Submitted:
21 September 2025
Posted:
22 September 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
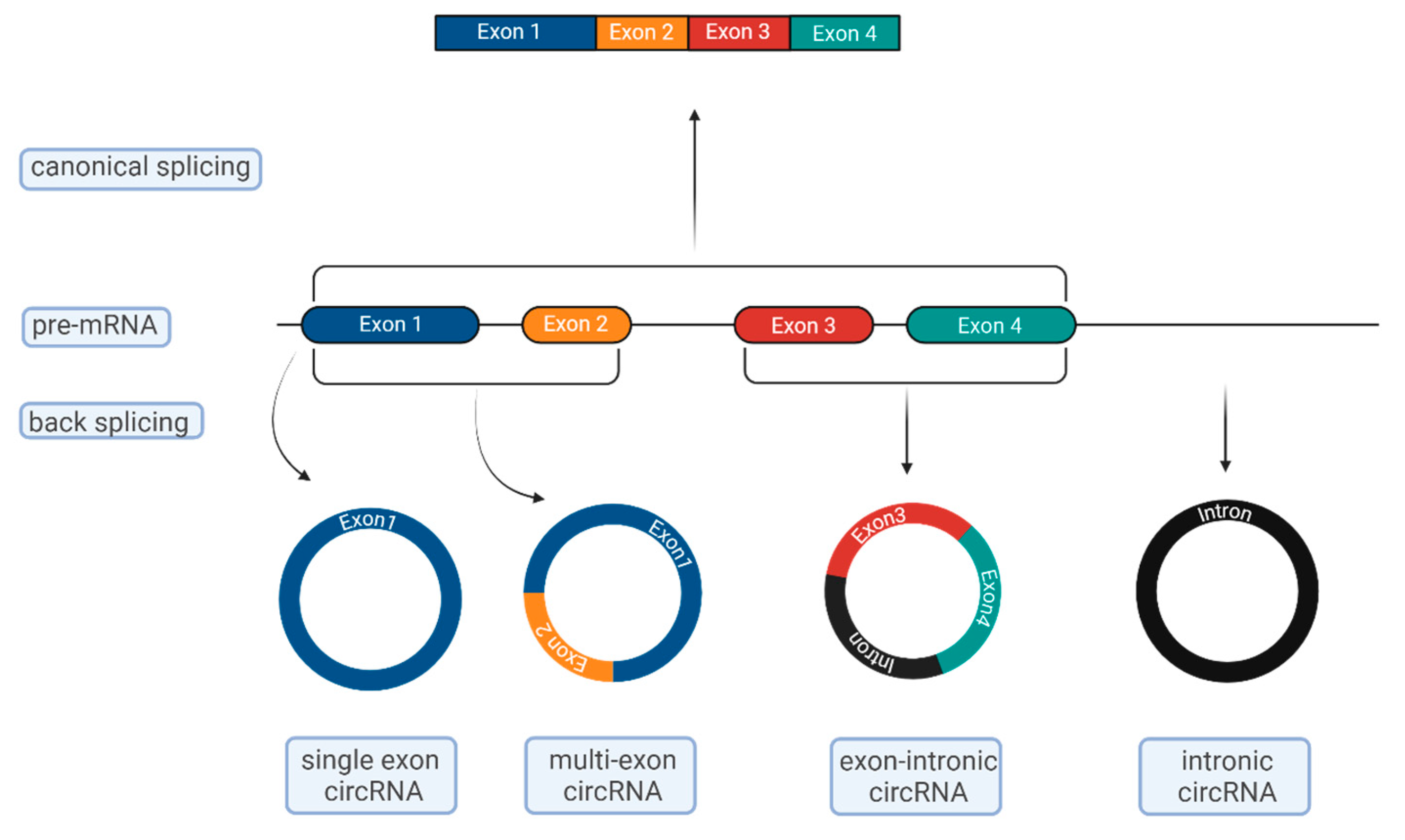
2. Biogenesis of CircRNAs
3. Mode of Action of CircRNAs
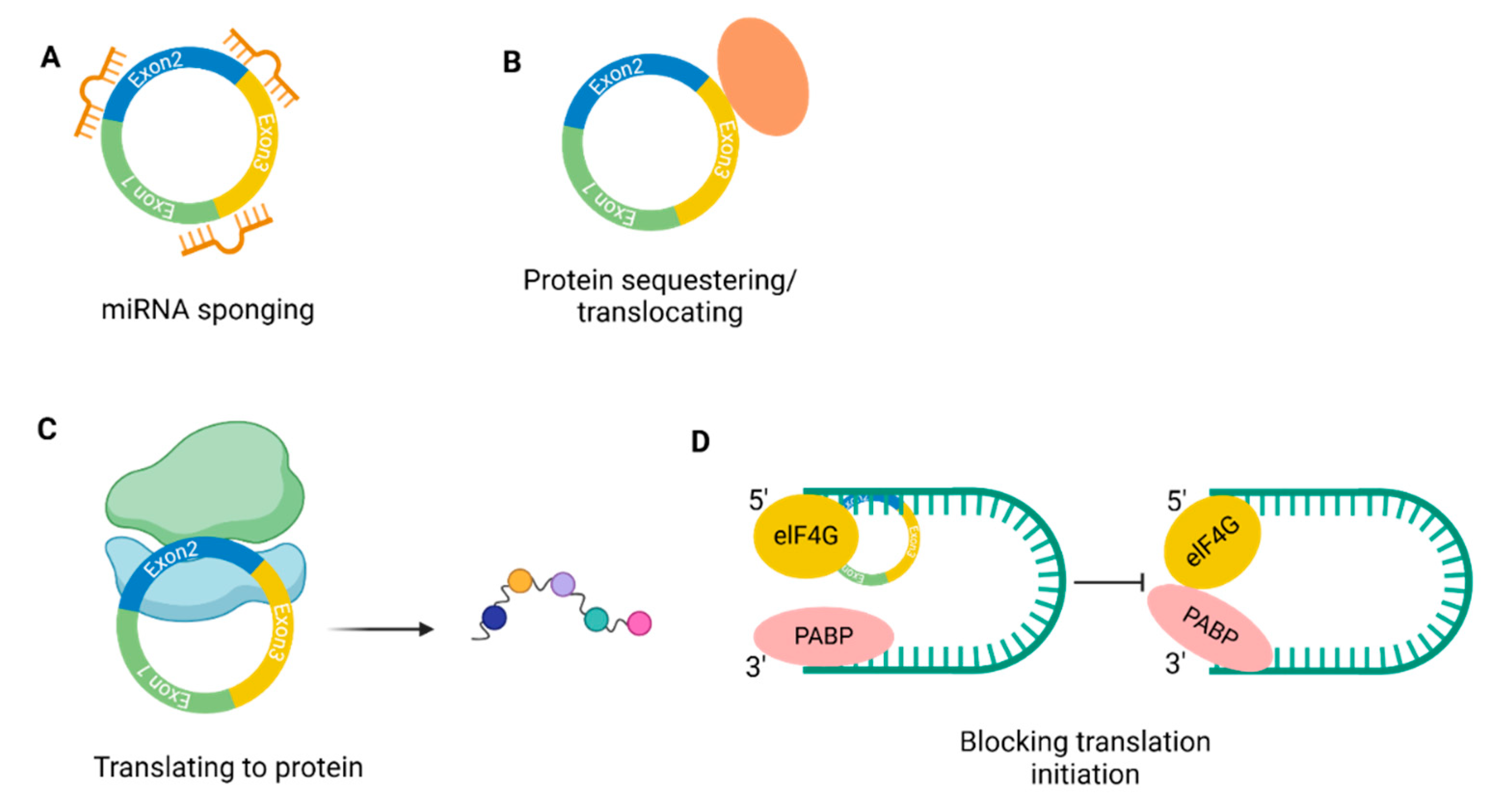
4. Regulatory Networks and the Control of Regulatory Networks
4.1. Regulation of Cell Proliferation by CircRNAs
4.2. Regulation of Epithelial-Mesenchymal Transition(EMT) and Cancer Progression by CircRNAs
4.3. Regulation of Pluripotency and Early Lineage Differentiation by CircRNAs
5. Endometrium – Cytology, Histology, Signaling Pathways
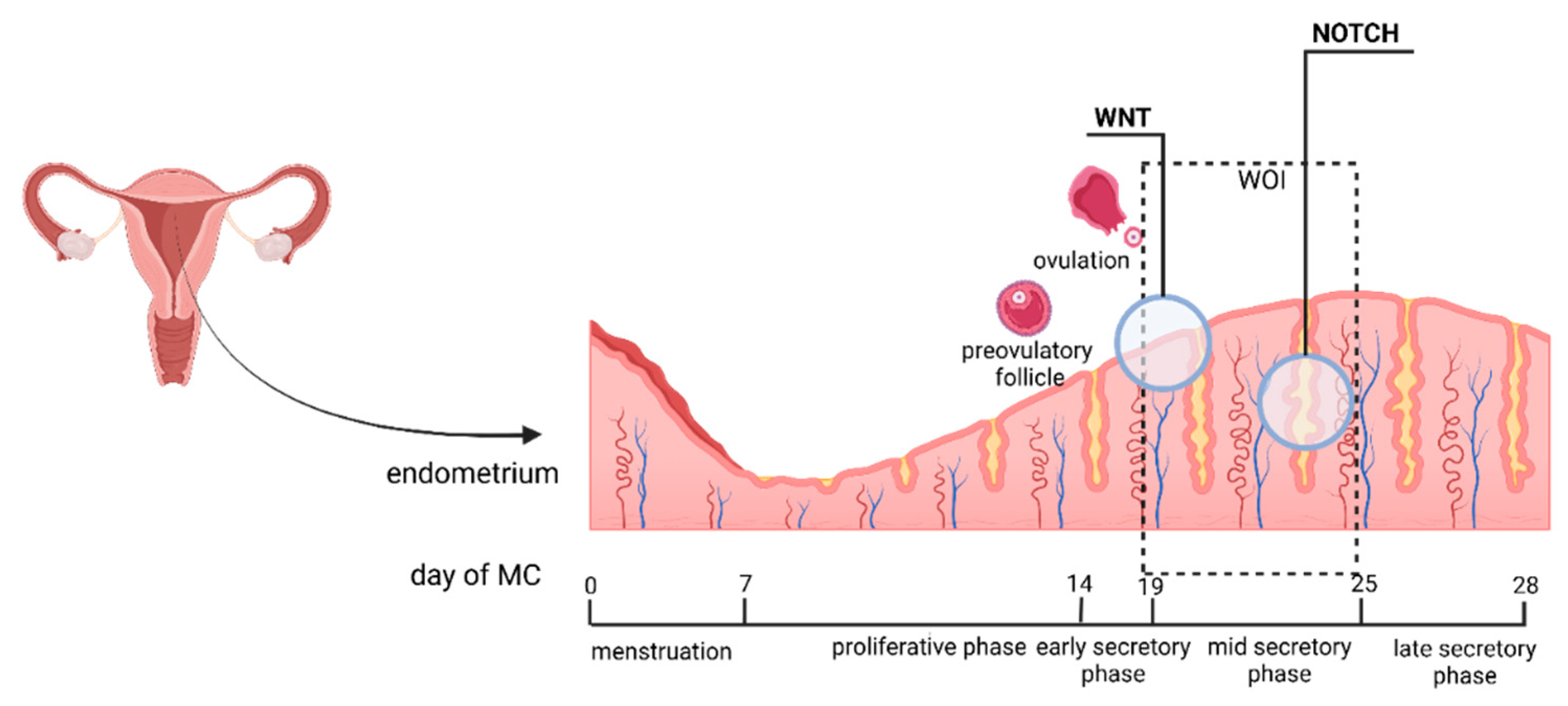
5.1. Endometrium and Endometrial Receptivity
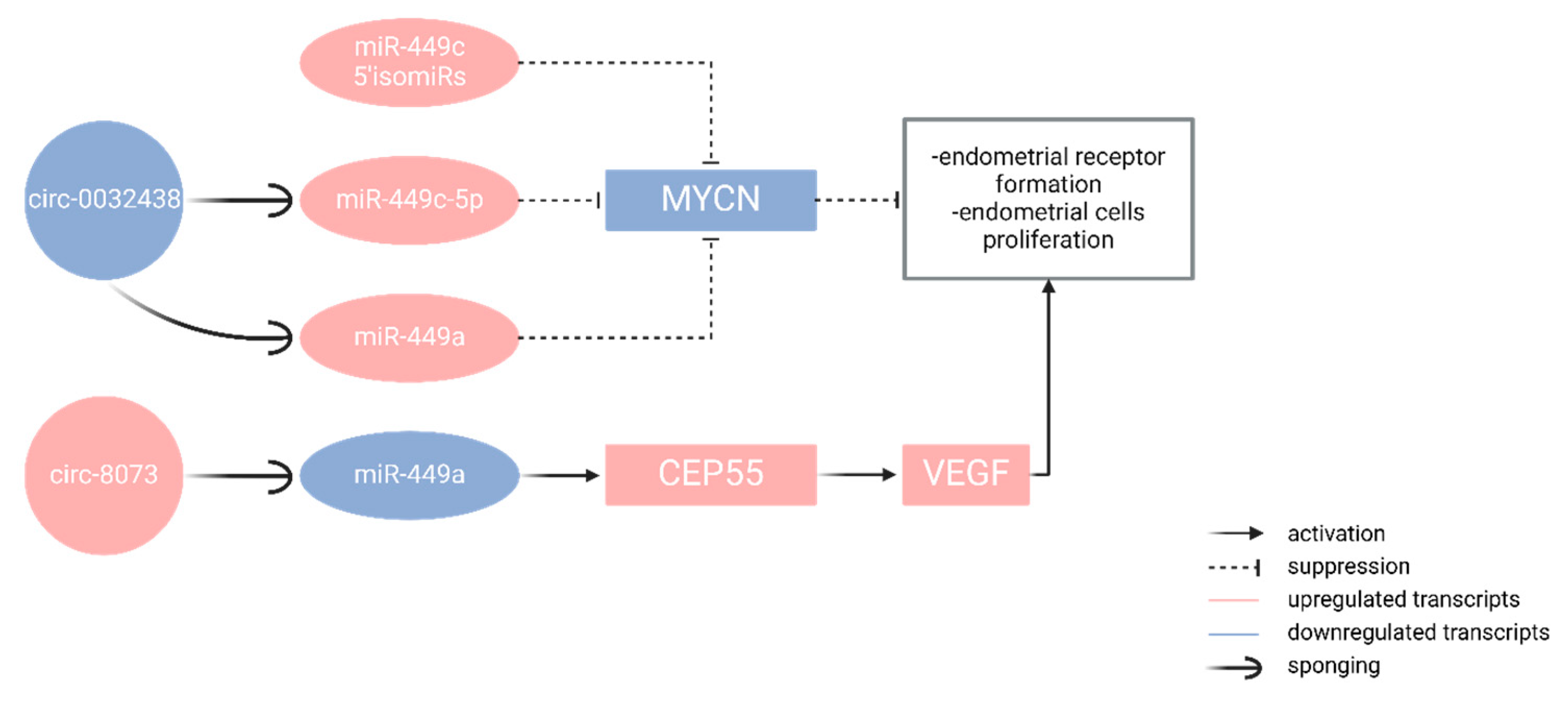
5.2. Repeated Implantation Failure and CircRNAs
| KEGG Pathway | HitGenes |
|---|---|
| HIF-1 signaling pathway | HIF1A, IGF1R, PIK3R1,MAP2K1,VEGFA |
| Focal adhesion | CDC42,IGF1R, PIK3R1,MAP2K1,VEGFA |
| VEGF signaling pathway | CDC42,PIK3R1,MAP2K1,VEGFA |
| Autophagy - animal | HIF1A, IGF1R, PIK3R1,MAP2K1 |
| PI3K-Akt signaling pathway | IGF1R, PIK3R1,MAP2K1,VEGFA |
| mTOR signaling pathway | IGF1R, PIK3R1,MAP2K1 |
6. Endometrial Cancer
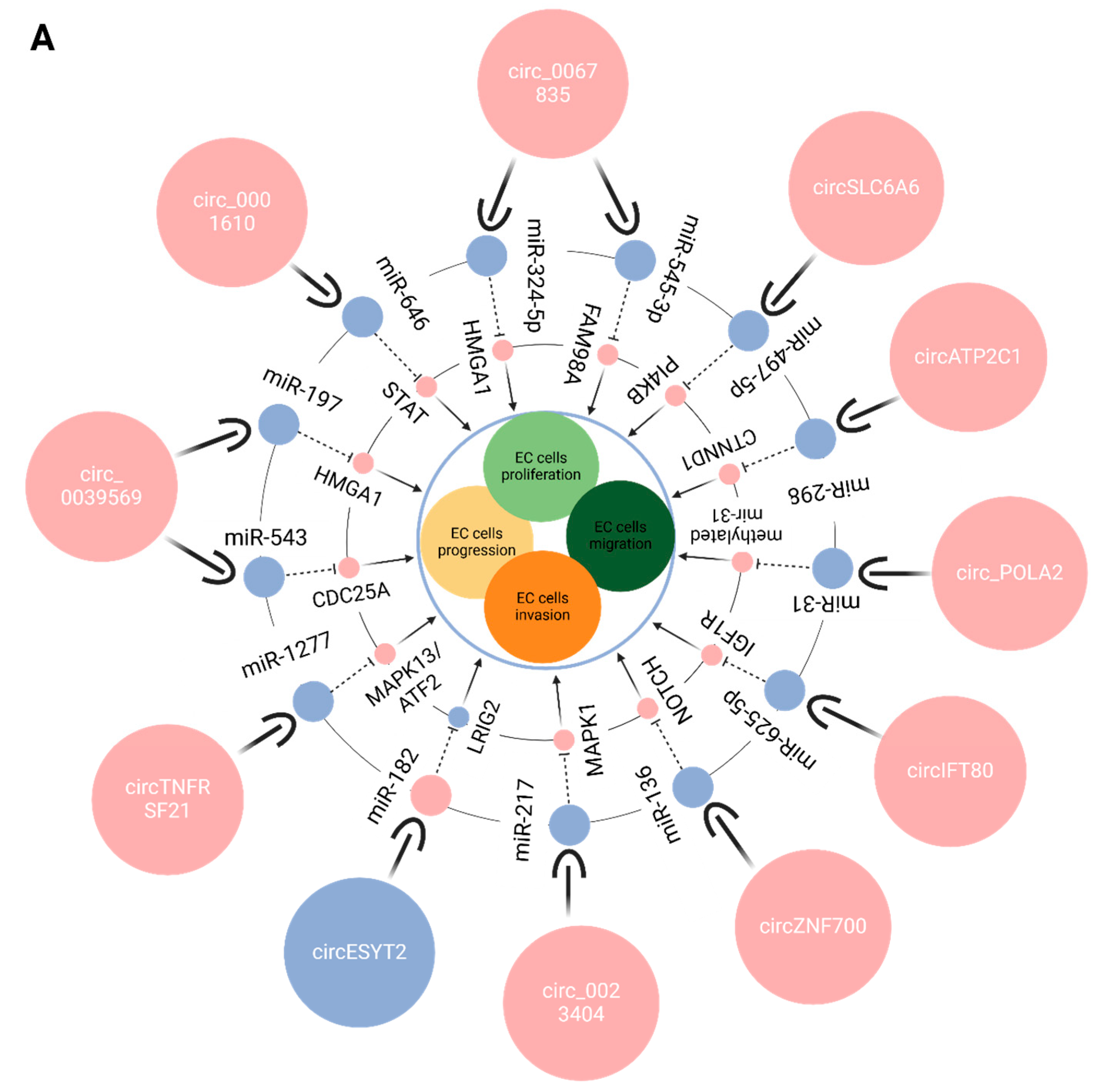
6.1. CircRNAs and EC
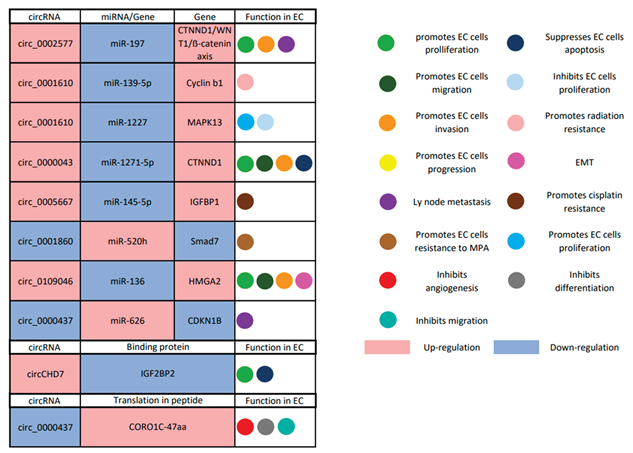 |
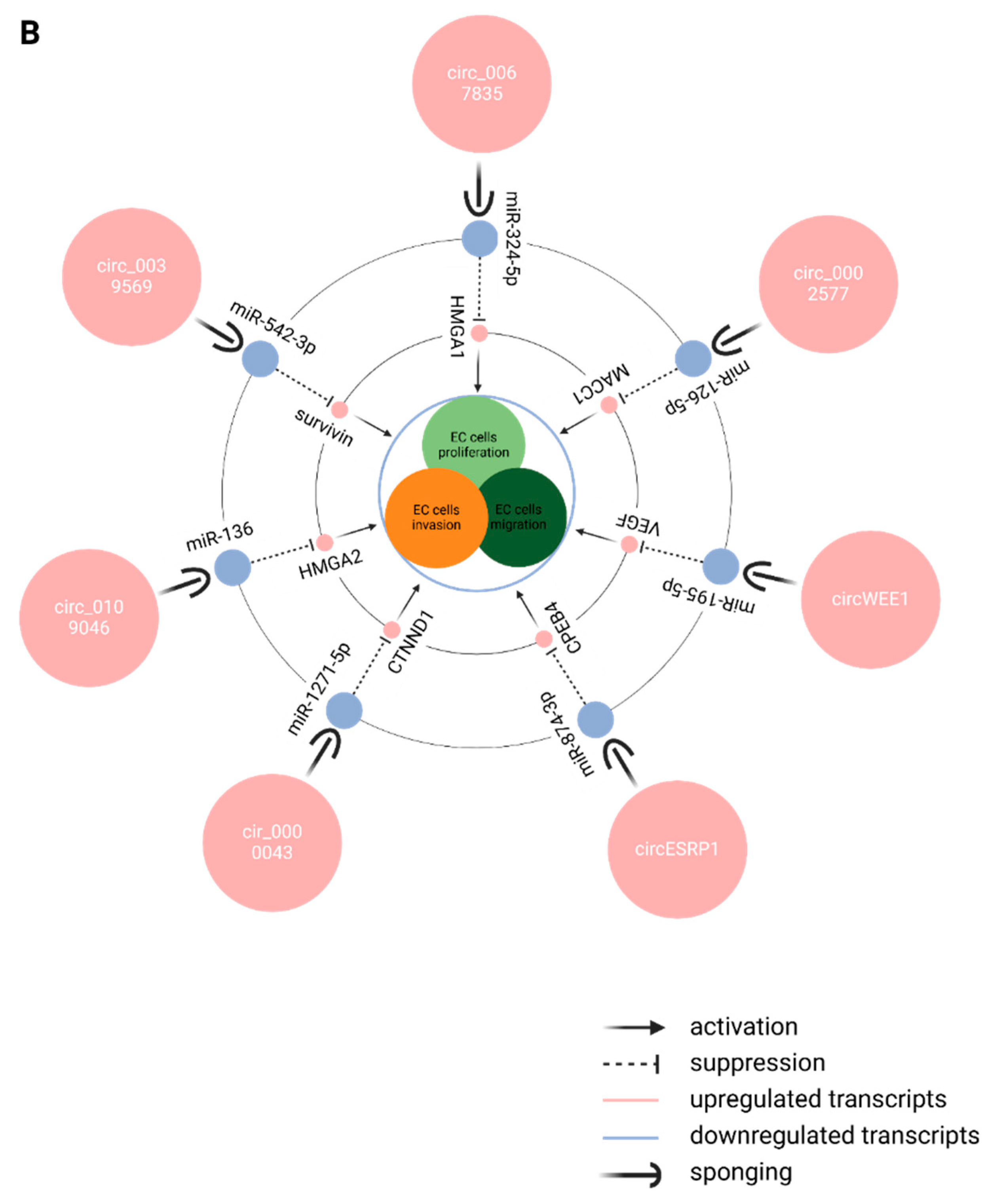
7. Discussion
In Vitro Attempts to Target CircRNAs
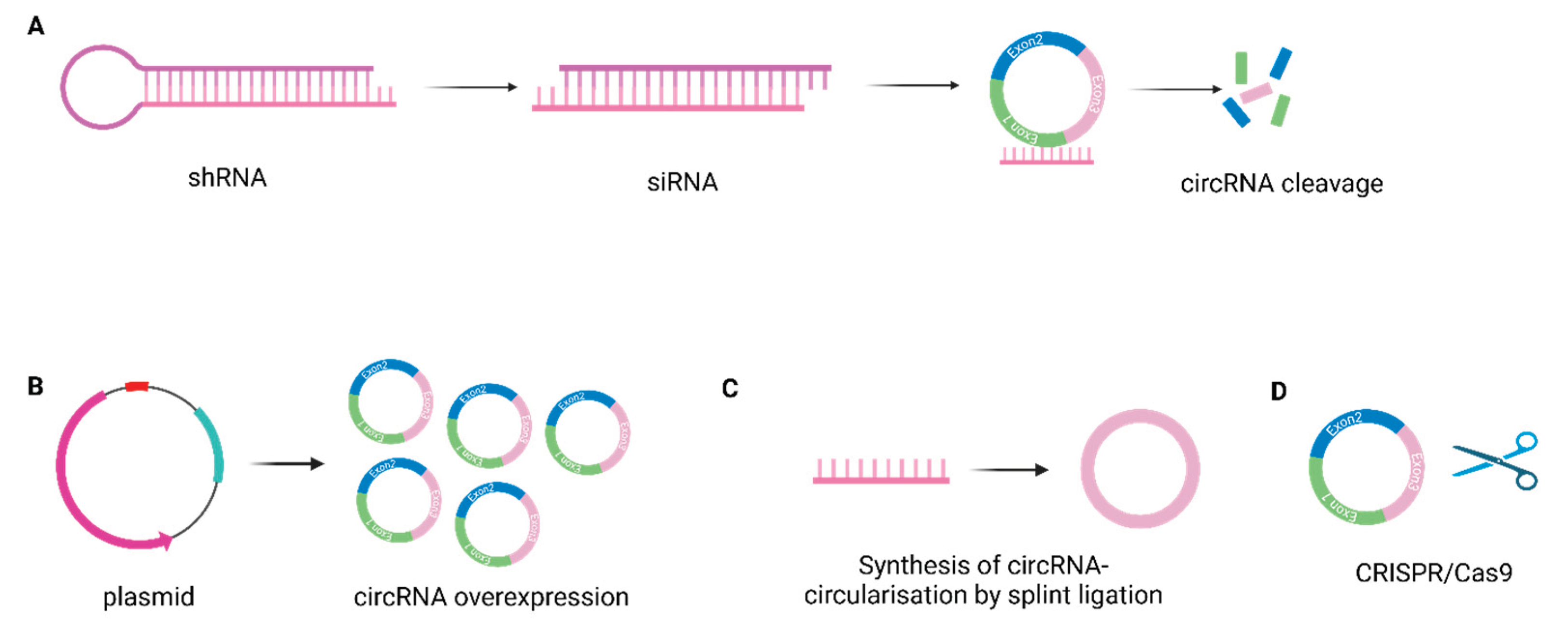
8. Conclusion
Supplementary Materials
Author Contributions
Conflict of Interest
References
- Chen, L. L., & Yang, L. (2015). Regulation of circRNA biogenesis. RNA biology, 12(4), 381-388. [CrossRef]
- Enuka, Y., Lauriola, M., Feldman, M. E., Sas-Chen, A., Ulitsky, I., & Yarden, Y. (2016). Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic acids research, 44(3), 1370-1383. [CrossRef]
- Guo, J. U., Agarwal, V., Guo, H., & Bartel, D. P. (2014). Expanded identification and characterization of mammalian circular RNAs. Genome biology, 15, 1-14. [CrossRef]
- Glažar, P., Papavasileiou, P., & Rajewsky, N. (2014). circBase: a database for circular RNAs. Rna, 20(11), 1666-1670. [CrossRef]
- Ghosal, S., Das, S., Sen, R., Basak, P., & Chakrabarti, J. (2013). Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Frontiers in genetics, 4, 283. [CrossRef]
- Zhao, R. T., Zhou, J., Dong, X. L., Bi, C. W., Jiang, R. C., Dong, J. F., ... & Zhang, J. N. (2018). Circular ribonucleic acid expression alteration in exosomes from the brain extracellular space after traumatic brain injury in mice. Journal of neurotrauma, 35(17), 2056–2066. [CrossRef]
- Ivanov, A., Memczak, S., Wyler, E., Torti, F., Porath, H. T., Orejuela, M. R., ... & Rajewsky, N. (2015). Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell reports, 10(2), 170–177. [CrossRef]
- Ashwal-Fluss, R., Meyer, M., Pamudurti, N. R., Ivanov, A., Bartok, O., Hanan, M., ... & Kadener, S. (2014). circRNA biogenesis competes with pre-mRNA splicing. Molecular cell, 56(1), 55–66. [CrossRef]
- Zhang, X. O., Wang, H. B., Zhang, Y., Lu, X., Chen, L. L., & Yang, L. (2014). Complementary sequence-mediated exon circularization. Cell, 159(1), 134–147. [CrossRef]
- Capel, B., Swain, A., Nicolis, S., Hacker, A., Walter, M., Koopman, P., ... & Lovell-Badge, R. (1993). Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell, 73(5), 1019–1030. [CrossRef]
- Kramer, M. C., Liang, D., Tatomer, D. C., Gold, B., March, Z. M., Cherry, S., & Wilusz, J. E. (2015). Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes & development, 29(20), 2168–2182. [CrossRef]
- Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., ... & Sharpless, N. E. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna, 19(2), 141–157. [CrossRef]
- Zhang, X. O., Wang, H. B., Zhang, Y., Lu, X., Chen, L. L., & Yang, L. (2014). Complementary sequence-mediated exon circularization. Cell, 159(1), 134–147. [CrossRef]
- Zhang, Y., Xue, W., Li, X., Zhang, J., Chen, S., Zhang, J. L., ... & Chen, L. L. (2016). The biogenesis of nascent circular RNAs. Cell reports, 15(3), 611–624. [CrossRef]
- Conn, S. J., Pillman, K. A., Toubia, J., Conn, V. M., Salmanidis, M., Phillips, C. A., ... & Goodall, G. J. (2015). The RNA binding protein quaking regulates formation of circRNAs. Cell, 160(6), 1125–1134. [CrossRef]
- Liu, D., Dredge, B. K., Bert, A. G., Pillman, K. A., Toubia, J., Guo, W., ... & Goodall, G. J. (2024). ESRP1 controls biogenesis and function of a large abundant multiexon circRNA. Nucleic acids research, 52(3), 1387–1403. [CrossRef]
- Bonczek, O., Wang, L., Gnanasundram, S. V., Chen, S., Haronikova, L., Zavadil-Kokas, F., & Vojtesek, B. (2022). DNA and RNA binding proteins: from motifs to roles in cancer. International Journal of Molecular Sciences, 23(16), 9329. [CrossRef]
- Stoll, L., Sobel, J., Rodriguez-Trejo, A., Guay, C., Lee, K., Venø, M. T., ... & Regazzi, R. (2018). Circular RNAs as novel regulators of β-cell functions in normal and disease conditions. Molecular metabolism, 9, 69-8. [CrossRef]
- Geng, H. H., Li, R., Su, Y. M., Xiao, J., Pan, M., Cai, X. X., & Ji, X. P. (2016). The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PloS one, 11(3), e0151753. [CrossRef]
- Zhang, J., Hu, H., Zhao, Y., & Zhao, Y. (2018). CDR1as is overexpressed in laryngeal squamous cell carcinoma to promote the tumour’s progression via miR-7 signals. Cell proliferation, 51(6), e12521.
- Han, S., Zhang, T., Kusumanchi, P., Huda, N., Jiang, Y., Liangpunsakul, S., & Yang, Z. (2020). Role of microRNA-7 in liver diseases: a comprehensive review of the mechanisms and therapeutic applications. Journal of Investigative Medicine, 68(7), 1208–1216. [CrossRef]
- Huang, G., Zhu, H., Shi, Y., Wu, W., Cai, H., & Chen, X. (2015). cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/β-catenin pathway. PloS one, 10(6), e0131225. [CrossRef]
- Wang, M., Chen, B., Ru, Z., & Cong, L. (2018). CircRNA circ-ITCH suppresses papillary thyroid cancer progression through miR-22-3p/CBL/β-catenin pathway. Biochemical and biophysical research communications, 504(1), 283–288. [CrossRef]
- Du, W. W., Yang, W., Chen, Y., Wu, Z. K., Foster, F. S., Yang, Z., ... & Yang, B. B. (2017). Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. European heart journal, 38(18), 1402–1412. [CrossRef]
- Du, W. W., Yang, W., Li, X., Awan, F. M., Yang, Z., Fang, L., ... & Yang, B. B. (2018). A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene, 37(44), 5829–5842. [CrossRef]
- Wang, Y., & Wang, Z. (2015). Efficient backsplicing produces translatable circular mRNAs. Rna, 21(2), 172–179. [CrossRef]
- Pamudurti, N. R., Bartok, O., Jens, M., Ashwal-Fluss, R., Stottmeister, C., Ruhe, L., ... & Kadener, S. (2017). Translation of circRNAs. Molecular cell, 66(1), 9–21. [CrossRef]
- Legnini, I., Di Timoteo, G., Rossi, F., Morlando, M., Briganti, F., Sthandier, O., ... & Bozzoni, I. (2017). Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Molecular cell, 66(1), 22–37. [CrossRef]
- Zhang, M., Zhao, K., Xu, X., Yang, Y., Yan, S., Wei, P., ... & Zhang, N. (2018). A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nature communications, 9(1), 4475. [CrossRef]
- Homem, C. C., Repic, M., & Knoblich, J. A. (2015). Proliferation control in neural stem and progenitor cells. Nature Reviews Neuroscience, 16(11), 647–659. [CrossRef]
- Shan, K., Liu, C., Liu, B. H., Chen, X., Dong, R., Liu, X., ... & Yan, B. (2017). Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation, 136(17), 1629–1642. [CrossRef]
- Shen, Y., Zhao, N., Zhao, N., Hu, X., He, X., Xu, Y., ... & Xu, X. (2022). Tumor-suppressive and oncogenic roles of microRNA-149-5p in human cancers. International Journal of Molecular Sciences, 23(18), 10823. [CrossRef]
- Zheng, Q., Bao, C., Guo, W., Li, S., Chen, J., Chen, B., ... & Huang, S. (2016). Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nature communications, 7(1), 11215. [CrossRef]
- Zhong, Z., Lv, M., & Chen, J. (2016). Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Scientific reports, 6(1), 30919. [CrossRef]
- Zhang, Z., Yang, T., & Xiao, J. (2018). Circular RNAs: promising biomarkers for human diseases. EBioMedicine, 34, 267–274. [CrossRef]
- Wu, Z., Shi, W., & Jiang, C. (2018). Overexpressing circular RNA hsa_circ_0002052 impairs osteosarcoma progression via inhibiting Wnt/β-catenin pathway by regulating miR-1205/APC2 axis. Biochemical and biophysical research communications, 502(4), 465–471. [CrossRef]
- Lamouille, S., Xu, J., & Derynck, R. (2014). Molecular mechanisms of epithelial–mesenchymal transition. Nature reviews Molecular cell biology, 15(3), 178–196. [PubMed]
- Dai, Y., Li, D., Chen, X., Tan, X., Gu, J., Chen, M., & Zhang, X. (2018). Circular RNA myosin light chain kinase (MYLK) promotes prostate cancer progression through modulating Mir-29a expression. Medical science monitor: international medical journal of experimental and clinical research, 24, 3462. [CrossRef]
- Zhang, X., Luo, P., Jing, W., Zhou, H., Liang, C., & Tu, J. (2018). circSMAD2 inhibits the epithelial–mesenchymal transition by targeting miR-629 in hepatocellular carcinoma. OncoTargets and therapy, 2853–2863. [CrossRef]
- Yu, C. Y., Li, T. C., Wu, Y. Y., Yeh, C. H., Chiang, W., Chuang, C. Y., & Kuo, H. C. (2017). The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nature communications, 8(1), 1149. [CrossRef]
- Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., ... & Rajewsky, N. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature, 495(7441), 333–338. [CrossRef]
- Li, H., Wei, X., Yang, J., Dong, D., Hao, D., Huang, Y., ... & Chen, H. (2018). circFGFR4 promotes differentiation of myoblasts via binding miR-107 to relieve its inhibition of Wnt3a. Molecular Therapy-Nucleic Acids, 11, 272–283. [CrossRef]
- Li, X., Zheng, Y., Zheng, Y., Huang, Y., Zhang, Y., Jia, L., & Li, W. (2018). Circular RNA CDR1as regulates osteoblastic differentiation of periodontal ligament stem cells via the miR-7/GDF5/SMAD and p38 MAPK signaling pathway. Stem cell research & therapy, 9, 1–14. [CrossRef]
- https://histology.siu.edu.
- Gray, C. A., Bartol, F. F., Tarleton, B. J., Wiley, A. A., Johnson, G. A., Bazer, F. W., & Spencer, T. E. (2001). Developmental biology of uterine glands. Biology of reproduction, 65(5), 1311–1323. [CrossRef]
- Fu, X. D. (2014). Non-coding RNA: a new frontier in regulatory biology. National science review, 1(2), 190–204. [CrossRef]
- Garcia-Alonso, L., Handfield, L. F., Roberts, K., Nikolakopoulou, K., Fernando, R. C., Gardner, L., ... & Vento-Tormo, R. (2021). Mapping the temporal and spatial dynamics of the human endometrium in vivo and in vitro. Nature genetics, 53(12), 1698–1711. [CrossRef]
- Nusse, R., & Varmus, H. (2012). Three decades of Wnts: a personal perspective on how a scientific field developed. The EMBO journal, 31(12), 2670–2684. [CrossRef]
- Komiya, Y., & Habas, R. (2008). Wnt signal transduction pathways. Organogenesis, 4(2), 68–75.
- Dexter, J. S. (1914). The analysis of a case of continuous variation in Drosophila by a study of its linkage relations. The American Naturalist, 48(576), 712–758. [CrossRef]
- Kumar, R., Juillerat-Jeanneret, L., & Golshayan, D. (2016). Notch antagonists: potential modulators of cancer and inflammatory diseases. Journal of medicinal chemistry, 59(17), 7719–7737. [CrossRef]
- Lowell, S., Jones, P., Le Roux, I., Dunne, J., & Watt, F. M. (2000). Stimulation of human epidermal differentiation by Delta–Notch signalling at the boundaries of stem-cell clusters. Current Biology, 10(9), 491–500. [CrossRef]
- Hellström, M., Phng, L. K., Hofmann, J. J., Wallgard, E., Coultas, L., Lindblom, P., ... & Betsholtz, C. (2007). Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature, 445(7129), 776–780.
- Bolós, V., Grego-Bessa, J., & De La Pompa, J. L. (2007). Notch signaling in development and cancer. Endocrine reviews, 28(3), 339–363.
- Orchard, M. D., & Murphy, C. R. (2002). Alterations in tight junction molecules of uterine epithelial cells during early pregnancy in the rat. Acta histochemica, 104(2), 149–155. [CrossRef]
- Murphy, C. R., Hosie, M. J., & Thompson, M. B. (2000). The plasma membrane transformation facilitates pregnancy in both reptiles and mammals. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology, 127(4), 433–439. [CrossRef]
- Whitby, S., Zhou, W., & Dimitriadis, E. (2020). Alterations in epithelial cell polarity during endometrial receptivity: a systematic review. Frontiers in Endocrinology, 11, 596324. [CrossRef]
- Cha, J., Sun, X., & Dey, S. K. (2012). Mechanisms of implantation: strategies for successful pregnancy. Nature medicine, 18(12), 1754–1767. [CrossRef]
- Ruiz-Alonso, M., Blesa, D., & Simón, C. (2012). The genomics of the human endometrium. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease, 1822(12), 1931–1942.
- Sebastian-Leon, P., Garrido, N., Remohí, J., Pellicer, A., & Diaz-Gimeno, P. (2018). Asynchronous and pathological windows of implantation: two causes of recurrent implantation failure. Human Reproduction, 33(4), 626–635. [CrossRef]
- Simon, A., & Laufer, N. (2012). Assessment and treatment of repeated implantation failure (RIF). Journal of assisted reproduction and genetics, 29, 1227–1239. [CrossRef]
- Altmäe, S., Martinez-Conejero, J. A., Esteban, F. J., Ruiz-Alonso, M., Stavreus-Evers, A., Horcajadas, J. A., & Salumets, A. (2013). MicroRNAs miR-30b, miR-30d, and miR-494 regulate human endometrial receptivity. Reproductive sciences, 20(3), 308–317. [CrossRef]
- Kim, A., Jung, H., Choi, W. J., Hong, S. N., & Kim, H. Y. (2014). Detection of endometrial and subendometrial vasculature on the day of embryo transfer and prediction of pregnancy during fresh in vitro fertilization cycles. Taiwanese Journal of Obstetrics and Gynecology, 53(3), 360–365. [CrossRef]
- Kasius, A., Smit, J. G., Torrance, H. L., Eijkemans, M. J., Mol, B. W., Opmeer, B. C., & Broekmans, F. J. (2014). Endometrial thickness and pregnancy rates after IVF: a systematic review and meta-analysis. Human reproduction update, 20(4), 530–541. [CrossRef]
- Yu, C. Y., & Kuo, H. C. (2019). The emerging roles and functions of circular RNAs and their generation. Journal of biomedical science, 26(1), 29. [CrossRef]
- Liu, L., Li, L., Ma, X., Yue, F., Wang, Y., Wang, L., ... & Zhang, X. (2018). Altered circular RNA expression in patients with repeated implantation failure. Cellular Physiology and Biochemistry, 44(1), 303–313. [CrossRef]
- Wang, A., & Chen, P. (2024). Comprehensive analysis of circRNA-miRNA-mRNA network related to angiogenesis in recurrent implantation failure. BMC Medical Genomics, 17(1), 193.
- Salmasi, S., Sharifi, M., & Rashidi, B. (2021). Ovarian stimulation and exogenous progesterone affect the endometrial miR-16-5p, VEGF protein expression, and angiogenesis. Microvascular Research, 133, 104074. [CrossRef]
- Lin, S. C., Wang, C. C., Wu, M. H., Yang, S. H., Li, Y. H., & Tsai, S. J. (2012). Hypoxia-induced microRNA-20a expression increases ERK phosphorylation and angiogenic gene expression in endometriotic stromal cells. The Journal of Clinical Endocrinology & Metabolism, 97(8), E1515–E1523. [CrossRef]
- Liu, X., Zhang, L., Liu, Y., Cui, J., Che, S., An, X., ... & Cao, B. (2018). Circ-8073 regulates CEP55 by sponging miR-449a to promote caprine endometrial epithelial cells proliferation via the PI3K/AKT/mTOR pathway. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 1865(8), 1130–1147. [CrossRef]
- Mladinov, D., Liu, Y., Mattson, D. L., & Liang, M. (2013). MicroRNAs contribute to the maintenance of cell-type-specific physiological characteristics: miR-192 targets Na+/K+-ATPase β1. Nucleic acids research, 41(2), 1273–1283. [CrossRef]
- He, X., Liu, N., Mu, T., Lu, D., Jia, C., Wang, S., ... & Ma, Y. (2020). Oestrogen induces epithelial-mesenchymal transition in endometriosis via circ_0004712/miR-148a-3p sponge function. Journal of cellular and molecular medicine, 24(17), 9658–9666. [CrossRef]
- Burke, W. M., Orr, J., Leitao, M., Salom, E., Gehrig, P., Olawaiye, A. B., ... & Society of Gynecologic Oncology Clinical Practice Committee. (2014). Endometrial cancer: a review and current management strategies: part II. Gynecologic oncology, 134(2), 393–402. [CrossRef]
- Jones, E. R., O’Flynn, H., Njoku, K., & Crosbie, E. J. (2021). Detecting endometrial cancer. The Obstetrician & Gynaecologist, 23(2), 103–112.
- Lu, K. H., & Broaddus, R. R. (2020). Endometrial cancer. New England Journal of Medicine, 383(21), 2053–2064.
- Gao, J., Fan, Y. Z., Gao, S. S., & Zhang, W. T. (2023). Circulating microRNAs as Potential Biomarkers for the Diagnosis of Endometrial Cancer: a Meta-Analysis. Reproductive Sciences, 30(2), 464–472. [CrossRef]
- McCluggage, W. G., Bosse, T., Gilks, C. B., Howitt, B. E., McAlpine, J. N., Nucci, M. R., ... & Parra-Herran, C. (2024). FIGO 2023 endometrial cancer staging: too much, too soon?. International Journal of Gynecologic Cancer, 34(1). [CrossRef]
- Vergote, I., & Matias-Guiu, X. (2023). New FIGO 2023 endometrial cancer staging validation. Welcome to the first molecular classifiers and new pathological variables!. European Journal of Cancer, 193. [CrossRef]
- Weinstein, J. N., Collisson, E. A., Mills, G. B., Shaw, K. R., Ozenberger, B. A., Ellrott, K., ... & Stuart, J. M. (2013). The cancer genome atlas pan-cancer analysis project. Nature genetics, 45(10), 1113–1120. [CrossRef]
- Yang, Y., Wu, S. F., & Bao, W. (2024). Molecular subtypes of endometrial cancer: Implications for adjuvant treatment strategies. International Journal of Gynecology & Obstetrics, 164(2), 436–459. [CrossRef]
- Selves, J., e Gloria, H. D. C., Brunac, A. C., Saffi, J., Guimbaud, R., Brousset, P., & Hoffmann, J. S. (2024). Exploring the basis of heterogeneity of cancer aggressiveness among the mutated POLE variants. Life Science Alliance, 7(1). [CrossRef]
- Kanopiene, D., Vidugiriene, J., Povilas Valuckas, K., Smailyte, G., Uleckiene, S., & Bacher, J. (2014). Endometrial cancer and microsatellite instability status. Open medicine, 10(1). [CrossRef]
- Jamieson, A., Thompson, E., Huvila, J., Leung, S., Lum, A., Helpman, L., ... & Mcalpine, J. (2021). OP008/# 194 P53ABN molecular subtype encompasses a morphologically diverse subset of endometrial cancers and identifies therapeutic opportunities to improve outcomes. International Journal of Gynecological Cancer, 31 (Suppl 4), A14–A14.
- Ribeiro-Santos, P., Martins Vieira, C., Viana Veloso, G. G., Vieira Giannecchini, G., Parenza Arenhardt, M., Müller Gomes, L., ... & Nogueira-Rodrigues, A. (2024). Tailoring endometrial cancer treatment based on molecular pathology: Current status and possible impacts on systemic and local treatment. International Journal of Molecular Sciences, 25(14), 7742. [CrossRef]
- Ye, F., Tang, Q. L., Ma, F., Cai, L., Chen, M., Ran, X. X., ... & Jiang, X. F. (2019). Analysis of the circular RNA transcriptome in the grade 3 endometrial cancer. Cancer Management and Research, 6215–6227. [CrossRef]
- Ruijtenberg, S., & van den Heuvel, S. (2016). Coordinating cell proliferation and differentiation: Antagonism between cell cycle regulators and cell type-specific gene expression. Cell cycle, 15(2), 196–212. [CrossRef]
- Dou, Y., Kawaler, E. A., Zhou, D. C., Gritsenko, M. A., Huang, C., Blumenberg, L., ... & Krek, A. (2020). Proteogenomic characterization of endometrial carcinoma. Cell, 180(4), 729–748. [CrossRef]
- Zong, Z. H., Liu, Y., Chen, S., & Zhao, Y. (2020). Circ_PUM1 promotes the development of endometrial cancer by targeting the miR-136/NOTCH3 pathway. Journal of cellular and molecular medicine, 24(7), 4127–4135. [CrossRef]
- Shen, Q., He, T., & Yuan, H. (2019). Hsa_circ_0002577 promotes endometrial carcinoma progression via regulating miR-197/CTNND1 axis and activating Wnt/β-catenin pathway. Cell Cycle, 18(11), 1229–1240. [CrossRef]
- Yuan, S., Zheng, P., Sun, X., Zeng, J., Cao, W., Gao, W., ... & Wang, L. (2021). Hsa_Circ_0001860 promotes Smad7 to enhance MPA resistance in endometrial cancer via miR-520h. Frontiers in Cell and Developmental Biology, 9, 738189. [CrossRef]
- Sun, G., Tian, J., Xiao, Y., & Zeng, Y. (2023). Circular RNA circ_0005667 promotes cisplatin resistance of endometrial carcinoma cells by regulating IGF2BP1 through miR-145-5p. Anti-Cancer Drugs, 34(7), 816–826. [CrossRef]
- Włodarczyk, K., Kuryło, W., Pawłowska-Łachut, A., Skiba, W., Suszczyk, D., Pieniądz, P., ... & Wertel, I. (2024). circRNAs in Endometrial Cancer—A Promising Biomarker: State of the Art. International Journal of Molecular Sciences, 25(12), 6387. [CrossRef]
- Koler, M., Achache, H., Tsafrir, A., Smith, Y., Revel, A., & Reich, R. (2009). Disrupted gene pattern in patients with repeated in vitro fertilization (IVF) failure. Human reproduction, 24(10), 2541–2548. [CrossRef]
- Mohamed, O. A., Jonnaert, M., Labelle-Dumais, C., Kuroda, K., Clarke, H. J., & Dufort, D. (2005). Uterine Wnt/β-catenin signaling is required for implantation. Proceedings of the National Academy of Sciences, 102(24), 8579–8584. [CrossRef]
- Inyawilert, W., Fu, T. Y., Lin, C. T., & Tang, P. C. (2015). Let-7-mediated suppression of mucin 1 expression in the mouse uterus during embryo implantation. Journal of Reproduction and Development, 61(2), 138–144. [CrossRef]
- Li, Q., Liu, W., Chiu, P. C., & Yeung, W. S. (2020). Mir-let-7a/g enhances uterine receptivity via suppressing Wnt/β-catenin under the modulation of ovarian hormones. Reproductive Sciences, 27, 1164–1174. [CrossRef]
- Zheng, Q., Zhang, D., Cui, X., Sun, J., Liang, C., Qin, H., ... & Yan, Q. (2017). MicroRNA-200c impairs uterine receptivity formation by targeting FUT4 and α1, 3-fucosylation. Cell Death & Differentiation, 24(12), 2161–2172. [CrossRef]
- Dong, L.; Zhang, L.; Liu, H.; Xie, M.; Gao, J.; Zhou, X.; Zhao, Q.; Zhang, S.; Yang, J. Circ_0007331 knock-down suppresses the progression of endometriosis via miR-200c-3p/HiF-1alpha axis. J. Cell Mol. Med., 2020, 24, 12656–12666. [Google Scholar] [CrossRef]
- Amjadi, F., Salehi, E., Zandieh, Z., Rashidi, M., Taleahmad, S., Aflatoonian, R., & Mehdizadeh, M. (2019). Comparative evaluation of NOTCH signaling molecules in the endometrium of women with various gynecological diseases during the window of implantation. Iranian Journal of Basic Medical Sciences, 22(4), 426. [CrossRef]
- Garcia-Alonso, L., Handfield, L. F., Roberts, K., Nikolakopoulou, K., Fernando, R. C., Gardner, L., ... & Vento-Tormo, R. (2021). Mapping the temporal and spatial dynamics of the human endometrium in vivo and in vitro. Nature genetics, 53(12), 1698–1711. [CrossRef]
- Sandbothe, M., Buurman, R., Reich, N., Greiwe, L., Vajen, B., Gürlevik, E., ... & Skawran, B. (2017). The microRNA-449 family inhibits TGF-β-mediated liver cancer cell migration by targeting SOX4. Journal of hepatology, 66(5), 1012–1021. [CrossRef]
- Nikolova, M., Naydenov, M., Glogovitis, I., Apostolov, A., Saare, M., Boggavarapu, N., ... & Yahubyan, G. (2021). Coupling miR/isomiR and mRNA expression signatures unveils new molecular layers of endometrial receptivity. Life, 11(12), 1391. [CrossRef]
- Schwab, M. (2004). MYCN in neuronal tumours. Cancer letters, 204(2), 179–187. [CrossRef]
- Baluapuri, A., Wolf, E., & Eilers, M. (2020). Target gene-independent functions of MYC oncoproteins. Nature Reviews Molecular Cell Biology, 21(5), 255–267. [CrossRef] [PubMed]
- Liu, X., Zhang, L., Liu, Y., Cui, J., Che, S., An, X., ... & Cao, B. (2018). Circ-8073 regulates CEP55 by sponging miR-449a to promote caprine endometrial epithelial cells proliferation via the PI3K/AKT/mTOR pathway. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 1865(8), 1130–1147. [CrossRef]
- Wang, S., Zhang, M., Zhang, T., Deng, J., Xia, X., & Fang, X. (2020). microRNA-141 inhibits TGF-β1-induced epithelial-to-mesenchymal transition through inhibition of the TGF-β1/SMAD2 signalling pathway in endometriosis. Archives of Gynecology and Obstetrics, 301, 707–714. [CrossRef]
- Peng, X., Zhu, Y., Wang, T., Wang, S., & Sun, J. (2023). Integrative analysis links autophagy to intrauterine adhesion and establishes autophagy-related circRNA-miRNA-mRNA regulatory network. Aging (Albany NY), 15(16), 8275.
- Cobellis, L., Caprio, F., Trabucco, E., Mastrogiacomo, A., Coppola, G., Manente, L., ... & De Luca, A. (2008). The pattern of expression of Notch protein members in normal and pathological endometrium. Journal of anatomy, 213(4), 464–472. [CrossRef]
- Jiang, N., Pan, W., Li, J., Cao, T., & Shen, H. (2020). Upregulated circular RNA hsa_circ_0008433 regulates pathogenesis in endometriosis via miRNA. Reproductive Sciences, 27, 2002–2017. [CrossRef]
- Rossi, J. J. (2008). Expression strategies for short hairpin RNA interference triggers. Human gene therapy, 19(4), 313–317. [CrossRef]
- Müller, S., & Appel, B. (2017). In vitro circularization of RNA. RNA biology, 14(8), 1018–1027.
- Ran, F. A., Hsu, P. D., Wright, J., Agarwala, V., Scott, D. A., & Zhang, F. (2013). Genome engineering using the CRISPR-Cas9 system. Nature protocols, 8(11), 2281–2308. [CrossRef]
- Wang, A. Z., Langer, R., & Farokhzad, O. C. (2012). Nanoparticle delivery of cancer drugs. Annual review of medicine, 63(1), 185–198.
- Shi, X., Wang, B., Feng, X., Xu, Y., Lu, K., & Sun, M. (2020). circRNAs and exosomes: a mysterious frontier for human cancer. Molecular therapy Nucleic acids, 19, 384–392. [CrossRef]
- El-Andaloussi, S., Lee, Y., Lakhal-Littleton, S., Li, J., Seow, Y., Gardiner, C., ... & Wood, M. J. (2012). Exosome-mediated delivery of siRNA in vitro and in vivo. Nature protocols, 7(12), 2112–2126. [CrossRef]
- Shtam, T. A., Kovalev, R. A., Varfolomeeva, E. Y., Makarov, E. M., Kil, Y. V., & Filatov, M. V. (2013). Exosomes are natural carriers of exogenous siRNA to human cells in vitro. Cell Communication and Signaling, 11, 1–10. [CrossRef]
- Setten, R. L., Rossi, J. J., & Han, S. P. (2019). The current state and future directions of RNAi-based therapeutics. Nature reviews Drug discovery, 18(6), 421–446. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





