington’s disease, drug addiction and schizophrenia |
[1,2,3,24], [9,35,36,37,38], [40,41,42,43], [44,46,47,48], [49,50,51,52], [53,54,55,57], [58,59,60,61] |
| Epinephrine (EP) |
C₉H₁₃NO₃ | Tegmental and medulla | Fight-or-flight response | N/A | [3,34,35], [37], [42,46,52,58] |
|
| Norepinephrine (NE) | C₈H₁₁NO₃ | Locus coeruleus of the midbrain, brain stem, limbic system, cerebral cortex, thalamus. |
Good feeling | depression | [2,3,34,35], [37,40,42,46,52,53,55,58] |
|
| Amino Acids | Glutamate (GL) | C₅H₉NO₄ | central nervous system (Brain and Spinal cord) | excitatory to other NTs, learning, memory, vision, |
epilepsy, schizophrenia, excitotoxicity, multiple sclerosis, amyotrophic lateral sclerosis, and Parkinson’s disease |
[21,46,58,62] |
| GABA | C₄H₉NO₂ | Hypothalamus, spinal cord, cerebellum, retina | excitatory/ inhibitory to other NTs | epilepsy, convulsions, sleep disorders and pain. | [46,58] |
|
| Choline | Acetylcholine (ACH) | C₇H₁₆NO₂⁺ | Basal nuclei and cortex, neuromuscular junctions, brain |
excitatory to other NTs, memory, learning | Alzheimer's disease, hallucination, tetanic muscle spasms |
[46,58] |
| Precursors | LEVODOPA | C₉H₁₁NO₄ | Oral drugs found in blood | Treatment of Parkinson’s disease | N/A | [2,3,33,41,45,46,58] |
3.2. Origin of Multiplexed Neurotransmitter Signals and Motivation of the Need for AI
3.3. Electrochemical Biosensors
3.3.1. Enhancement of Biosensors’ Selectivity
- Enzymes: Enzymes serve as biorecognition elements. They are NT-specific and catalyze a reaction with the target analyte. The resulting product is directly detected by the sensors at lower potentials, which enhances the selectivity for neurotransmitter detection.
- Antibodies or Antigens. They also serve as bio-recognition elements on immunobiosensors. They bind specifically to the target analyte, and the resulting complex byproduct is detected.
- DNA: DNA strands are used to detect complementary DNA sequences or specific genetic material when used on DNA biosensors.
- Microbes: Microbes are functionalized on microbial biosensors where the whole cells or parts of cells are used to detect analytes. These biosensors can be employed for environmental monitoring.
- Light: Optical biosensors use light-based techniques for detection, such as fluorescence, luminescence, or surface plasmon resonance for specific NT detection. Light of different frequencies are used for the selective NT detection.
- Pressure: Piezoelectric Biosensors measure changes in mass on the sensor surface, typically using a quartz crystal microbalance during NT detection. Here, pressure is used as the discriminative parameter for the selective detection of the NTs in a complex mixture.
3.3.2. Enhancement of Biosensors’ Sensitivity
3.3.3. Challenges Faced by Enhancing Biosensors’ Performance
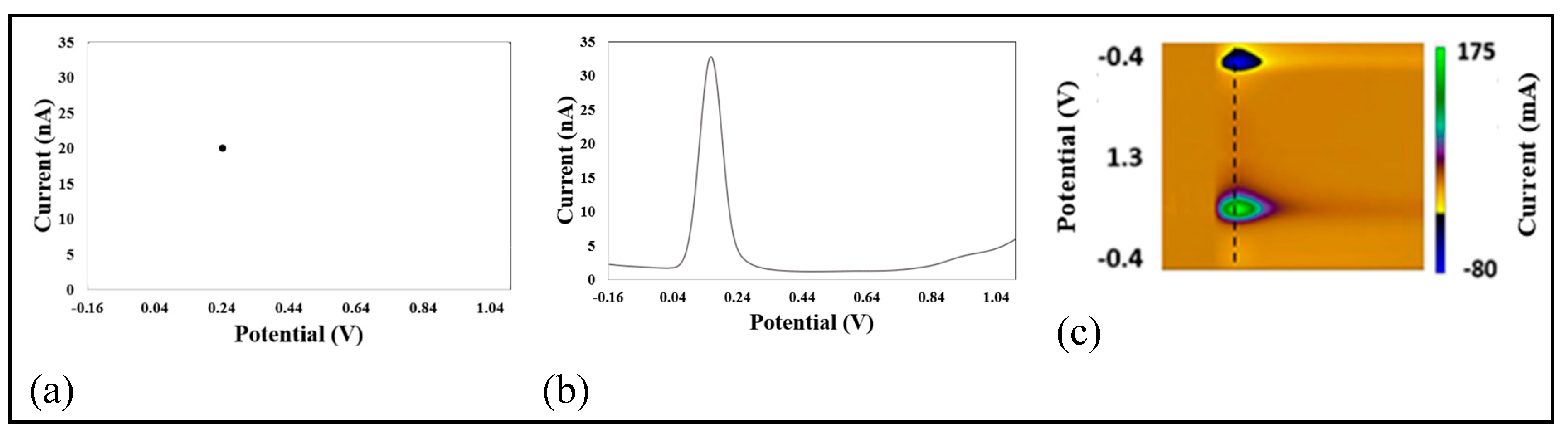
3.4. Transduced Signal Output from Biosensors and Voltammetric Measurement Techniques
3.5. Artificial Intelligence (AI)
3.5.1. Supervised Learning
3.5.2. Unsupervised Learning
3.5.3. Semi Supervised Learning
3.5.4. Reinforcement Learning
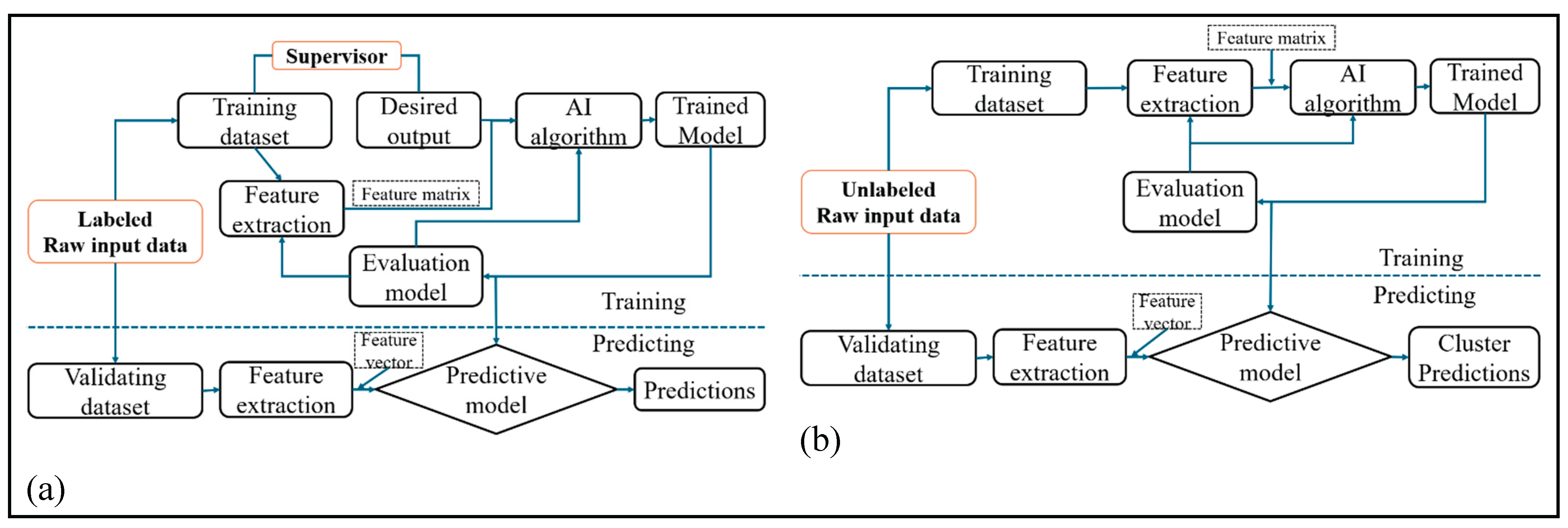
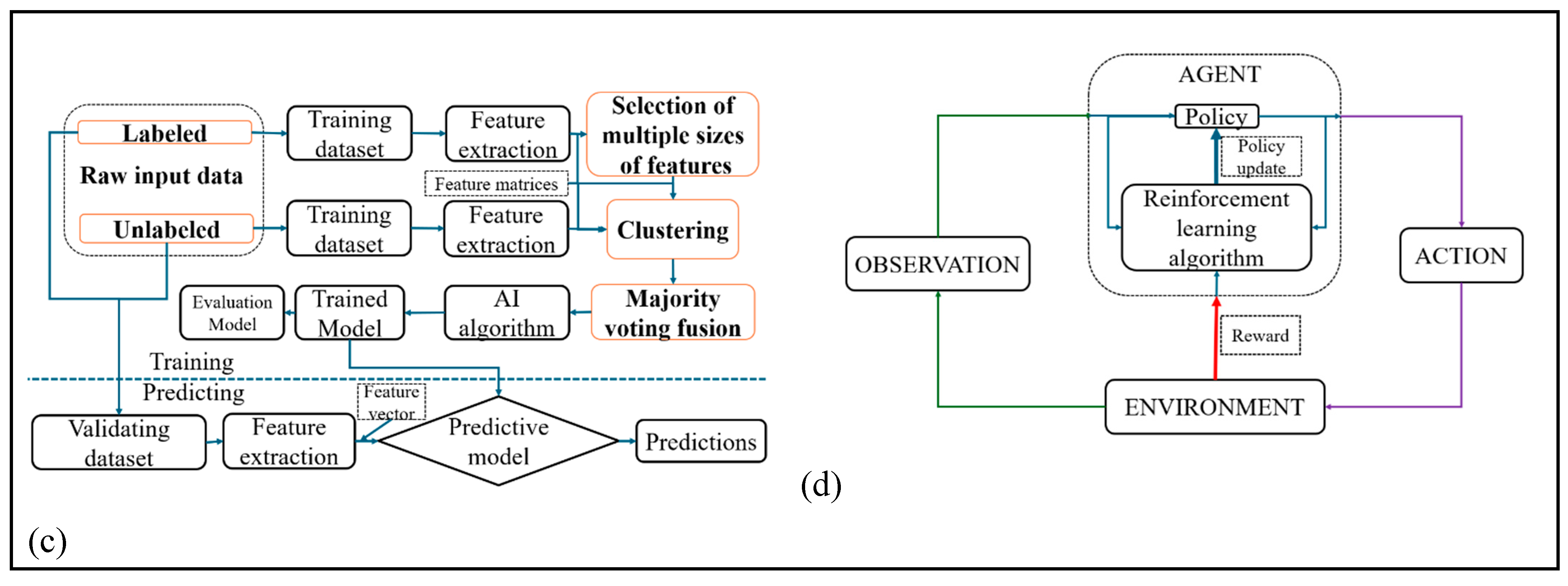
| Learning type | definition | Type of dada | Goal | Example of tasks | Learning signal | Common algorithm | Human effort for labeling | Challenges | References |
|---|---|---|---|---|---|---|---|---|---|
| Supervised | Learning from fully labeled data | Labeled data | Predict outcomes (classification, regression) | Image classification, spam detection, price prediction | Direct supervision (ground-truth labels) | Linear regression, decision trees, SVM, neural nets | High | Requires large, labeled datasets | All 30 except [3,35,37] |
| Semi supervised | Learning from a small amount of labeled and unlabeled data | Mostly unlabeled data with some labeled examples | Improve learning using a few labeled data points | Text classification with limited labels | Weak supervision (partial labels) | Self-training, co-training, label propagation | Moderate | Making good use of limited labeled data | N/A |
| Unsupervised | Learning from completely unlabeled data | Unlabeled data | Discover hidden patterns or structure | Clustering, dimensionality reduction | No supervision (structure learning) | K-means, PCA, DBSCAN, autoencoders | None | Interpreting unsupervised results | [3,35,37] |
| Reinforcement | Learning through interaction with an environment | States, actions, rewards | Maximize cumulative reward over time | Game playing, robotics, recommendation systems | Reward signal (positive/negative feedback) | Q-learning, SARSA, Deep Q-Networks (DQN), policy gradients | None (but requires environmental simulation) | Balancing exploration vs. exploitation | N/A |
3.5.5. Machine Learning Algorithms
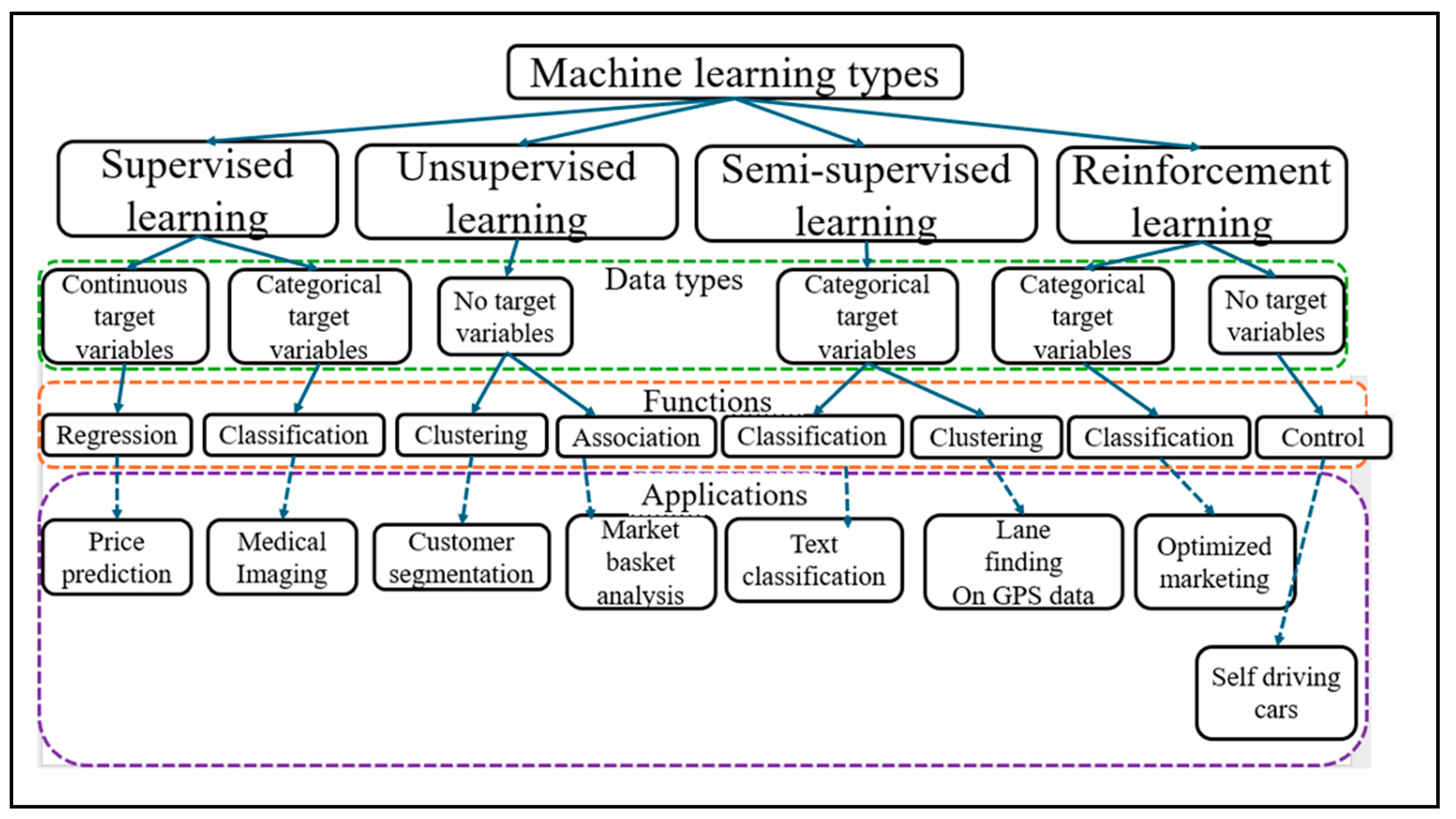
3.5.6. Pattern Recognition (PR)
- Detection: Identifying and categorizing different NTs.
- Quantification: Predicting the concentrations of known NTs.
- Simultaneous detection and quantification: Determining both the types and quantities of NTs. The word “estimation” is also used interchangeable with Simultaneous detection and quantification throughout this paper.
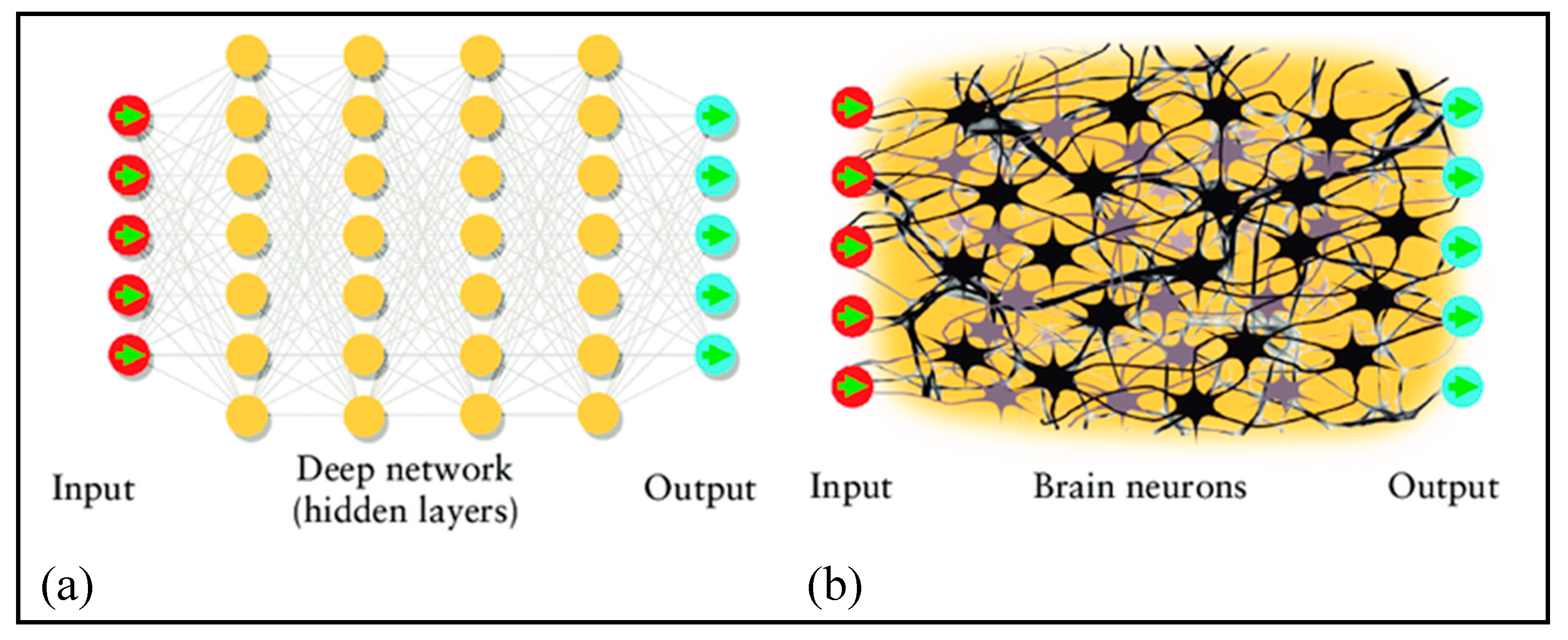
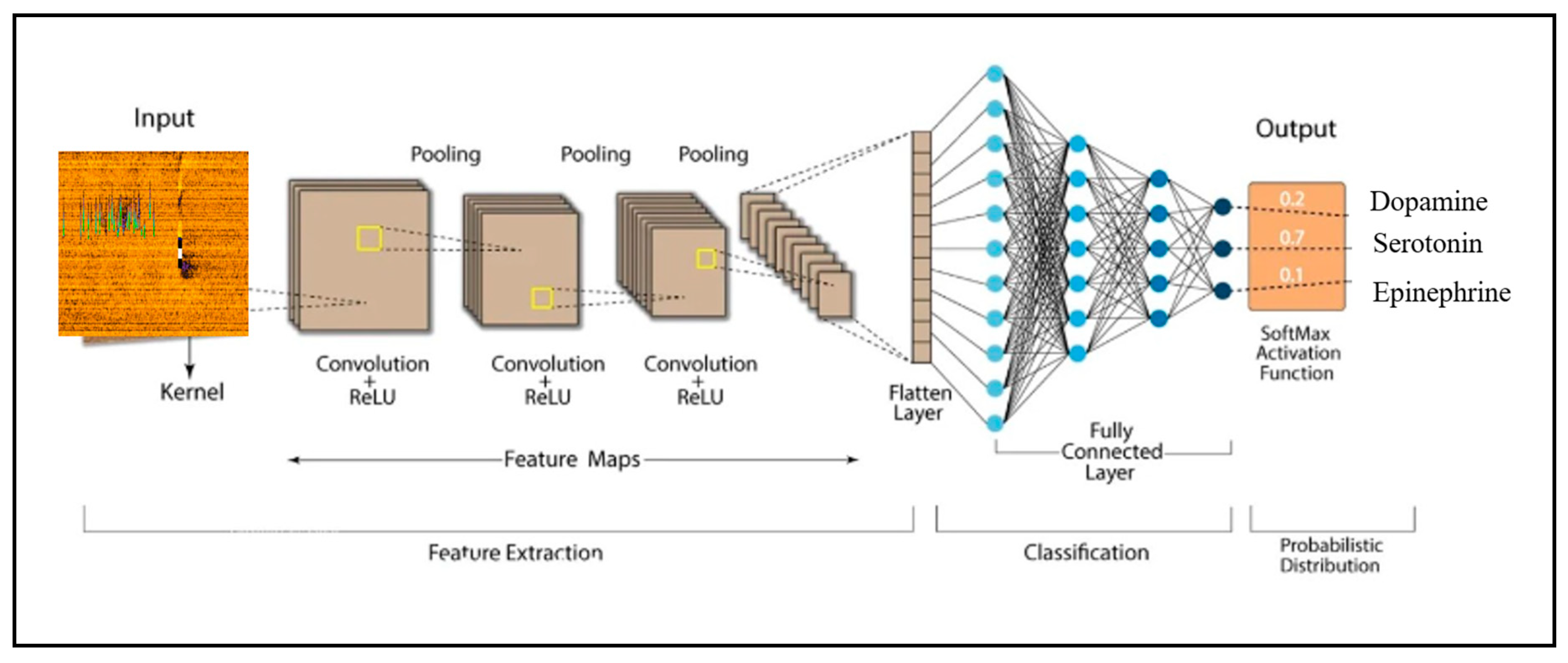
3.5.7. Deep Learning
- Clarity of Aim
- b.
- The data
- c.
- Factors and Levels
- d.
- The procedure and experimental design
- e.
- Performing the experiment
- f.
- Performance metrics
- ➢
- TP = True Positives (correctly predicted positives). Here, the model said positive, and it's true.
- ➢
- TN = True Negatives (correctly predicted negatives). Here, the model said negative, and it's true.
- ➢
- FP = False Positives (incorrectly predicted positives). Here, the model said positive, but it's false.
- ➢
- FN = False Negatives (incorrectly predicted negatives). Here, the model said negative, but it's false.
- ➢
- yi= actual or true value
- ➢
- = predicted value
- ➢
- n = number of data points
4. Survey Outcome
4.1. Detection of Neurotransmitters
4.1.1. Application of Conventional Machine Learning Algorithms
- linear discriminant analysis
- b.
- Support vector machines
- c.
- Randomforest
- d.
- Hierarchical clustering algorithm
- d.
- Embedded machine learning
4.1.2. Application of Deep Learning Algorithms
4.2. Quantification (Prediction of Concentrations) of Neurotransmitters
- Linear Regression
- 2.
- Quadratic regression
- 3.
- Bayessian linear regression
- 4.
- Kernelized ridge regression
4.3. Simultaneous Detection and Quantification of Neurotransmitters
4.3.1. Application of Principal Component Regression and Partial Least Squared Regression
| Scope | Summary | References |
|---|---|---|
| NT detection | These are classification processes that group NTs one at a time into distinct categories without prior knowledge of their types or concentrations in the biological fluid being studied. | [2,3,9,35,37,38,40,44,47,48,49,50,52,56,59,60,61] |
| NT quantification | These are regression processes used to quantify NT concentrations one at a time, based on prior knowledge of the types of NTs present in the biological fluid being studied | [21,24,34,36,45,46,47,50,52,53,56] |
| Simultaneous Detection and quantification of NTs | These are combined classification and regression processes in which multiple NTs are simultaneously categorized and their concentrations quantified, without prior knowledge of the types or concentrations of NTs present in the biological fluid being studied | [1,33,42,43,54,55,57,58,111] |
| Algorithm | Learning type | Summary | References |
|---|---|---|---|
| LDA | Supervised | Reduces data dimensions while maximizing class separation by finding the feature combinations that best distinguish between categories | [2,3,35,37], [49] |
| SVM | Supervised | Finds the optimal boundary (hyperplane) to separate classes by maximizing the margin between different class data points for better generalization. | [38,55] |
| RF | Supervised and ensemble | Build multiple decision trees and combine their outputs for more accurate, performant, robust and stable predictions. | [38,52,55], [56] |
| GBM and CATBOOST | Supervised and ensemble | build models sequentially, where each new model corrects errors made by the previous ones and combines many weak learners (usually decision trees) to create a strong predictive model. | [24,41,48] |
| XGBOOST | Supervised | an optimized version of gradient boosting that is faster and more efficient through advanced regularization, parallel processing, and handling of missing values | [40,48,49] |
| HCA | Unsupervised | builds a hierarchy of clusters by either merging or splitting data points based on similarity and creates a dendrogram to visualize the nested grouping of data. | [2,3,35], [37,55] |
| LR | Supervised | Models the relationship between one independent variable and one dependent variable by fitting a straight line and predicts the dependent variable based on the linear relationship with the independent variable. | [46,50,51,52,55,56] |
| MLR | Supervised | Models the relationship between two or more independent variables and one dependent variable, fitting a linear equation to predict the outcome based on the combined effect of all input variables. | [35,37] |
| QR | Supervised | Models the relationship between the independent variable(s) and the dependent variable using a second-degree polynomial (a quadratic equation) | [46] |
| BLR | Supervised | Incorporates Bayesian inference to estimate the distribution of model parameters, providing a probabilistic approach to linear regression and offering not just point estimates but also uncertainty estimates for the model's predictions | [36] |
| KRR | Supervised | Combines Ridge Regression with the kernel trick to model nonlinear relationships and maps input features into a higher-dimensional space to perform linear regression in that space, enabling it to capture complex patterns. | [36] |
| PCR | Supervised and dimensionality reduction | Combines Principal Component Analysis (PCA) for dimensionality reduction with linear regression. It uses the principal components (uncorrelated features) as inputs to predict the target variable. | [1,33,41], [42,111] |
| PLSR | Supervised and dimensionality reduction | Reduces predictors to a smaller set of uncorrelated components while maximizing the covariance between predictors and the response variable | [1,21,34,41], [46,57,111] |
| DL | Supervised, Semi supervised, unsupervised or reinforcement learning | Uses multi-layered neural networks to automatically learn complex patterns from large amounts of data | [9,35,37], [42,43], [45,47,53], [58,59,60,61] |
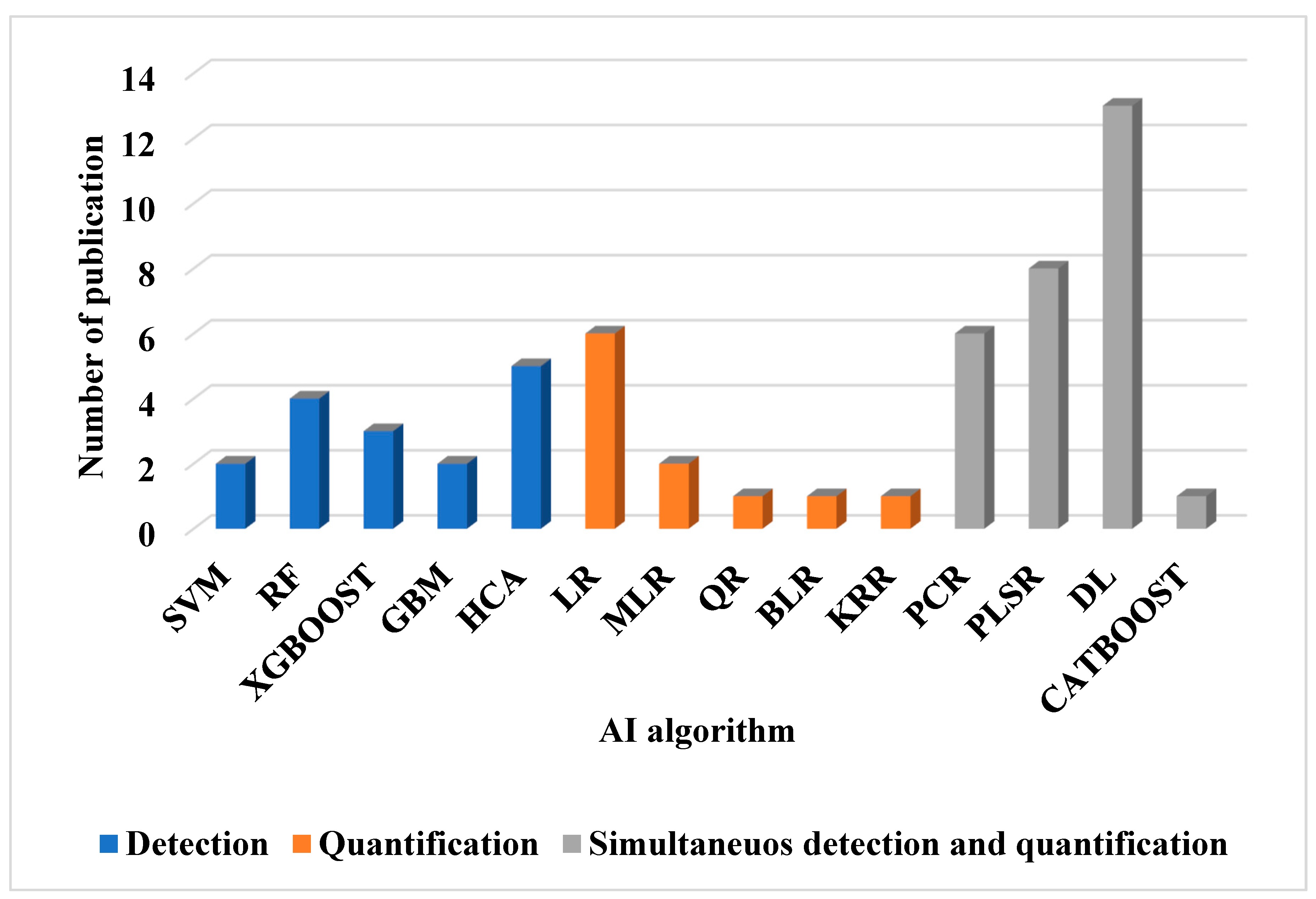
4.3.2. Application of Deep Learning and Artificial Neural Networks
4.3.3. Application of Embedded Machine Learning
4.4. AI Algorithms and Voltammetric Techniques for NT Detection
4.5. Limitations of the Existing Studies Employing AI Algorithms Trained on Voltammetric Data
- Elevated detection thresholds for NT estimation. Sazonova et al. [1] extended the estimation thresholds of NT concentrations beyond the actual concentration ranges, resulting in wider confidence intervals. Consequently, the predicted NT concentrations often fail to reflect the true levels present in complex biological matrices, thereby compromising the reliability of these estimations.
- Limited discrimination of NTs during simultaneous NT quantification. Although the proposed computational models demonstrate high performance metrics, they are frequently unable to fully discriminate between different NT species. Accurate and complete discrimination is essential for comprehensive profiling of individual NTs and is a critical requirement for identifying biomarkers associated with neurodegenerative diseases.
- Restricted NT species coverage and simplified mixtures of NT species. Existing studies generally investigate only a limited subset of NTs, even though biological fluids naturally contain a vast array of NT species in dynamic equilibrium. For a more representative analysis, it is necessary to evaluate extended NT species libraries and more complex mixtures, which better reflect physiological conditions.
- Resource-intensive computational models. Many of the AI-based approaches proposed for automatic NT detection and quantification are computationally intensive, resulting in slow processing times and high resource demands. These limitations hinder their applicability in real-time or near-real-time settings. To overcome this, model compression techniques such as knowledge distillation or transfer learning can be employed to develop lightweight, computationally efficient alternatives (e.g., simplified linear models).
- Model overfitting and bias-related performance issues. Numerous AI models exhibit significant overfitting, as indicated by disproportionately high training accuracy compared to validation performance. This can be attributed to the high-dimensionality, noise, and variance inherent in biological datasets. These issues cause models to learn noise patterns along with meaningful signals. Effective mitigation strategies include robust data preprocessing, noise reduction, and advanced feature engineering to isolate and prioritize relevant features from complex datasets.
4.6. Tools Used for the Automatic Detection and Quantification of Neurotransmitters
5. Discussion
5.1. Challenges Faced in the Automatic Detection and Quantification of Neurotransmitters
5.2. Potential Application of the Automatic Detection and Quantification of Neurotransmitters
5.2.1. Estimation of Neurotransmitters in Cerebrospinal Fluid from Brain Samples
5.2.2. Estimation of Neurotransmitters from Urine Samples
5.2.3. Estimation of Neurotransmitters from Blood Samples
6. Conclusion
References
- N. Sazonova, J. Njagi, Z. Marchese, M. Ball, S. Andreescu, and S. Schuckers, “Detection and prediction of concentrations of neurotransmitters using voltammetry and pattern recognition,” Ann. Int. Conf. of the IEEE Eng in Med. And Biol. Soci., pp. 3493–3496, 2009.
- S. Abbasi-Moayeda, M. R. Hormozi-Nezhada and M. Maazac, “A multichannel single-well sensor array for rapid and visual discrimination of catecholamine neurotransmitters” Sensors & Actuators: B. Chemical 296 (2019) 126691.
- W. Xiaotong, L. Yuling, L. Yan, Lei Tan, W. Jinyi, W. Zixuan, M. Zhong and Y. Liang “Colorimetric nanozyme sensor array for the pattern recognition of monoamine neurotransmitters using dendritic mesoporous silica embedded with metal nanoparticles” Sensors & Actuators: B. Chemical 369 (2022) 132287.
- Bo Si and Edward Song “Recent Advances in the Detection of Neurotransmitters” MDPI, Chemosensors 4, Jan 2018.
- H. Jiayi, E.Spanolios, C.E.Froehlich,C.L.Wouters and C.L.Haynes, “Recent Advances in the Development and Characterization of Electrochemical and Electrical Biosensors for Small Molecule Neurotransmitters”, ACS Sens. 2023, 8, 1391−1403.
- Yi Su, S.Bian, and M.Sawan, “ Real-time in vivo detection techniques for neurotransmitters: a review,” Royal Society of Chemistry, Analyst. 2020.145.6193.
- D.Yifan,L.Shihua & T.Yang, “Real-Time Monitoring of Neurotransmitters in the Brain of Living Animals”, ACS Appl. Mater. Interfaces 2023, 15, 138−157.
- G.F. Giordano, L.F.Ferreira, I.R.S.Bezerra, J.A.Barbosa, J.N.Y.Costa, G.J.C.Pimentel & R.S.Lima, “Machine learning toward high-performance electrochemical sensors”, Analytical and Bioanalytical Chemistry (2023) 415:3683–3692.
- J. I. F De Oliveira, M. C. Faleiros, D. C. Ferreira, M. Veerappan, and S. N. Khaled, “Empowering Electrochemical Biosensors with AI:Overcoming Interference for Precise Dopamine Detection in Complex Samples” Adv. Intell. Syst. 2023, 5, 2300227.
- J.Njagi, M.M. Chernov, J. C. Leiter, and S. Andreescu, “Amperometric Detection of Dopamine in Vivo with an Enzyme Based Carbon Fiber Microbiosensor” Anal. Chem. 2010, 82, 989–996.
- E.Dumitrescu, A.Deshpande, K. N. Wallace, and S. Andreescu, “Time-Dependent Monitoring of Dopamine in the Brain of LiveEmbryonic Zebrafish Using Electrochemically Pretreated Carbon Fiber Microelectrodes”. [CrossRef]
- N.Chauhan,, S. Soni, P. Agrawal,Y.P.S. Balhara, U. Jain, “Recent advancement in nanosensors for neurotransmitters detection: Present and future perspective”, Process Biochemistry 91 (2020) 241–259.
- Singh, A.Sharma,, A. Ahmed, A. K. Sundramoorthy, H. Furukawa, S. Arya and A. Khosla, “Recent Advances in Electrochemical Biosensors: Applications, Challenges, and Future Scope”, Biosensors 2021, 11, 336. [CrossRef]
- Nissar, M. I. Izharuddin, W. A.Mir & T.A.Shaikh, “Machine Learning Approaches for Detection and Diagnosis of Parkinson’s Disease - A Review” 7th Int. Conf on Ad .Comp& Com. Sys (ICACCS),2021.
- Rana, A. Dumka , R. Singh , M. Rashid , N. Ahmad & M. K. Panda, “An Efficient Machine Learning Approach for Diagnosing Parkinson’s Disease by Utilizing Voice Features”, Electronics 2022, 11, 3782. [CrossRef]
- R.Mathur, V. Pathak and D. Bandil, “Parkinson Disease Prediction Using Machine Learning Algorithm”, Advances in Intelligent Systems and Computing 841. [CrossRef]
- R. S. Stadena, I.Moldoveanua,J.F.van Stadena, “Pattern recognition of neurotransmitters using multimode sensing”, Journal of Neuroscience Methods 229 (2014) 1–7.
- S. Banerjee, S. McCracken, Md. F. Hossain and G. Slaughter “Electrochemical Detection of Neurotransmitters,” MDPI, Biosensors 2020, 10, 101.
- S. D. Niyonambaza, P. Kumar, P. Xing,J .Mathault, P.D. Koninck, E. Boisselier, M. Boukadoum and A.Miled, “A Review of Neurotransmitters Sensing Methods for Neuro-Engineering Research” MDPI, Applied Sciences 2019.
- T.Shiravand & A.Azadbakht, “Impedimetric biosensor based on bimetallic AgPt nanoparticle-decorated carbon nanotubes as highly conductive film surface”, Jour. Solid State Electrochem (2017) 21:1699–1711. [CrossRef]
- L. Martens,N. B. Kroemer, V. Teckentrup, L. Colic, N. Palomero-Gallagher, L. Meng and M. Walter “Localized Prediction of Glutamate from Whole-Brain Functional Connectivity of the Pregenual Anterior Cingulate Cortex” The Journal of Neuroscience, November 18, 2020, 40(47):9028–9042.
- L. Zhang, H. Cai, J. Pei, H. Li, L. Cao, W. Zhu, R. Dang and Y. Deng, “Simultaneous detection of multiple neurotransmitters and their metabolites in rat brain homogenates and microdialysates by LC-MS/MS”. Royal Society of Chemistry, Anal Methods 2020.
- Yangguang, A.M. Buchanan, C. E. Witt and P. Hashemi, “Frontiers in electrochemical sensors for neurotransmitter detection: towards measuring neurotransmitters as chemical diagnostics for brain disorders” Royal Society of Chemistry, Anal Methods 2019.11.2738.
- Nchouwat et al, “Simultaneous Detection and Quantification of Age-Dependent Dopamine Release”, preprint 2025. [CrossRef]
- S.K. Arumugasamy, G. Chellasamy, S. Gopi, S. Govindaraju and K. Yun, “Current advances in the detection of neurotransmitters by nanomaterials: An update,” Trends in Analytical Chemistry,2002.
- N.Desai, D. Rana, S. Salave, R. Gupta, P. Patel, B, Karunakaran,A. Sharma, G. Jyotsnendu, D. Benival and N. Kommineni, “Chitosan: A Potential Biopolymer in Drug Delivery and Biomedical Applications”, Pharmaceutics 2023, 15, 1313. [CrossRef]
- J. Ding and Y. Guo, “Recent Advances in Chitosan and its Derivatives in Cancer Treatment”, Frontiers in Pharmacology, Volume 13, Article 888740, April 2022.
- Aranaz, A. R. Alcántara, M. C. Civera, C. Arias, B. Elorza, A. H. Caballero, and N.Acosta,” Chitosan: An Overview of Its Properties and Applications, Polymers 2021, 13, 3256. [CrossRef]
- S. Naskar, S. Sharma, K. Ketousetuo “Chitosan-based nanoparticles: An overview of biomedical applications and its preparation” Journal of Drug Delivery Science and Technology 49 (2019) 66–81.
- Y. Jiang and W. Jayne, “Recent development in chitosan nanocomposites for surface-based biosensor applications” Electrophoresis 2019, 40, 2084–2097.
- X. Jin, X.Fengna , L. Deshui, Q. Wu and L.Xianfu, “The effect of the chitosan membrane properties on the enzyme adsorption and performance for the construction of horseradish peroxidase biosensors”, Carbohydrate Polymers 85 (2011) 786–791.
- D.L.Robinson, B.J. Venton, M.L. Heien and R.M. Wightman, “Detecting subsecond dopamine release with fast-scan cyclic voltammetry in vivo”, Clinical Chemistry. 49 (10):1763–73. October 2003. PMID 14500617. [CrossRef]
- M. Salimian, M. R. Sohrabi and S. Mortazavinik “Application of net analyte signal and principal component regression for rapid simultaneous determination of Levodopa and carbidopa in commercial pharmaceutical formulation and breast (human) milk sample using spectrophotometric method” Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy 283 (2022) 121741.
- Dowek, M. Berge, P. Prognon, L. François-Xavier, E. Larquet, A. Tfayli, L. M. M. Minh and E. Caudron “Discriminative and quantitative analysis of norepinephrine and epinephrine by surface-enhanced Raman spectroscopy with gold nanoparticle suspensions” Analytical and Bioanalytical Chemistry (2022) 414:1163–1176.
- S. Jafarinejad, A. Bigdeli, M. Ghazi-Khansari, P. Sasanpour and M. R. Hormozi-Nezhad, “Identification of Catecholamine Neurotransmitters Using a Fluorescent Electronic Tongue” ACS Chem. Neurosci. 2020, 11, 25−33.
- Kallabis, P. Beyerlein, F. Lisdat “Quantitative determination of dopamine in the presence of interfering substances supported by machine learning tools” Bioelectrochemistry 157 (2024) 108667.
- S. Jafarinejad, M. Ghazi-Khansari, FGhasemi, P.Sasanpour & M. R.Hormozi-Nezhad “Colorimetric Fingerprints of Gold Nanorods for Discriminating Catecholamine Neurotransmitters in Urine Samples” Science Report 15-8- 2017.
- K.S. Siamak, N. Ouassil, S.J. Yang, and M. P. Landry, “Identifying Neural Signatures of Dopamine Signaling with Machine Learning”, ACS Chem. Neurosci. 2023, 14, 2282−2293.
- M.H.M. Facure, G. Gahramanova, D.Zhang,T. Zhang,C. E. Shuck, L.A. Mercante, D. S. Correa, Y. Gogotsi, “All-MXene electronic tongue for neurotransmitters detection”, Biosensors and Bioelectronics 262 (2024) 116526.
- Yuki Komoto et al “Time-resolved neurotransmitter detection in mouse brain tissue using an artificial intelligence-nanogap” Scientific Reports (2020) 10:11244.
- J. Kim, Y. Oh, C. Park, Y. M. Kang, H. Shin, I. Y. Kim, and Jang, “Comparison study of partial least squares regression analysis and principal component analysis in fast-scan cyclic voltammetry,” International Journal of Electrochemical Science, vol. 14, no. 7, pp. 5924–5937, 2019.
- Hoseok, H. Shin, H. U. Cho, C.D. Blaha, M. L. Heien, O. Yoonbae , K. H. Lee, and D. P. Jang,”Neurochemical Concentration Prediction Using Deep Learning vs Principal Component Regression in Fast Scan Cyclic Voltammetry: A Comparison Study” ACS Chem. Neurosci. 2022, 13, 2288−2297.
- S. Kang, Y. Jeong and Ji-Woong Choi, “Simultaneous Estimation of Tonic Dopamine and Serotonin withHigh Temporal Resolution In Vitro Using Deep Learning” 45th Annual International Conference of the IEEE Engineering,2023.
- S.Rantataro,, I.Parkkinen, M. Airavaara, & T. Laurila,”Real-time selective detection of dopamine and serotonin at nanomolar concentration from complex in vitro systems”,Biosensors and Bioelectronics 241 (2023) 115579.
- Anna Marie Buchanan et al “Serotonin as a biomarker of toxin-induced Parkinsonism” Molecular Medicine (2024) 30:33.
- Simon Bellemare-Rousseau et al “FPGA-based Prediction System for Neurotransmitter Concentration Measurement from Spectrophotometry Data” IEEE INT CONF.2020.
- Doyun Kim et al “An Automated Cell Detection Method for TH-positive Dopaminergic Neurons in a Mouse Model of Parkinson’s Disease Using Convolutional Neural Networks” Experimental Neurobiology 2023.
- Jian Lv et al “Amperometric Identification of Single Exosomes and Their Dopamine Contents Secreted by Living Cells” Anal. Chem. 2023, 95, 11273−11279.
- Andrea Di Credico et al “Machine learning identifies phenotypic profile alterations of human dopaminergic neurons exposed to bisphenols and perfluoroalkyls” Science Report (2023) 13:21907.
- Arijit Pal, Souvik Biswas, Koel Chaudhury, and Soumen Das “Paper Sensor Modified with MoS2 for Detection of Dopamine Using a Machine-Intelligent Web App Interface” ACS Appl. Mater. Interfaces 2023, 15, 43060−43074.
- S. Kapur, G. Remington, “Serotonin-dopamine interaction and its relevance to schizophrenia,” Am J. Psychiatry, vol. 153, pp.466-76, 1996.
- V. Kammarchedu and A.Ebrahimi “Advancing Electrochemical Screening of Neurotransmitters Using a Customizable Machine Learning-Based Multimodal System” Chemical and biological sensors 2023.
- Bang, Yi Luo, L. S. Barbosa, K. T. Kishida, M. R. Witcher, and P. R. Montague“Noradrenaline tracks emotional modulation of attention in human amygdala”Current Biology 33, 1–8, NOV,20,2023.
- Sajeet et al “A Machine Learning Approach for Simultaneous Electrochemical Detection of Dopamine and Serotonin in an Optimized Carbon Thread-based Miniaturized Device” IEEE Sensor Journal Apr, 17, 2024.
- 55. Abhinav Goyal et al “Resolution of tonic concentrations of highly similar neurotransmitters using voltammetry and deep learning”, Molecular Psychiatry, 2024. [CrossRef]
- K. Unger, J.P. Keller, M. Altermatt, V. Gradinaru, L.L. Looger and L.Tian, “Directed Evolution of a Selective and Sensitive Serotonin Sensor via Machine Learning”, Cell 183, 1986–2002, December 23, 2020.
- C.S. Movassaghi, K. A. Perrotta, H.Yang, R.Iyer, X. Cheng, M. Dagher, M. A.Fillol and A. M. Andrews, “Simultaneous serotonin and dopamine monitoring across timescales by rapid pulse voltammetry with partial least squares regression”, Analytical and Bioanalytical Chemistry (2021) 413:6747–6767.
- Zhang, S. Shailja, C. Borba, Y. Miao, M.Goebel, R.Ruschel, K. Ryan, W. Smith, and B. S. Manjunath, “Automatic classification and neurotransmitter prediction of synapses in electron microscopy”, Biological Imaging (2022), 2: e6.
- G.H. G. Matsushita, C. da Cunha and A.Sugi, “Automatic Identification of Phasic Dopamine Release”, 25th International Conference on Systems, Signals and Image Processing (IWSSIP), 2018.
- G.H.G. Matsushita, A.H. Sugi, Y. M.G. Costa, A.Gomez-A,C.Da Cunha, and L. S. Oliveira, “Phasic dopamine release identification using convolutional neural network” Computers in Biology and Medicine 114 (2019) 103466.
- Y. Xue, J.Wenliang, Y. Jiang, y. Ping, and L.Mao, “Deep Learning for Voltammetric Sensing in a Living Animal Brain”, Angew. Chem. Int. Ed. 2021, 60, 23777 – 23783.
- Chun-Hung Chang, L. Chieh-Hsin and L. Hsien-Yuan “Machine Learning and Novel Biomarkers for the Diagnosis of Alzheimer’s Disease” MDPI, Int. J. Mol. Sci. 2021, 22, 2761.
- Stepen et al, “At home adaptive dual target deep brain stimulation in Parkinson’s disease with proportional control”, Brain, Volume 147, Issue 3, March 2024, Pages 911–922,.
- Satoka H. Fujimoto1 et al “Deep brain stimulation induces white matter remodeling and functional changes to brain-wide networks”.
- D.M Herz, M.J Frank, H. Tan and S. Groppa “Subthalamic control of impulsive actions: insights from 3 deep brain stimulation in Parkinson’s disease”, Brain, awae184. [CrossRef]
- Izabely de dos Reis Paula et al “Deep brain stimulation of the subthalamic nucleus under general anesthesia versus local anesthesia in the treatment of Parkinson’s disease: a meta-analysis of randomized clinical trials”, Neurosurgical Review (2024) 47:346.
- K.A. Johnson, M.S. Okun, K.W. Scangos, H.S. Mayberg and C. de Hemptinne, “Deep brain stimulation for refractory major depressive disorder: a comprehensive review”, Molecular Psychiatry (2024) 29:1075–1087. [CrossRef]
- Alik S. Widge, “Closing the loop in psychiatric deep brain stimulation: physiology, psychometrics, and plasticity”, Neuropsychopharmacology (2024) 49:138–149. [CrossRef]
- Nanditha Rajamani et al “Deep brain stimulation of symptom-specific networks in Parkinson’s disease”, Nature Communications | (2024) 15:4662.
- B. Davidson, L. Milosevic, L. Kondrataviciute, L. V. Kalia and S. K. Kalia,” Neuroscience fundamentals relevant to neuromodulation: Neurobiology of deep brain stimulation in Parkinson's disease”, Neurotherapeutics 21 (2024) e00348.
- Chudzik, A.Sledzianowski and A. W. Przybyszewski, “Machine Learning and Digital Biomarkers Can Detect Early Stages of Neurodegenerative Diseases” Sensors 2024, 24, 1572. [CrossRef]
- Nishanth Gopinath “Artificial intelligence and neuroscience: An update on fascinating relationships”, Process Biochemistry 125 (2023) 113–120.
- Z.Zhou , Tailin Xu and X. Zhang, “Empowerment of AI algorithms in biochemical sensors”, Trends in Analytical Chemistry 173 (2024) 117613.
- C.Rivera, J. J. Swerdlow, R. L. Summerscales,P. P. T. Uppala,R. M. Filho, M. R. C. Neto and H. J. Kwon, “Data-Driven Modeling of Smartphone-Based Electrochemiluminescence Sensor Data Using Artificial Intelligence”, Sensors 2020, 20, 625. [CrossRef]
- Y.Yang, F. Xu, J.Chen, C. Tao, Y. Li, Q. Chen, S. Tang, H. K. Lee and W. Shen, “Artificial intelligence-assisted smartphone-based sensing for bioanalytical applications: A review”, Biosensors and Bioelectronics 229 (2023) 115233.
- P.Puthongkham, S.Wirojsaengthonga and A.Suea-Ngamd, “Machine learning and chemometrics for electrochemical sensors: moving forward to the future of analytical chemistry”, Analyst, 2021, 146, 6351.
- B. Kitchenham and S. Charters, ‘‘Guidelines for performing systematic literature reviews in software engineering,’ School Comput. Sci. Math., Keele Univ., Keele, U.K., Tech. Rep. EBSE-2007-01, 2007.
- B. Kitchenham, ‘‘Procedures for performing systematic reviews,’ Dept. Comput. Sci., Keele Univ., Keele, U.K., Tech. Rep. 0400011T.1, 2004, pp. 1–26, vol. 33.
- R. M. Wightman, L.J. May and A.C. Michael,” Detection of dopamine dynamics in the brain” ANALYTICAL CHEMISTRY, VOL. 60, NO. 13, JULY 1, 1988 • 769 A.
- Y. Wang, Y.Li, L.Tang, J.Lu and J.Li,” Application of graphene-modified electrode for selective detection of dopamine”, Electrochemistry Communications 11 (2009) 889–892.
- M. Sajid, N. Baig, and K. Alhooshani,” Chemically modified electrodes for electrochemical detection of dopamine: Challenges and opportunities”, Trends in Analytical Chemistry 118 (2019) 368e385.
- P.A. Rasheed and Jae-Seung Lee, “Recent advances in optical detection of dopamine using nanomaterials”, Microchim Acta (2017) 184:1239–1266.
- Y.R.Kima, S.Bonga, Y.Kanga, Y.Yanga, R. K. Mahajanb, J.S. Kimc and H. Kima, “Electrochemical detection of dopamine in the presence of ascorbic acid using graphene modified electrodes”, Biosensors and Bioelectronics 25 (2010) 2366–2369.
- X.Liu, and J.Liu, “Biosensors and sensors for dopamine detection”, VIEW. 2021;2:20200102.
- J. Stewart, J. Hendry, and L. Dennany, “Whole Blood Electrochemiluminescent Detection of Dopamine”, Anal. Chem. 2015, 87, 11847−11853.
- L. Wu, L. Feng, J. Ren and X. Qu,” Electrochemical detection of dopamine using porphyrin-functionalized graphene”, Biosensors and Bioelectronics 34 (2012) 57–62.
- 87. X.Zhang,X.Chen, S. Kai, H. Wang, J. Yang, F.Wu and Z. Chen, “Highly Sensitive and Selective Detection of Dopamine Using One-Pot Synthesized Highly Photoluminescent Silicon Nanoparticles”, Anal. Chem. 2015, 87, 3360−3365.
- J.Zhao, L.Zhao, C. Lan, and S. Zhao, “Graphene quantum dots as effective probes for label-free fluorescence detection of dopamine”, Sensors and Actuators B 223 (2016) 246–251.
- Jaekyung Kim et al “Automatic and Reliable Quantification of Tonic Dopamine Concentrations In Vivo Using a Novel Probabilistic Inference Method”, ACS Omega 2021, 6, 6607−6613.
- .
- K. E. Dunham1 and B. J. Venton, “Electrochemical and biosensor techniques to monitor neurotransmitter changes with depression”, Analytical and Bioanalytical Chemistry (2024) 416:2301–2318.
- K. J. Ressler and C.B. Nemeroff, “The Role of Serotonergic and Noradrenergic Systems in Depression and Anxiety Disorder,” Depr. Anxi., vol. 12 (Suppl 1), pp.2-19, 2000.
- Niyonambaza S.D., Kumar P., Xing P., Mathault J., De Koninck P., Boisselier E., Boukadoum M., Miled A. “A Review of Neurotransmitters Sensing Methods for Neuro-Engineering Research” Appl. Sci. 2019; 9:4719.
- Linan-Rico A., Ochoa-Cortes F., Beyder A., Soghomonyan S., Zuleta-Alarcon A., Coppola V., Christofi F.L. Mechanosensory Signaling in Enterochromaffin Cells and 5-HT Release: Potential Implications for Gut Inflammation. Front. Neurosci. 2016;10:564. [CrossRef]
- Camilleri M. Serotonin in the gastrointestinal tract. Curr. Opin. Endocrinol. Diabetes Obes. 2009;16:53–59. [CrossRef]
- Reigstad C.S., Salmonson C.E., Rainey J.F., III, Szurszewski J.H., Linden D.R., Sonnenburg J.L., Farrugia G., Kashyap P.C. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J. 2015;29:1395–1403. [CrossRef]
- Murley A.G., Rowe J.B. Neurotransmitter deficits from frontotemporal lobar degeneration. Brain. 2018;141:1263–1285. [CrossRef]
- Iovino L., Tremblay M.E., Civiero L. Glutamate-induced excitotoxicity in Parkinson’s disease: The role of glial cells. J. Pharmacol. Sci. 2020;144:151–164. [CrossRef]
- Le Gall L., Anakor E., Connolly O., Vijayakumar U.G., Duddy W.J., Duguez S. Molecular and Cellular Mechanisms Affected in ALS. J. Pers. Med. 2020;10:101. [CrossRef]
- Strandwitz P. Neurotransmitter modulation by the gut microbiota. Brain Res. 2018;1693:128–133. [CrossRef]
- Koshal P., Jamwal S., Kumar P. Glucagon-like Peptide-1 (GLP-1) and neurotransmitters signaling in epilepsy: An insight review. Neuropharmacology. 2018;136:271–279. [CrossRef]
- W. Song, L. S. Seung, M. Savini, L. Popp, L. C.Vicki and L. Segatori “ Ceria Nanoparticles Stabilized by Organic Surface Coatings Activate the Lysosome-Autophagy System and Enhance Autophagic Clearance” American Chemical Society,2014.
- S. Boonkaew, A. Dettlaff, M.Sobaszek, R. Bogdanowicz, M. Jönsson-Niedziółka,” Electrochemical determination of neurotransmitter serotonin using boron/nitrogen co-doped diamond-graphene nanowall-structured particles”, Journal of Electroanalytical Chemistry 926 (2022) 116938.
- N. Alyamni, J. L. Abot and A. G. Zestos, “Advances in Voltammetric Methods for the Measurement of Biomolecules” ECS Sens. Plus 3 027001: 2024. [CrossRef]
- S.Bindra and· R. Jain,” Artificial intelligence in medical science: a review”, Irish Journal of Medical Science (1971 -) (2024) 193:1419–1429.
- H.A. Younis, , T. A. E.Eisa,, M.Nasser,T.M.Sahib,A.A. Noor,O. M. Alyasiri, S. Salisu, I.M. Hayder, and H. A. Younis,” A Systematic Review and Meta-Analysis of Artificial Intelligence Tools in Medicine and Healthcare: Applications, Considerations, Limitations, Motivation and Challenges”, Diagnostics 2024, 14, 109. [CrossRef]
- Grzybowski, K. Jin and W. Hongkang Wu, “Challenges of artificial intelligence in medicine and dermatology” Clinics in Dermatology (2024) 42, 210–215.
- Morris Gordon et al “A scoping review of artificial intelligence in medical education: BEME Guide No. 84”, MEDICAL TEACHER 2024, VOL. 46, NO. 4, 446–470.
- 2021; 109. Bo Yin,, Hong Lin Zhai, Bing Qiang Zhao, Ke Xin Bi, Jia Ying Mi, “Chemometrics-assisted simultaneous voltammetric determination of multiple neurotransmitters in human serum”, Bioelectrochemistry 139 (2021) 107739.
- T. W. Bernklau, B. Righetti, L. S. Mehrke, and S. N. Jacob,” Striatal dopamine signals reflect perceived cue–action–outcome associations in mice”, Nature Neuroscience, 10 Jan, 2024. [CrossRef]
- Nchouwat et al “Pattern Recognition ofNeurotransmitters: Complexity Reduction for Serotonin and Dopamine”,Biosensors 2025, 15(4), 209. [CrossRef]
- Im-Fong Ip, Yi-Shan Wang, and Chia-Chen Chang, “Aptamer-based detection of serotonin based on the rapid in situ synthesis of colorimetric gold nanoparticles,” Nanotechnology Reviews 2023; 12: 20220514.
- M. Dagher, Ka.A. Perrotta, S. A. Erwin, A. Hachisuka, R. Iyer, S.C. Masmanidis, H.Yang, and A. M. Andrews,” Optogenetic Stimulation of Midbrain Dopamine Neurons Produces Striatal Serotonin Release,” ACS Chem. Neurosci. 2022, 13, 946−958.
- P.Lachance, D. Gauvreau, É. Boisselier, M.Boukadoum and A. Miled, “Breaking Barriers: Exploring Neurotransmitters through In Vivo vs. In Vitro Rivalry”, Sensors 2024, 24, 647. [CrossRef]
- Tan Kian Hua, “ A Short Review on Machine Learning” Authorea,Oct 04,2022.
- .
- J. Apon, Md. S. Abid, K. A. Morshed, M. M. Nishat, F. Faisal &N.N. I. Moubarak,” Power System Harmonics Estimation using Hybrid Archimedes Optimization Algorithm-based Least Square method,” 13th ICTS, Oct 2021.
- M.R Kabir, M.M Muhaimin, M.A Mahir, M.M Nishat, F. Faisal &N.N.I. Moubarak,” Procuring MFCCs from Crema-D Dataset for Sentiment Analysis using Deep Learning Models with Hyperparameter Tuning,” IEEE, RAAICON, Dec 2021.
- A.A Rahman, M.I Siraji, L.I Khalid, F. Faisal, M.M Nishat, M.R Islam,N.N.I Moubarak, “Detection of Mental State from EEG Signal Data: An Investigation with Machine Learning Classifiers,” 14th Int, Conf, KST, Jan 2022.
- N.N.I Moubarak, N.M.M Omar, VN Youssef, “Smartphone-sensor-based human activities classification for forensics: a machine learning approach” Journal of Electrical Systems and Information Technology 11 (1), 33.
- S. Battaglia, C. Nazzi and J. F. Thayer, “Genetic differences associated with dopamine and serotonin release mediate fear-induced bradycardia in the human brain” Translational Psychiatry (2024) 14:24. [CrossRef]
- F. S. Manciu, M. Manciu, J.D. Ciubuc, E.M. Sundin,K. Ochoa,M. Eastman, W. G. Durrer, J.Guerrero, B.Lopez, M. Subedi and K.E. Bennet,” Simultaneous Detection of Dopamine and Serotonin—A Comparative Experimental and Theoretical Study of Neurotransmitter Interactions” Biosensors 2019, 9, 3. [CrossRef]
- Jahangiri, H. A. Rakha, “Applying machine learning techniques to transportation mode recognition using mobile phone sensor data,” IEEE transactions on intelligent transportation systems ,2406/2417, 16 -May,2015.
- https://www.parkinson.org/understanding-parkinsons/statistics?utm_source=google&utm_medium=adgrant&utm_campaign=Info&utm_term=parkinson%27s%20statistics&gad_source=1&gclid=Cj0KCQjw2ou2BhCCARIsANAwM2HRDW60Eer6uiH8VlfM4apLoohXrzhLcsfi_BqRmUOzK-De0RVNjOUaAgVEEALw_wcB.
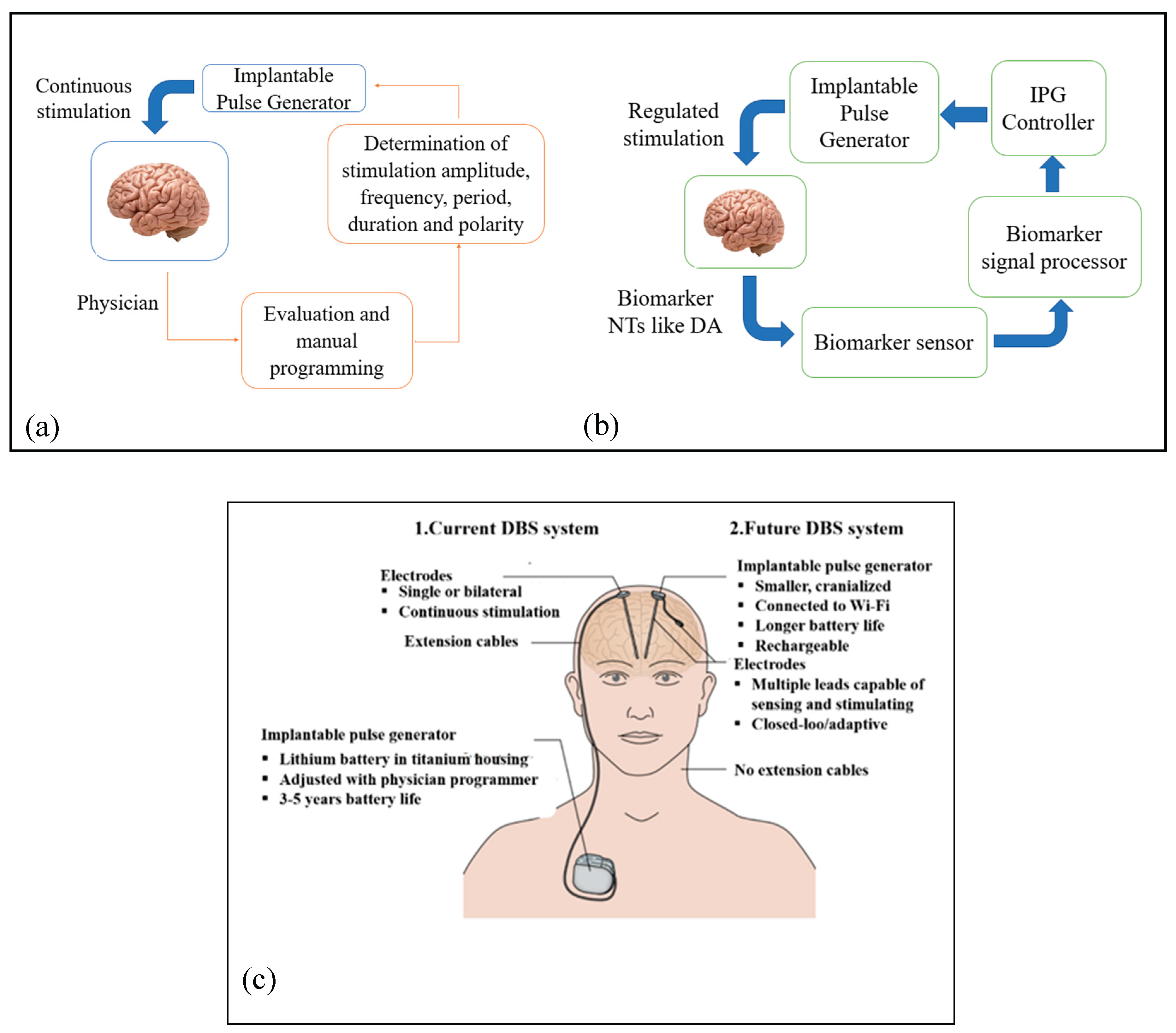
- M. Parastarfeizabadi and A. Z. Kouzani,”Advances in closed-loop deep brain stimulation devices”,Journal of NeuroEngineering and Rehabilitation (2017) 14:79.
- W. Bouthour , P. Mégevand , J.Donoghue, C.Lüscher,and N. Birbaumer and P. Krack.”Biomarkers for closed-loop deep brain stimulation in Parkinson disease and beyond”Nature Review Neurology. 2019 Jun;15(6):343-352. [CrossRef]
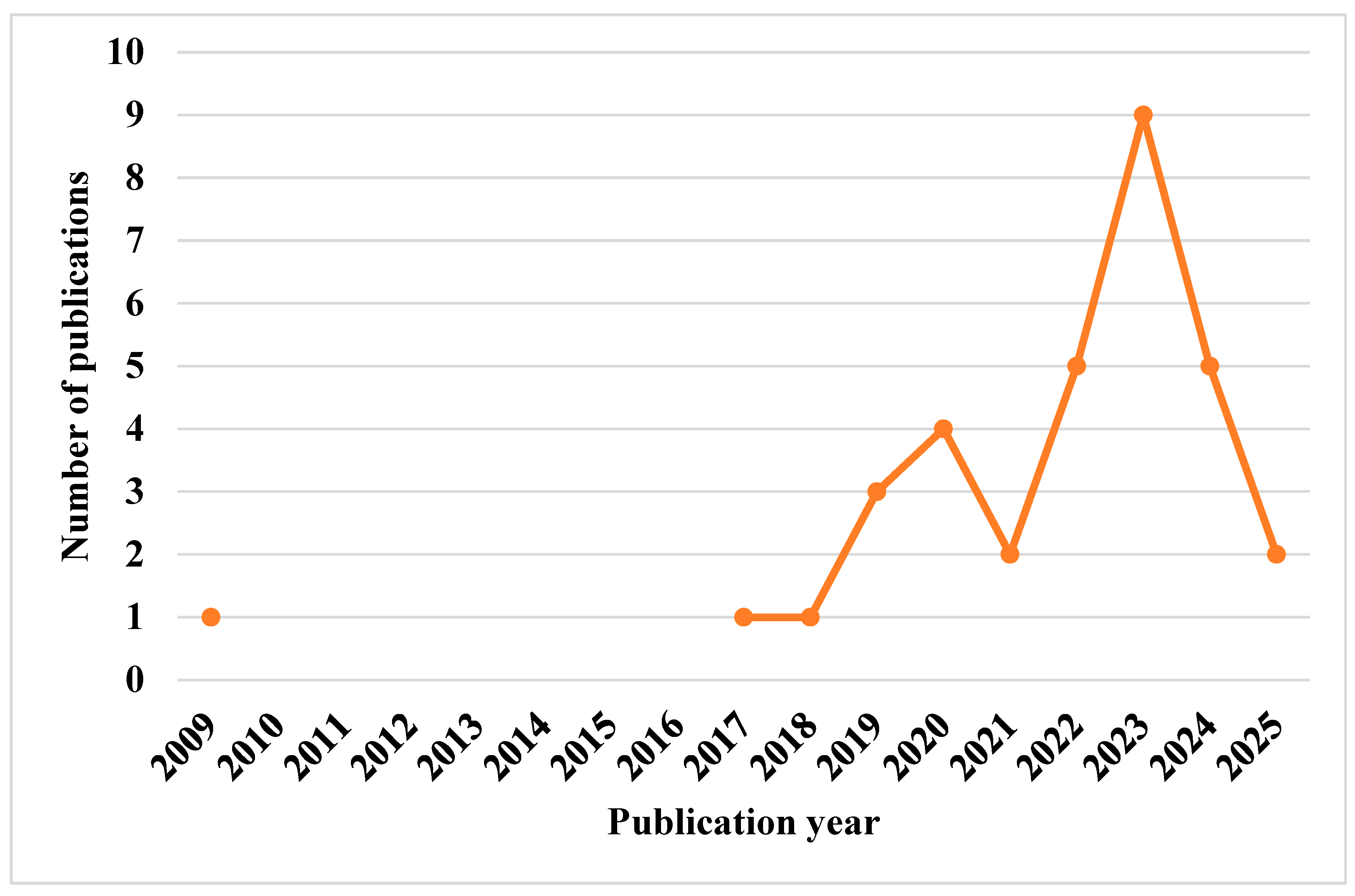
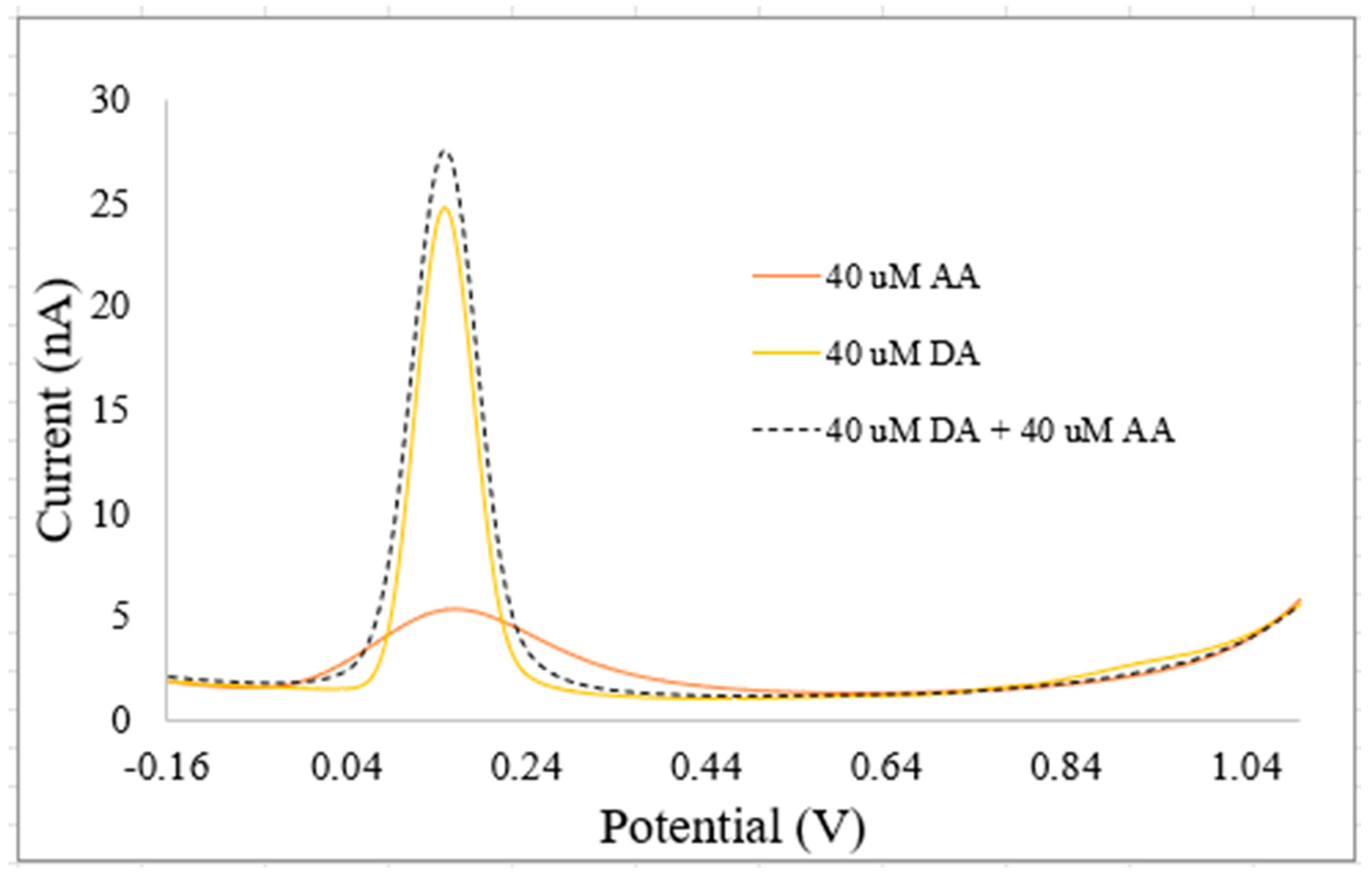
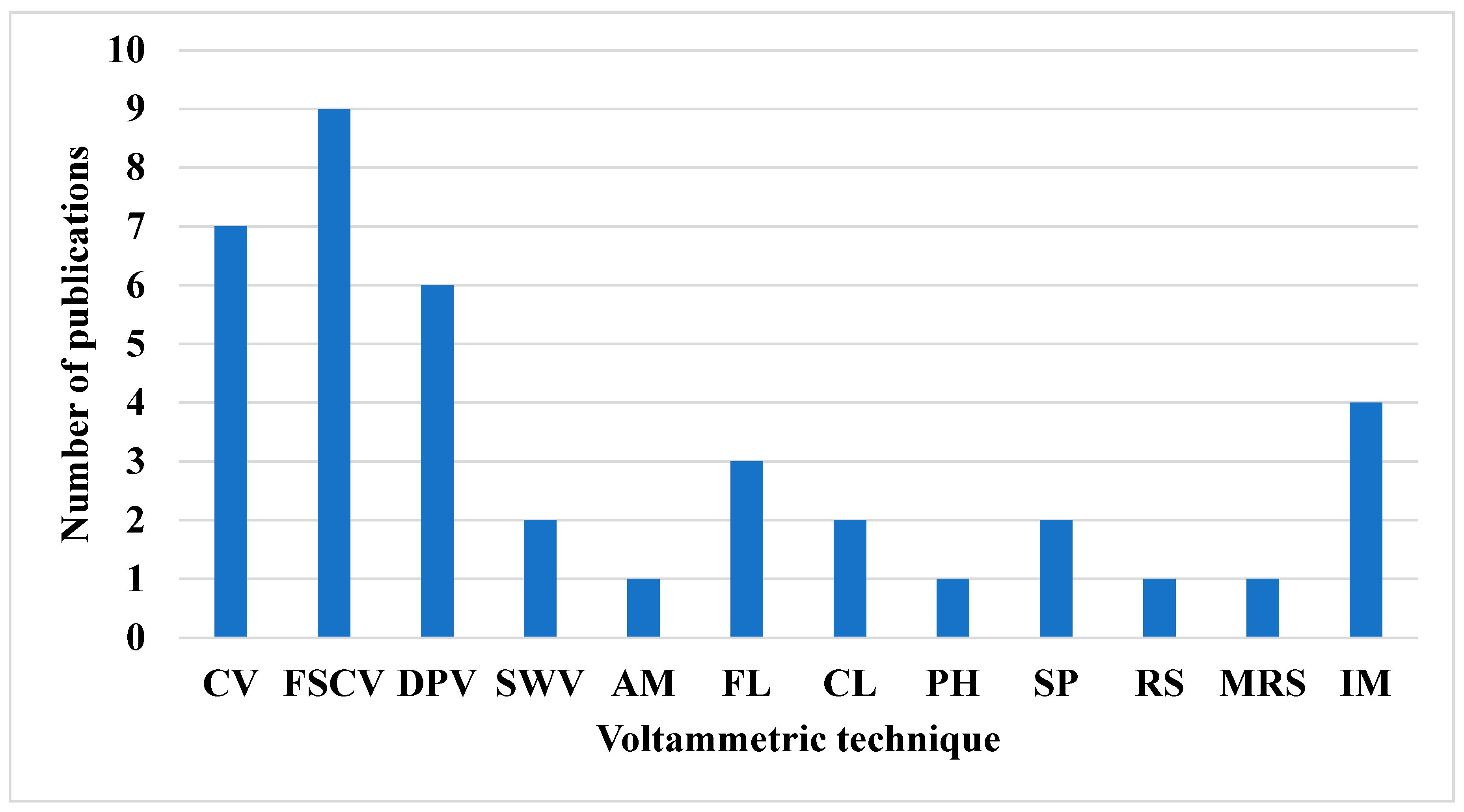
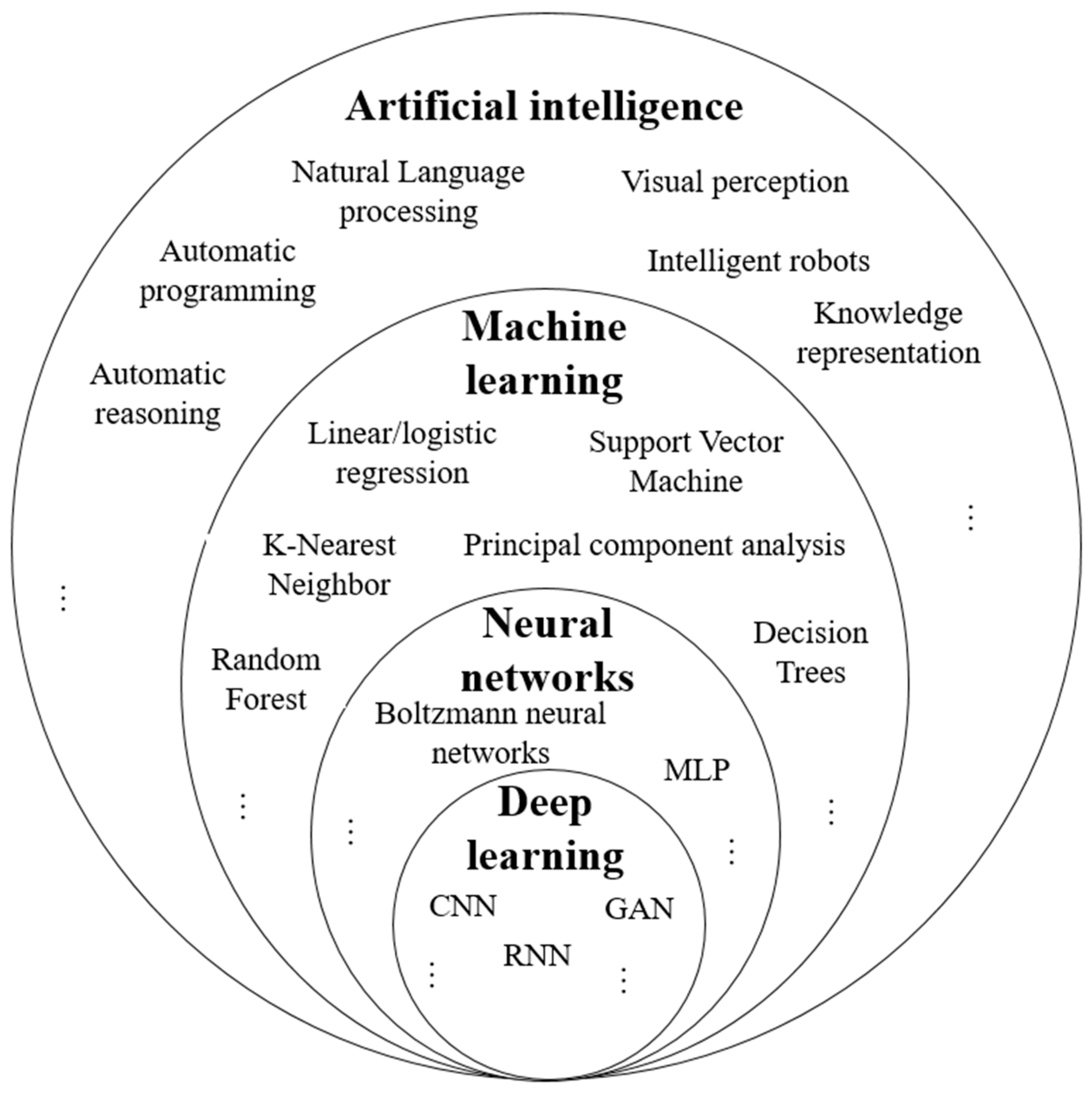
| Paper | Summary | Scope | |||
|---|---|---|---|---|---|
| NT Analysis | Voltammetric sensing techniques | Simultaneous NT detection | Application of AI | ||
| Bo Si et Al [4] | Summarized recent NT sensing techniques and explored prospects for their simultaneous detection | ✓ | |||
| Yi Su et al [6] | Analyzed current in-vivo NT detection techniques, focusing on real-time brain disorder diagnosis | ✓ | |||
| Saikat et al [18] | Surveyed electrochemical techniques for real-time, high-temporal-resolution NT sensing. | ✓ | ✓ | ✓ | |
| Shimwe et al [19] | Reviewed advances in NT detection, covering in-vivo sampling, imaging, electrochemical, nano-sensing, spectrometric, and analytical methods for both in vitro and in vivo use | ✓ | ✓ | ✓ | |
| Yangguang et al [23] | Highlighted key technical advances and in-vivo NT studies that may aid brain disorder diagnostics | ✓ | ✓ | ||
| Shiva et al [25] | Examined how NTs interact with nanomaterials, their role in diagnostics, detection techniques, and prospects for simultaneous detection and use in biological samples | ✓ | ✓ | ||
| Pathath et al [82] | Summarized recent nanomaterial-based optical methods for dopamine detection, covering its clinical relevance and advances in spectroscopic techniques. | ✓ | |||
| Xixian et al [84] | Discussed dopamine detection via molecular recognition methods and recent advances using nanomaterials and molecularly imprinted polymers | ✓ | |||
| Dunham et al [91] | Reviewed electrochemical techniques for NT detection and studied depression mechanisms. | ✓ | ✓ | ||
| This review | Reviews the use of AI for the automated detection and quantification of multiple NTs | ✓ | ✓ | ✓ | ✓ |
| Detection technique | Summary | Reference |
|---|---|---|
| CV | Does redox processes by sweeping the potential of a working electrode linearly back and forth while measuring the resulting current | [40,44,50,52,54,55,56,57], |
| FSCV | It operates by applying a rapidly varying voltage to a microelectrode, enabling real-time monitoring of NT dynamics | [41,42,43,45,53,59,60,61] |
| DPV | Enhances electrode sensitivity by superimposing small voltage pulses onto a linearly increasing potential and measuring the current just before and after each pulse | [1,36,50,52,54] |
| SWV | Applies a symmetrical square wave potential on top of a staircase potential and measures the difference in current at the end of each forward and reverse pulse. | [9,52] |
| AM | Applies a constant potential to an electrode and measuring the resulting current proportional to the concentration of an electroactive species over time. | [48] |
| FL | Measures the intensity of fluorescent light emitted by a substance after it has absorbed light or other electromagnetic radiation | [2,35,56] |
| CL | Measures the concentration of a substance by detecting the intensity of its color, typically using a colorimeter to quantify light absorption at a specific wavelength | [3,37] |
| PH | Measures the intensity of light, typically in the visible spectrum, and is used to determine the concentration of substances based on the amount of light absorbed or transmitted through a sample | [51] |
| SP | Measures how much light a substance absorbs across a specific range of wavelengths, typically using a spectrophotometer. | [33,46] |
| RS | Uses the scattering of monochromatic light, usually from a laser, to study vibrational, rotational, and other low-frequency modes in molecules | [34] |
| MRS | Measures the magnetic properties of atomic nuclei to provide information about the chemical environment and structure of molecules in a sample | [21] |
| IM | Captures visual representations of NT signaling using specialized techniques like microscopy, MRI, CT scans, or fluorescence. | [38,47,49,58] |
| Method | Type | Description | Pros | Cons | Examples |
|---|---|---|---|---|---|
| Statistical tests | Filter | Selects features based on statistical scores | Fast and model independent | Ignore the interactions between features and is univariate | Chi-square, ANOVA, Mutual information |
| Recursive Feature Elimination (RFE) | Wrapper | Recursively removes the least important features based on model performance | Considers feature interactions and is model specific | Computationally expensive and prone to overfitting | RFE with SVM or RF |
| Forward/Backward Selection | Wrapper | Iteratively adds/removes features based on model score | Considers feature combinations |
Slow with many features, model-specific | Stepwise Regression |
| Lasso Regression | Embedded | Uses regularization to shrink less important features’ coefficients to zero | Integrated with model and handles multicollinearity | Only linear relationships with biased coefficients | L1 regularization |
| Tree-based Feature importance | embedded | Uses importance-scores from models like decision trees | Captures non*linear relationships and is fast with trees | Model specific and sometimes less interpretable | Random Forest, XGBoost |
| Filter+ Wrapper or Embedded | Hybrid | Combines speed of filter with accuracy of wrapper/embedded methods | Balanced trade-off between performance and efficiency | More complex implementation and tuning of parameters is required | SelectKBest + RFE |
| Method | Type | Description | Pros | Cons | Use Cases | References |
|---|---|---|---|---|---|---|
| Principal Component Analysis (PCA) | Linear | Projects data into directions of maximum variance | Unsupervised, fast and reduces redundancy | Assumes linearity, hard to interpret components | Preprocessing image compression | [1,21,24,33,37,42,46,48,57] |
| Linear Discriminant Analysis (LDA) | Linear | Finds axes that maximize class separation | Supervised, good for classification | Only works with labeled data, assumes normality | Face recognition, pattern classification | [2,3,35,37,49] |
| t-distributed Stochastic Neighbor Embedding (t-SNE) | Non-linear | Preserves local structure for visualization | Captures complex patterns and great for 2D/3D plots | Slow, non-deterministic, not suitable for downstream ML models | High-dimensional data visualization | [35] |
| Uniform Manifold Approximation and Projection (UMAP) | Non-linear | Similar to t-SNE but faster and preserves more global structures | Fast, scalable, preserves global and local structures | Complex tuning, not always interpretable | Bioinformatics, NLP embeddings | [56] |
| Isomap | Manifold learning | Preserves geodesic distances on a manifold | Captures non-linear structures | Sensitive to noise, slow on large datasets | 3D shape analysis and visualization | N/A |
| Neural Autoencoder | Autoencoder | Learns low-dimensional representations via neural networks | Learns non-linear features and is customizable | Requires more data and tuning, it is a black-box so less interpretable | Image denoising, anomaly detection | [9,35,43] |
| Truncated SVD | Matrix Factorization | Factorizes a matrix into low-rank approximations | Efficient, interpretable, works with sparse data | Assumes linearity, less powerful for non-linear data | Text data recommender systems | [43] |
| Technique | Description | Use case | Pros | Cons | References |
|---|---|---|---|---|---|
| Hold-Out Validation | Splits dataset into training and test sets, usually in a ratio 80:20 | Quick checks large datasets | Simple and fast | Performance depends on split and high variance | [24,52,55,58] |
| K-Fold Cross-Validation | Splits data into K parts, trains on k-1, tests on 1 and repeats k times | General-purpose model evaluation | Reduces variance, uses data efficiently | Computationally expensive | [1,2,9,24,33,34,35,40,46,47,49,53] |
| Stratified K-Fold Cross-Validation | Same as K-Fold but preserves class ratios | Classification with imbalanced classes | Fairer evaluation in class imbalance | More complex than regular K-Fold | [24,111] |
| Leave-One-Out Cross-Validation | Special case of K-Fold where k=n (n is the number of samples) | Adapted for small datasets | Very low bias | Very high variance and high computation cost. | [38,52] |
| Nested Cross-Validation | Inner loop for model tuning, outer loop for evaluation | Hyperparameter tuning with fair evaluation. | Avoid overfitting during tuning. | Very computationally expensive. | [56] |
| Bootstrap | Sample data with replacement, evaluate across many resamples. | Adapted for small datasets, estimating confidence intervals. | Good variance estimation. | Biased estimates, complex interpretation. | N/A |
| Grid Search Cross-Validation | Exhaustively tries combinations of hyperparameters with cross validation. | Hyperparameter tuning. | Systematic and thorough. | Computationally intensive and doesn't scale well. | [24,59,60] |
| Random Search Cross-Validation | Randomly samples hyperparameter combinations. | Tuning with large search spaces. | More efficient than grid search. | May miss optimal values. | [38] |
| Bayesian Optimization | Probabilistically selects promising hyperparameter values. | Advanced hyperparameter tuning. | More efficient and informed than grid/random search. | More complex implementation. | [24,111] |
| Automated ML (AutoML) | Uses meta-learning or optimization to automate model selection. | Users with limited ML expertise. | Handles selection, tuning, and ensemble techniques. | Less control; can be a black box. | [9,35,43] |
| Metric | Formula | Focus | Range | Applied when | Sensitive to | References |
|---|---|---|---|---|---|---|
| Accuracy |
|
Overall correct predictions | [0,1] | Classes are balanced and errors are equally weighted | Class imbalance | [1,2,3,9,33,35,38,48,49,50,58,59,60] |
| Precision | Correctness of positive predictions | [0,1] | False positives weigh more | False positives | [46] | |
| Recall | Finds actual positives | [0,1] | False negative weigh more | False negatives | [47] | |
| F1-Score |
|
Balance between precision and recall | [0,1] | Classes are uneven with imbalanced datasets | Both FP and FN | [40,45,47,59,60] |
| Misclassification error | 1-Accuracy |
Overall error | [0,1] | An idea of the rate of error is needed | Class imbalance | N/A |
| Metric | Name | Formula | Focus | Range | Sensitiveness to outliers | Interpretation | References |
|---|---|---|---|---|---|---|---|
| MSE | Mean Squared Error | Penalize large errors more heavily | [0, ∞] | Yes | Penalizes large errors with less intuitive units as error is given as the square of the units of targets | [24,33,54] | |
| RMSE | Root Mean Square Error | Penalize large errors more heavily | [0, ∞] | Yes | Penalizes large errors more heavily with more intuitive units as it gives error in the same units as the targets | [34,42,52,54,55] | |
| MAE | Mean Absolute Error | Robust and easy-to-understand average error | [0, ∞] | No | Less interpretable but error given with the same units as the targets | [36,54] | |
| R2 | Coefficient of Determination | Explains variance of the data | [-∞,1] | Can be | Does not penalize large errors and explains how will models fits predictions | [1,21,24,33,54] |
| Study | ML Algorithm | NTs | Conc. range | Sensing Technique | Dataset measurements | Max. Acc.% | Type of study |
|---|---|---|---|---|---|---|---|
| Sazanova et al [1] | PCR, PLSR | DA, SE | 0-100 (uM) |
DPV |
216 Cross validation |
100 (with extended true values) | Simultaneous detection and quantification |
| Jose et al [9] | TinyML (DL) | AA, UA, DA, AA/DA, UA/DA, AA/UA/ DA | 0-500 (uM) |
SWV |
5492 (augmented) 80:20 split |
98.1 | Detection |
| Hoseok et al [42] | DL, PCR | DA, SE, EP, NE | 0-700 (nM) |
FSCV |
36 000 (augmented) 50:50 split |
96.23 | Simultaneous detection and quantification |
| Nchouwat et al [111] | PCR, PLSR | DA, SE | 0-100 (uM) | DPV | 216 Cross validation |
98 (with true values) | Simultaneous detection and quantification |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





