1. Introduction
Humans and other mammals express two IMPDH isoforms, IMPDH1 and IMPDH2, each comprising 514 amino acids and sharing 84% sequence identity. Both isoforms contain a catalytic domain that binds substrates and a regulatory Bateman domain, which modulates enzymatic activity through allosteric interactions with the catalytic core [
1]
. The IMPDH regulatory (Bateman) domain contains ATP- and GTP-binding sites. IMPDH exists as a constitutive tetramer, and nucleotide binding promotes reversible dimerization of the regulatory domains, leading to octamer formation [
2]
. IMPDH1 is constitutively expressed in normal lymphocytes but is highly upregulated in a subset of small cell lung cancers (SCLC) [
3]. IMPDH2 is overexpressed in hematological malignancies, including human leukemic cell lines and BCR-ABL–positive acute myelogenous leukemia [
4], in chronic myelogenous leukemia [
5] and other cancers, such as triple-negative breast cancer [
6],prostate cancer [
7,
8], kidney cancer [
9], nasopharyngeal carcinoma [
10], in a subset of small-cell lung cancers [
3,
11], in non-small cell lung cancer [
12] and in glioblastoma [
13,
14] and brain metastases [
15]
. Proteomic profiling of colorectal cancer plasma identified IMPDH2 as a potential biomarker [
16]
. Whereas protein tyrosine phosphorylation makes up only 2.5%, it has significant effects on nearly every aspect of cellular physiology [
17]
. Our previous work demonstrated that ALK and SRC kinase-mediated tyrosine phosphorylation of ACLY regulates its function [
18,
19]
. Although IMPDH2 overexpression in solid tumors is well documented, its post-translational regulation, particularly by tyrosine phosphorylation remains poorly understood. Here, we report for the first time that IMPDH2 is phosphorylated on a critical tyrosine residue by oncogenic kinases, as demonstrated through in vitro kinase assays and mass spectrometry-based phosphoproteomic analysis.
IMPDH1 and IMPDH2 pose a major challenge for isoform-specific drug development due to their 84% sequence identity. To overcome this, we analyzed sequence differences and hypothesized that the two isoforms may possess distinct phosphoinositide (PI) binding sites. PIs are lipid second messengers critical for membrane trafficking, metabolism, growth, signaling, and autophagy. Their phosphorylation generates seven distinct species, including PI3P, a key marker of endosomal and autophagic membranes. PI3P is recognized by FYVE and PX domain-containing proteins, suggesting that isoform-specific PI interactions could be leveraged for selective targeting of IMPDH isoforms [
20,
21]. FDA-approved IMPDH inhibitors—such as Mycophenolic acid (MPA), Mycophenolate mofetil (MMF), Ribavirin, and Mizoribine target the catalytic domains of both IMPDH1 and IMPDH2 and are used clinically for immunosuppressive and antiviral therapy [
22]. These inhibitors also promote the formation of Rods and Rings (RRs), or IMPDH filaments, which are non-membrane-bound intracellular polymeric structures [
23,
24,
25].
Our study shows that mycophenolic acid (MPA) induces the expression of catalytically inactive IMPDH2 and promotes filament formation in T-cell and B-cell lymphomas, as well as in solid tumors. This filamentous assembly may contribute to the toxicity and off-target effects of current IMPDH inhibitors, emphasizing the need for more selective therapeutics. We identify IMPDH2 as a direct substrate of the oncogenic kinases ALK and SRC, and demonstrate that PI3P binding specifically inhibits IMPDH2 activity but not that of IMPDH1. A self-derived IMPDH1 peptide from the Y233 domain also suppresses IMPDH2 function. Based on these findings and structural modeling, we performed in silico screening and discovered a novel allosteric inhibitor, comp-10, which specifically targets the allosteric domain and is distinct from current IMPDH inhibitors, thereby opening avenues for future therapeutic advancements
2. Materials and Methods
Reagents and Antibodies
Recombinant human IMPDH1 (Catalog # 8904-DH) and IMPDH2 (catalog# Catalog # 8349-DH) proteins were obtained from R&D Systems. IMPDH activity assay kit was from Biovision (Catalog#K495) or abcam (catalog#ab283395). PIP Strips – Lipid-Protein Interaction Assay (catalog#P-6001), PIP Arrays – Lipid-Protein Interaction Assay (catalog# P-6001), PI(3]P Beads (catalog#P-B003A), PI(4,5)P2 diC4 (catalog#P-4504) and PI(3)P diC8 (catalog#P-3008A) were purchased from Echelon Biosciences. All other reagents are from Sigma-Aldrich. Cell Proliferation Reagent WST-1 (Millepore-sigma), CellTiter 96® Non-Radioactive Cell Proliferation Assay (MTT) kit (catalog#G4001) from Promega. HA Tag Monoclonal Antibody (Thermo Fisher Scientific Cat#26183, RRID: AB_10978021), Pierce IP Lysis Buffer (Thermo Fisher Scientific Cat#87787), Pierce Anti-HA Magnetic Beads (Thermo Fisher Scientific Cat# 88837, RRID: AB_2861399) Halt Protease and Phosphatase Inhibitor (Thermo Fisher Scientific Cat#78440), ALK Recombinant Human Protein (Thermo Fisher Scientific, Cat# PV3867) and SRC Recombinant Human Protein (Thermo Fisher Scientific, Cat# P3044). Mizoribine (Cat #S1384), Ribavarin (Cat #S2504), mycophenolic acid (Cat# S2487), Mycophenolate mofetil (Cat#S1501), certinib (Cat#S7083). All these small-molecule inhibitors were purchased from Selleckchem. IMPDH1 (RRID:AB_2878992, Cat# 22092-1-AP), IMPDH2 (RRID:AB_2127351, Cat# 12948-1-AP) and GAPDH (RRID:AB_2107436, Cat# 60004-1-Ig) from Proteintech, HA-Tag (C29F4) Rabbit (RRID:AB_1549585, Cat# 3724S), Phospho-ALK (Tyr1604) Antibody (RRID:AB_331047, Cat# 3341S), ALK (D5F3®) XP® Rabbit (RRID:AB_11127207, Cat# 3633S) from Cell Signaling Technology.
Plasmids and Lentivirus Products
Custom Plasmid Preparation: IMPDH2-HA tagged_pLenti_MS2-P65-HSF1_mCherry, synthesized at GenScript, Piscataway, NJ. pLenti-EGFP-2xFYVE (Plasmid #136996, pLenti-EGFP-2xFYVE was a gift from Ken-Ichi Takemaru (Addgene plasmid Cat#136996;
http://n2t.net/addgene:136996;RRID:Addgene_136996), 3rd Gen. Packaging Mix & Lentifectin Combo Pack from Applied Biological System (ABM good# LV053-G074), Lenti-X™ Concentrator from TAKARA (catalog# 631231).
MCL, DLBCL, and ALCL Cell Lines
MCL, MCL-RL cells were derived from a patient with MCL at the University of Pennsylvania, Philadelphia, PA. JeKo-1, Maver Rec-1, Granta519, DLBCL, OCI-LY1, OCI-LY4, OCI-LY8, and TOLEDO; SUDHL-1, JB6, Karpas 299, SUP-M2, L82, and SR786 cell lines were derived from ALK+ALCL patients and cultured as described in earlier [
26]. Human CD4+ cells transduced with NPM-ALK, NA1, were created by our group as described earlier [
27]. The cell lines were regularly tested for Mycoplasma contamination using Mycoplasma detection kits from Thermo Fisher Scientific. All cell lines were routinely tested for Mycoplasma contamination using Mycoplasma detection kits (Thermo Fisher Scientific) and were authenticated. Cells were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (Pen/Strep) in a humidified incubator at 37°C with 5% CO
2.
ALK Inhibitor (ALKi) Resistant ALCL Cell Lines
ALKi crizotinib-resistant cell lines described earlier, KARPAS299 CR06 (resistant up to 600 nM of crizotinib and cultured at that concentration) and SUPM2 CR03 (resistant up to 300 nM of crizotinib and cultured at that concentration), and ALKi lorlatinib-resistant cell lines described earlier, KARPAS LR1000 (resistant up to 1000 nM of lorlatinib and cultured at 300 nM of lorlatinib) and SUPM2 LR1000 (resistant up to 1000 nM of lorlatinib and cultured at 300 nM of lorlatinib). All of these cell lines were obtained from Drs. Carlo Gambacorti-Passerini and Luca Mologni, University of Milano-Bicocca, Italy [
28]. The cell lines were grown in RPMI medium supplemented with 10% FBS and 1% penicillin/streptomycin in a humidified incubator at 37°C with 5% CO2.
IMPDH1 and IMPDH2 PIP Strip and PIP Array Binding Assay
For initial experiments, human recombinant IMPDH1 and IMPDH2 proteins were diluted to 1 µg/ml in 3% BSA and dissolved in PBS buffer. Next, the IMPDH2 concentration was further diluted to 100 ng/ml. The diluted proteins were subjected to a PIP strip lipid-protein interaction assay as described earlier. Based on IMPDH2’s unique binding to PI3P compared to IMPDH1, we further validated IMPDH2’s binding to PI3P on a PIP array coated with various concentrations.
IMPDH1 and IMPDH2 Activity Assay in the Presence of PIP2 (PI(4,5)P2) and PI3P (PI(3)P diC8)
Activity assays on human recombinant IMPDH1 and IMPDH2 were conducted using a Synergy H1 microplate reader from BioTek, 96-well plate readers, and a commercially available kit from Biovision, Inc. According to the assay kit instructions, the phospholipids PIP2 and PI3P were diluted in activity assay buffer and preincubated with recombinant IMPDH1 or IMPDH2 at concentrations ranging from 50 µM to 200 µM for 15 minutes. After this incubation, a substrate solution was added to the wells to initiate enzyme activity. The activity was continuously measured every minute until 30 to 60 minutes later triplicate.
In-Vitro Kinase Assay and LC-MS/MS Phosphoproteomics Analysis
We performed an in-vitro kinase assay on human recombinant IMPDH2 in the presence of active anaplastic lymphoma kinase (ALK) and SRC kinase. We subjected it to LC-MS/MS analysis as we described in our previous study [
18].
Structure-Based In-Silico Screening of IMPDH Inhibitors
The National Cancer Institute (NCI) maintains a database of compounds, known as the Mechanistic Set VI, which comprises 811 compounds derived from the 37,836 open compounds tested in the NCI human tumor 60-cell line screen. This mechanistic diversity set was chosen to represent a broad range of growth inhibition patterns in the NCI60 cell line screen based on the GI50 activity of the compounds. Compounds tested in the NCI-60 cell line screen were clustered using the FASTCLUS procedure in the SAS statistical package. This algorithm is based on MacQueen’s k-means algorithm, which minimizes the sum of squared distances from the cluster means. The procedure resulted in 1272 clusters. A single representative compound from each cluster, for which an adequate supply of material was available, was chosen. Some clusters are not represented in the set, as insufficient material was available. The database of compounds was downloaded. We subjected the compounds in the database with the following filter: 1) 100 <= Molecular Weight =< 500 2) Only one molecule, 3) 2 <= Number of rotatable bonds =< 6 4) Lipenski druglike =1 and no chiral centers. This resulted in 283 compounds for in silico screening. We generated ~15,200 energetically favorable conformations for in-silico screening. We docked these conformations into the GTP binding site of IMPDH2 and ranked them in order based on a scoring function. We selected 100 top-scoring compound-IMPDH2 complexes and selected 38 as the unique compounds based on National Service Center (NSC) identity. We got 15 compounds from the National Cancer Institute (NCI) and tested them for enzyme activity inhibition against recombinant human IMPDH2 in 96-well plate screening.
Cell Proliferation Assay
For the cell proliferation assay, the aforementioned cell lines were plated in 96-well plates at a density of 20,000 to 40,000 cells per well in standard RPMI medium, with DMSO as a control or with the respective drug concentrations in 100 µL of medium. After 48 hours in culture, cell proliferation was assessed using the WST-1 or MTT assay method following the manufacturer’s protocol.
Statistics and Reproducibility
The student’s t-test was used to analyze differences in Western blot densitometric values and activity assays to assess differences in cell growth and colony formation. p-values equal to or less than 0.05 were considered statistically significant without being adjusted for multiple comparisons. The statistical analysis was performed using GraphPad Prism 9.0. software and NIH ImageJ software for the densitometric quantitation of western blot data.
Data Availability
We confirm that all relevant data and methods are included in the main Article and the Supplementary Information section.
4. Discussion
Our study provides the first direct evidence that oncogenic tyrosine kinases (TKs), specifically ALK and SRC, phosphorylate IMPDH2, a key enzyme in purine biosynthesis. Although tyrosine phosphorylation accounts for a small part of total post-translational modifications, it plays a vital role in regulating metabolism, proliferation, and survival. While IMPDH2 overexpression is common in cancers, the functional significance of its phosphorylation has largely remained unexplored. We show that phosphorylation at the conserved Y233 residue within the IMPDH2 allosteric domain controls its enzymatic activity, directly linking TK signaling to metabolic reprogramming in cancer. This has important therapeutic implications, especially given the rise of resistance to FDA-approved ALK inhibitors such as crizotinib, ceritinib, alectinib, brigatinib, and lorlatinib. Notably, crizotinib is also approved for NPM-ALK-positive ALCL and neuroblastoma, but resistance continues to be a major clinical challenge.
Targeting downstream effectors like IMPDH2 could offer an alternative way to overcome resistance. We show that inhibiting IMPDH reduces the growth of both ALK inhibitor–sensitive and –resistant ALCL cells, highlighting IMPDH2 as a promising therapeutic target. Since ALK (EML4-ALK) and SRC play a role in other cancers, this approach may have wider relevance. We also identify PI3P as a natural lipid regulator of IMPDH2, binding specifically to basic residues within the CBS domains and inhibiting its enzymatic activity. Y233, found in this regulatory domain, is different from the active-site Cys-331 targeted by traditional inhibitors like mycophenolic acid (MPA). These results emphasize the potential of allosteric modulation as a new approach for inhibiting IMPDH2.
Using a structure-based computational screening approach, we identified Comp-10, a first-in-class allosteric IMPDH inhibitor. Unlike MPA, which induces filament formation (rods and rings) and paradoxically increases IMPDH protein levels, Comp-10 reduces IMPDH1/2 expression and prevents filament assembly. Comp-10 demonstrated superior efficacy in inhibiting the growth and colony formation of ALK-positive and MCL cells, including drug-resistant models, while sparing non-malignant cells like OVCAR3. The advantages of allosteric inhibitors, including reducing IMPDH1/2 protein levels and rods/ring formation, decrease toxicity and highlight the therapeutic potential of Comp-10 promise. With drug development timelines spanning 10–15 years and costs exceeding $2 billion, computational strategies like ours offer a cost-effective and accelerated path to drug discovery.
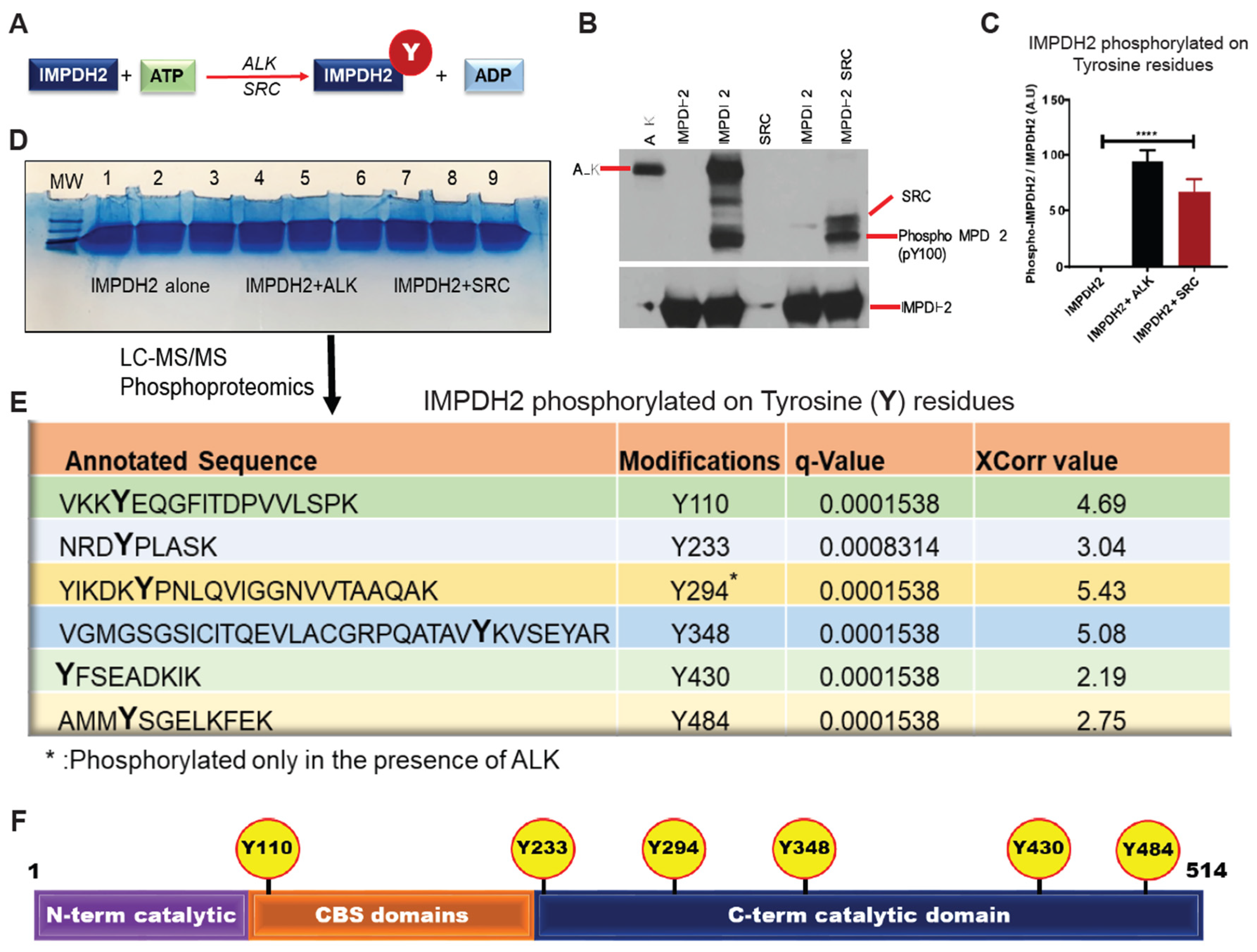
Figure 1.
Oncogenic kinases ALK and SRC phosphorylate IMPDH2 at specific tyrosine residues.A, Schematic of the in vitro kinase assay used to evaluate tyrosine phosphorylation of IMPDH2 by ALK and SRC kinases. B, Western blot analysis of in vitro kinase reactions using ALK and SRC with recombinant IMPDH2, probed with phospho-tyrosine-specific and total IMPDH2 antibodies. C, Densitometric quantification of tyrosine-phosphorylated IMPDH2 compared to total IMPDH2 levels, demonstrating enhanced phosphorylation in the presence of ALK and SRC. D, Large-scale in vitro kinase assay products were resolved by NuPAGE and analyzed by LC-MS/MS phosphoproteomics to identify phosphorylation sites. E, Identification of key tyrosine residues on IMPDH2 phosphorylated by ALK and SRC, as determined by mass spectrometry. F, Schematic of IMPDH2 structural domains highlighting novel ALK- and SRC-mediated phosphorylation sites.
Figure 1.
Oncogenic kinases ALK and SRC phosphorylate IMPDH2 at specific tyrosine residues.A, Schematic of the in vitro kinase assay used to evaluate tyrosine phosphorylation of IMPDH2 by ALK and SRC kinases. B, Western blot analysis of in vitro kinase reactions using ALK and SRC with recombinant IMPDH2, probed with phospho-tyrosine-specific and total IMPDH2 antibodies. C, Densitometric quantification of tyrosine-phosphorylated IMPDH2 compared to total IMPDH2 levels, demonstrating enhanced phosphorylation in the presence of ALK and SRC. D, Large-scale in vitro kinase assay products were resolved by NuPAGE and analyzed by LC-MS/MS phosphoproteomics to identify phosphorylation sites. E, Identification of key tyrosine residues on IMPDH2 phosphorylated by ALK and SRC, as determined by mass spectrometry. F, Schematic of IMPDH2 structural domains highlighting novel ALK- and SRC-mediated phosphorylation sites.
Figure 2.
IMPDH2 Y233 phosphorylation regulates PI3P binding. A, Schematic of PIP-lipid strip assay showing membranes coated with 15 distinct phospholipid species and incubated with HA peptide as a negative control. B, Recombinant human IMPDH1 (1.0 µg) incubated on PIP-lipid strips in 3% BSA/PBS overnight at 4°C; binding detected by Western blot using IMPDH1 antibody. C, Recombinant human IMPDH2 (1.0 µg) incubated under identical conditions and probed with IMPDH2 antibody. D, Control binding experiment using a reduced amount (0.1 µg) of IMPDH1 and probing with IMPDH2 antibody to assess specificity. E, Densitometric quantification of PI3P binding to IMPDH2 and IMPDH1, demonstrating preferential binding of PI3P to IMPDH2. F, Schematic of membrane-lipid arrays containing eight phosphoinositide species used to assess phosphoinositide-specific interactions. G, H, HA-tagged human IMPDH2 constructs expressed in HEK293T cells, with or without co-expression of SRC kinase; PI3P-bound IMPDH2-HA was detected by Western blot using anti-HA antibody. I, Live-cell imaging of HEK293T cells co-transfected with IMPDH2-mCherry and EGFP-FYVE (PI3P biosensor), revealing co-localization of IMPDH2 with PI3P in the presence of Y233 phosphorylation. All graphs represent mean ± SD from three biological replicates (n = 3). Statistical significance was assessed using unpaired t-tests: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Figure 2.
IMPDH2 Y233 phosphorylation regulates PI3P binding. A, Schematic of PIP-lipid strip assay showing membranes coated with 15 distinct phospholipid species and incubated with HA peptide as a negative control. B, Recombinant human IMPDH1 (1.0 µg) incubated on PIP-lipid strips in 3% BSA/PBS overnight at 4°C; binding detected by Western blot using IMPDH1 antibody. C, Recombinant human IMPDH2 (1.0 µg) incubated under identical conditions and probed with IMPDH2 antibody. D, Control binding experiment using a reduced amount (0.1 µg) of IMPDH1 and probing with IMPDH2 antibody to assess specificity. E, Densitometric quantification of PI3P binding to IMPDH2 and IMPDH1, demonstrating preferential binding of PI3P to IMPDH2. F, Schematic of membrane-lipid arrays containing eight phosphoinositide species used to assess phosphoinositide-specific interactions. G, H, HA-tagged human IMPDH2 constructs expressed in HEK293T cells, with or without co-expression of SRC kinase; PI3P-bound IMPDH2-HA was detected by Western blot using anti-HA antibody. I, Live-cell imaging of HEK293T cells co-transfected with IMPDH2-mCherry and EGFP-FYVE (PI3P biosensor), revealing co-localization of IMPDH2 with PI3P in the presence of Y233 phosphorylation. All graphs represent mean ± SD from three biological replicates (n = 3). Statistical significance was assessed using unpaired t-tests: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
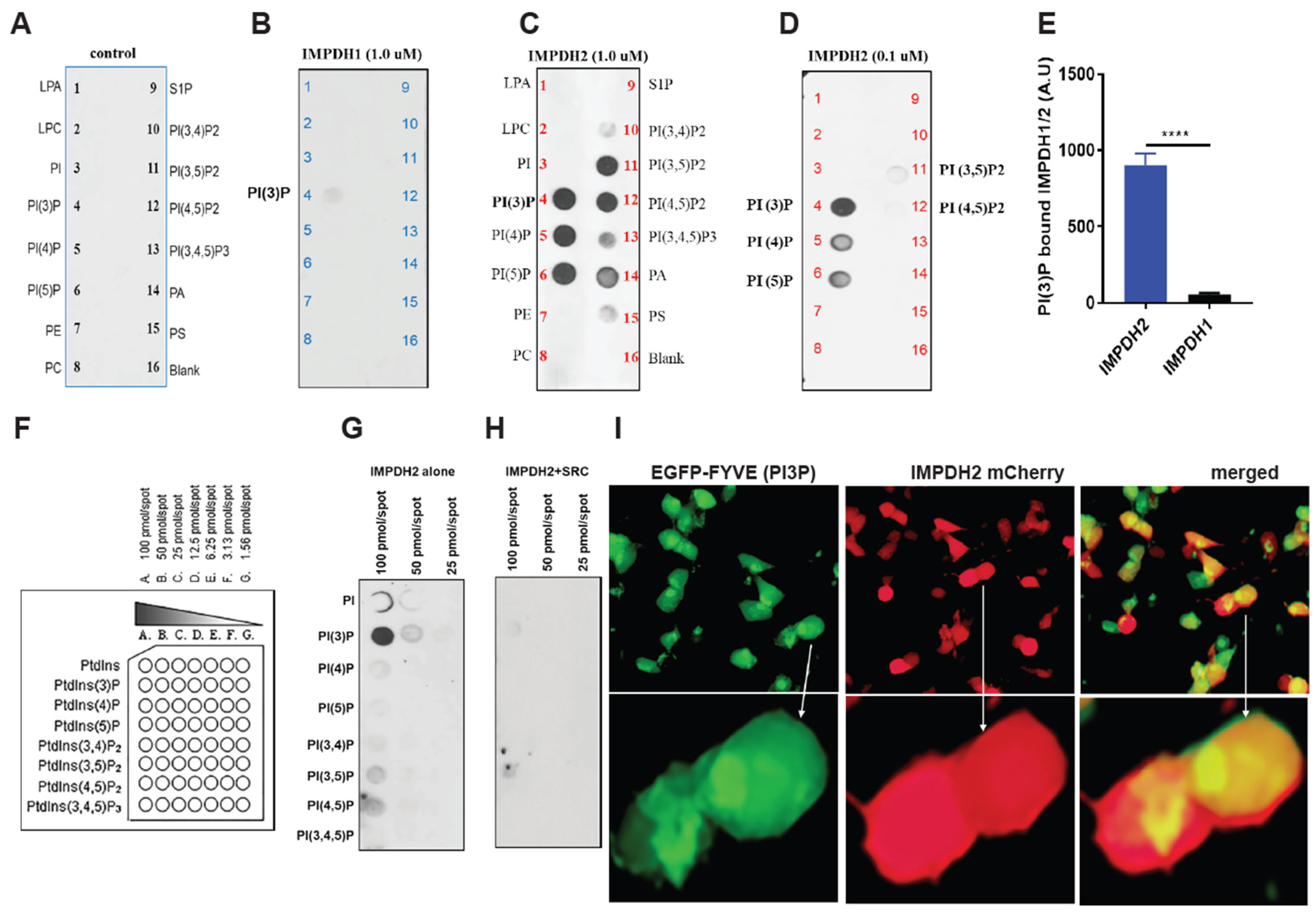
Figure 3.
PI3P binding inhibits IMPDH2 enzymatic activity. A, B, Chemical structures of phospholipids PI3P and PIP2 used in this study. C, Recombinant IMPDH2 was pre-incubated with increasing concentrations of synthetic PI3P; enzymatic activity was measured and showed dose-dependent inhibition. D, Recombinant IMPDH1 was similarly pre-incubated with PI3P to assess its sensitivity to PI3P-mediated inhibition. E, Comparative enzymatic activity of recombinant IMPDH1 and IMPDH2 pre-incubated with 200 µg synthetic PI3P or MPA (mycophenolic acid, a known IMPDH inhibitor) as a positive control. F, Recombinant IMPDH1 was pre-incubated with increasing concentrations of synthetic PIP2; no significant inhibition of enzymatic activity was observed. G, Recombinant IMPDH2 pre-incubated with synthetic PI3P confirms reproducible, dose-dependent inhibition of enzymatic activity. All graphs display mean ± SD from three biological replicates (n = 3). Statistical significance was calculated using unpaired t-tests: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Figure 3.
PI3P binding inhibits IMPDH2 enzymatic activity. A, B, Chemical structures of phospholipids PI3P and PIP2 used in this study. C, Recombinant IMPDH2 was pre-incubated with increasing concentrations of synthetic PI3P; enzymatic activity was measured and showed dose-dependent inhibition. D, Recombinant IMPDH1 was similarly pre-incubated with PI3P to assess its sensitivity to PI3P-mediated inhibition. E, Comparative enzymatic activity of recombinant IMPDH1 and IMPDH2 pre-incubated with 200 µg synthetic PI3P or MPA (mycophenolic acid, a known IMPDH inhibitor) as a positive control. F, Recombinant IMPDH1 was pre-incubated with increasing concentrations of synthetic PIP2; no significant inhibition of enzymatic activity was observed. G, Recombinant IMPDH2 pre-incubated with synthetic PI3P confirms reproducible, dose-dependent inhibition of enzymatic activity. All graphs display mean ± SD from three biological replicates (n = 3). Statistical significance was calculated using unpaired t-tests: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
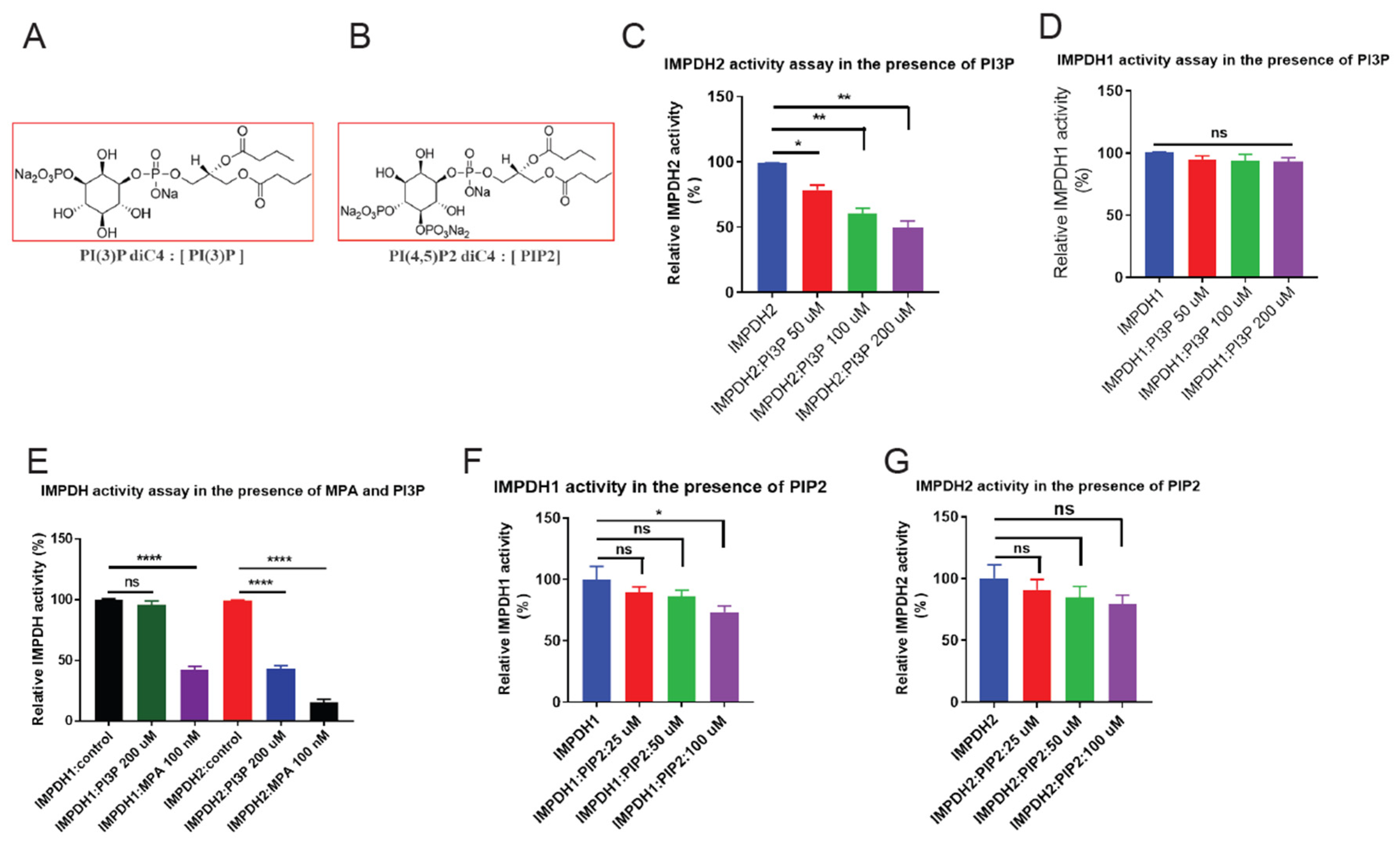
Figure 4.
Structure-based virtual screening identifies a novel small-molecule inhibitor of human IMPDH2. A, Ribbon diagram of the CBS (cystathionine β-synthase) domain of human inosine monophosphate dehydrogenase 2 (IMPDH2), illustrating the secondary structural elements. B, Surface representation of the GTP-binding pocket (highlighted in yellow) within the IMPDH2 structure, showing the targeted allosteric site for compound docking. C, Schematic overview of the in silico screening pipeline, including structure-based virtual screening, docking, and filtering steps leading to hit identification. D, Chemical structures of 15 top-ranked compounds identified from virtual screening; compound 10 (comp-10) was selected for further validation based on docking scores and predicted binding interactions. E, Molecular docking pose of comp-10 within the GTP-binding pocket of human IMPDH2 (based on the PDB reference structure), revealing key interactions with the binding site residues. F, In vitro enzymatic activity assay of recombinant IMPDH2 in the presence of comp-10, showing concentration-dependent inhibition and determination of the half-maximal inhibitory concentration (IC50). G, Enzymatic assay using the clinically established IMPDH inhibitor mycophenolic acid (MPA) as a positive control for benchmarking IC50 and assay reproducibility. H–J, Inhibition of endogenous IMPDH activity by comp-10 in human lymphoma cell lines: H, anaplastic large cell lymphoma (ALCL); I, mantle cell lymphoma (MCL); and J, diffuse large B-cell lymphoma (DLBCL). Comp-10 treatment significantly reduced IMPDH enzymatic activity across all tested cell lines. All the graphs show mean ± SD (n = 3 biological replicates), and all statistical analyses were conducted with unpaired t-tests: ∗p<0.05,∗∗p< 0.01,∗∗∗p< 0.001,∗∗∗∗p< 0.0001.
Figure 4.
Structure-based virtual screening identifies a novel small-molecule inhibitor of human IMPDH2. A, Ribbon diagram of the CBS (cystathionine β-synthase) domain of human inosine monophosphate dehydrogenase 2 (IMPDH2), illustrating the secondary structural elements. B, Surface representation of the GTP-binding pocket (highlighted in yellow) within the IMPDH2 structure, showing the targeted allosteric site for compound docking. C, Schematic overview of the in silico screening pipeline, including structure-based virtual screening, docking, and filtering steps leading to hit identification. D, Chemical structures of 15 top-ranked compounds identified from virtual screening; compound 10 (comp-10) was selected for further validation based on docking scores and predicted binding interactions. E, Molecular docking pose of comp-10 within the GTP-binding pocket of human IMPDH2 (based on the PDB reference structure), revealing key interactions with the binding site residues. F, In vitro enzymatic activity assay of recombinant IMPDH2 in the presence of comp-10, showing concentration-dependent inhibition and determination of the half-maximal inhibitory concentration (IC50). G, Enzymatic assay using the clinically established IMPDH inhibitor mycophenolic acid (MPA) as a positive control for benchmarking IC50 and assay reproducibility. H–J, Inhibition of endogenous IMPDH activity by comp-10 in human lymphoma cell lines: H, anaplastic large cell lymphoma (ALCL); I, mantle cell lymphoma (MCL); and J, diffuse large B-cell lymphoma (DLBCL). Comp-10 treatment significantly reduced IMPDH enzymatic activity across all tested cell lines. All the graphs show mean ± SD (n = 3 biological replicates), and all statistical analyses were conducted with unpaired t-tests: ∗p<0.05,∗∗p< 0.01,∗∗∗p< 0.001,∗∗∗∗p< 0.0001.
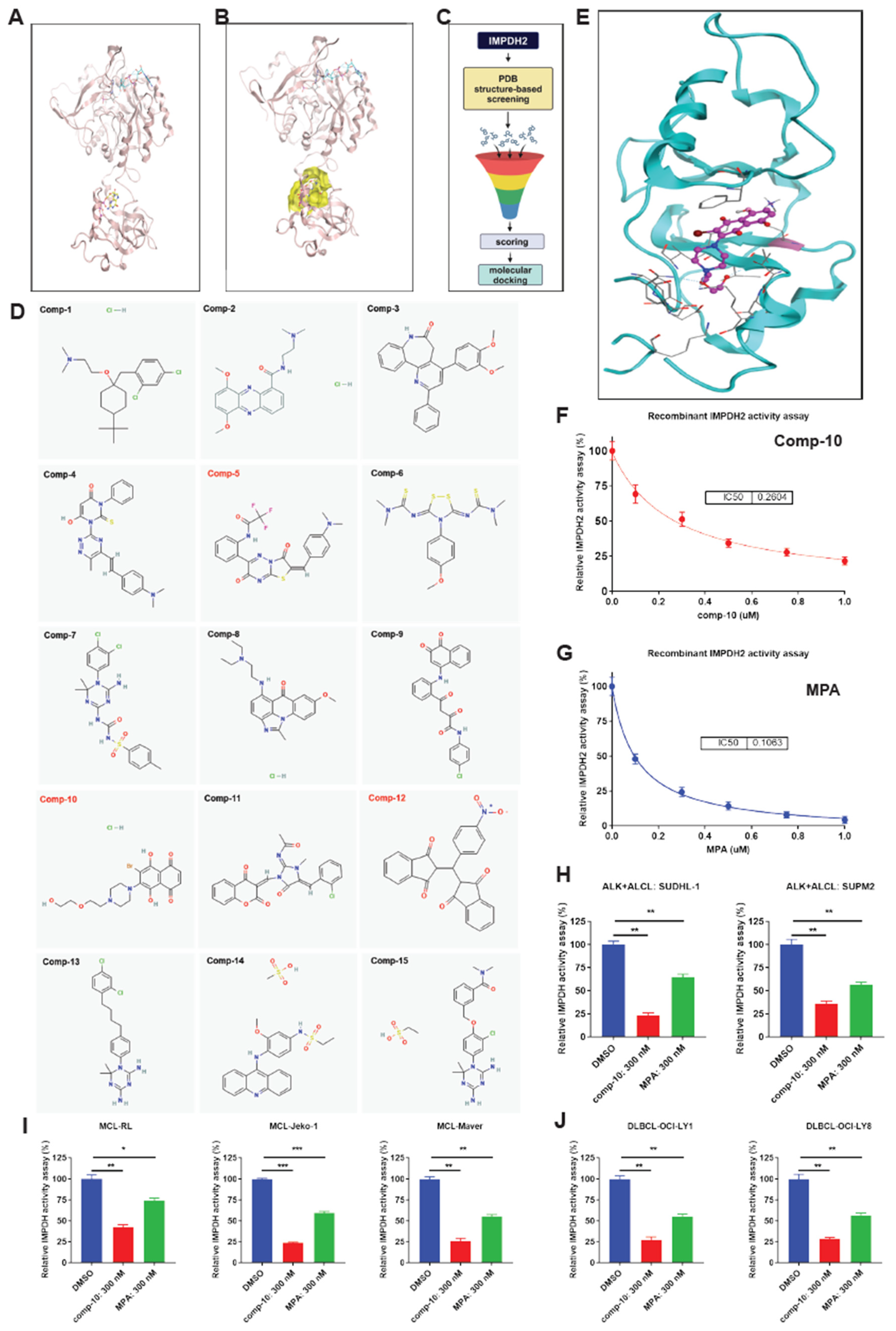
Figure 5.
Comp-10 downregulates IMPDH1/2 expression and prevents filament (rod/ring) formation in ALCL and MCL models. A, Western blot analysis of IMPDH1 and IMPDH2 expression in ALK inhibitor (ALKi)-sensitive ALCL cell lines (SUPM2 and L82) following 48-hour treatment with Comp-10 (300 nM) or mycophenolic acid (MPA). B, C, Densitometric quantification of IMPDH1 and IMPDH2 protein bands from panel A, confirming significant downregulation following Comp-10 treatment. D, Western blot of ALKi-resistant ALCL cell lines (Karpas299 and SUPM2-CR) treated with Comp-10 and MPA, showing consistent downregulation of IMPDH1/2 by Comp-10 and upregulation by MPA. E, F, Densitometric analysis of protein expression in resistant ALCL lines (panel D) supports differential regulation by Comp-10 and MPA. G, Quantitative RT–PCR analysis of IMPDH2 mRNA expression in MCL cell line Maver treated with Comp-10, MPA and MF. H, Western blot analysis of BTK inhibitor (BTKi) sensitive and resistant MCL cell lines treated with Comp-10 and MPA. I, Densitometric quantification of Western blots shown in panel H, confirming significant downregulation of IMPDH1/2 protein levels by Comp-10 across multiple MCL models. J–L, Light microscopy of SUDHL-1 cells expressing HA–GFP–IMPDH2 shows distinct patterns of filament formation: DMSO-treated control cells (J) lack filaments; MPA-treated cells (K) display prominent rod/ring structures; and Comp-10-treated cells (L) show no rod/ring or filament formation. Densitometric values of the Western blots (N=2). All the graphs show mean ± SD (n = 3 biological replicates), and all statistical analyses were conducted with unpaired t-tests:∗p<0.05,∗∗p< 0.01,∗∗∗p< 0.001,∗∗∗∗p< 0.0001.
Figure 5.
Comp-10 downregulates IMPDH1/2 expression and prevents filament (rod/ring) formation in ALCL and MCL models. A, Western blot analysis of IMPDH1 and IMPDH2 expression in ALK inhibitor (ALKi)-sensitive ALCL cell lines (SUPM2 and L82) following 48-hour treatment with Comp-10 (300 nM) or mycophenolic acid (MPA). B, C, Densitometric quantification of IMPDH1 and IMPDH2 protein bands from panel A, confirming significant downregulation following Comp-10 treatment. D, Western blot of ALKi-resistant ALCL cell lines (Karpas299 and SUPM2-CR) treated with Comp-10 and MPA, showing consistent downregulation of IMPDH1/2 by Comp-10 and upregulation by MPA. E, F, Densitometric analysis of protein expression in resistant ALCL lines (panel D) supports differential regulation by Comp-10 and MPA. G, Quantitative RT–PCR analysis of IMPDH2 mRNA expression in MCL cell line Maver treated with Comp-10, MPA and MF. H, Western blot analysis of BTK inhibitor (BTKi) sensitive and resistant MCL cell lines treated with Comp-10 and MPA. I, Densitometric quantification of Western blots shown in panel H, confirming significant downregulation of IMPDH1/2 protein levels by Comp-10 across multiple MCL models. J–L, Light microscopy of SUDHL-1 cells expressing HA–GFP–IMPDH2 shows distinct patterns of filament formation: DMSO-treated control cells (J) lack filaments; MPA-treated cells (K) display prominent rod/ring structures; and Comp-10-treated cells (L) show no rod/ring or filament formation. Densitometric values of the Western blots (N=2). All the graphs show mean ± SD (n = 3 biological replicates), and all statistical analyses were conducted with unpaired t-tests:∗p<0.05,∗∗p< 0.01,∗∗∗p< 0.001,∗∗∗∗p< 0.0001.
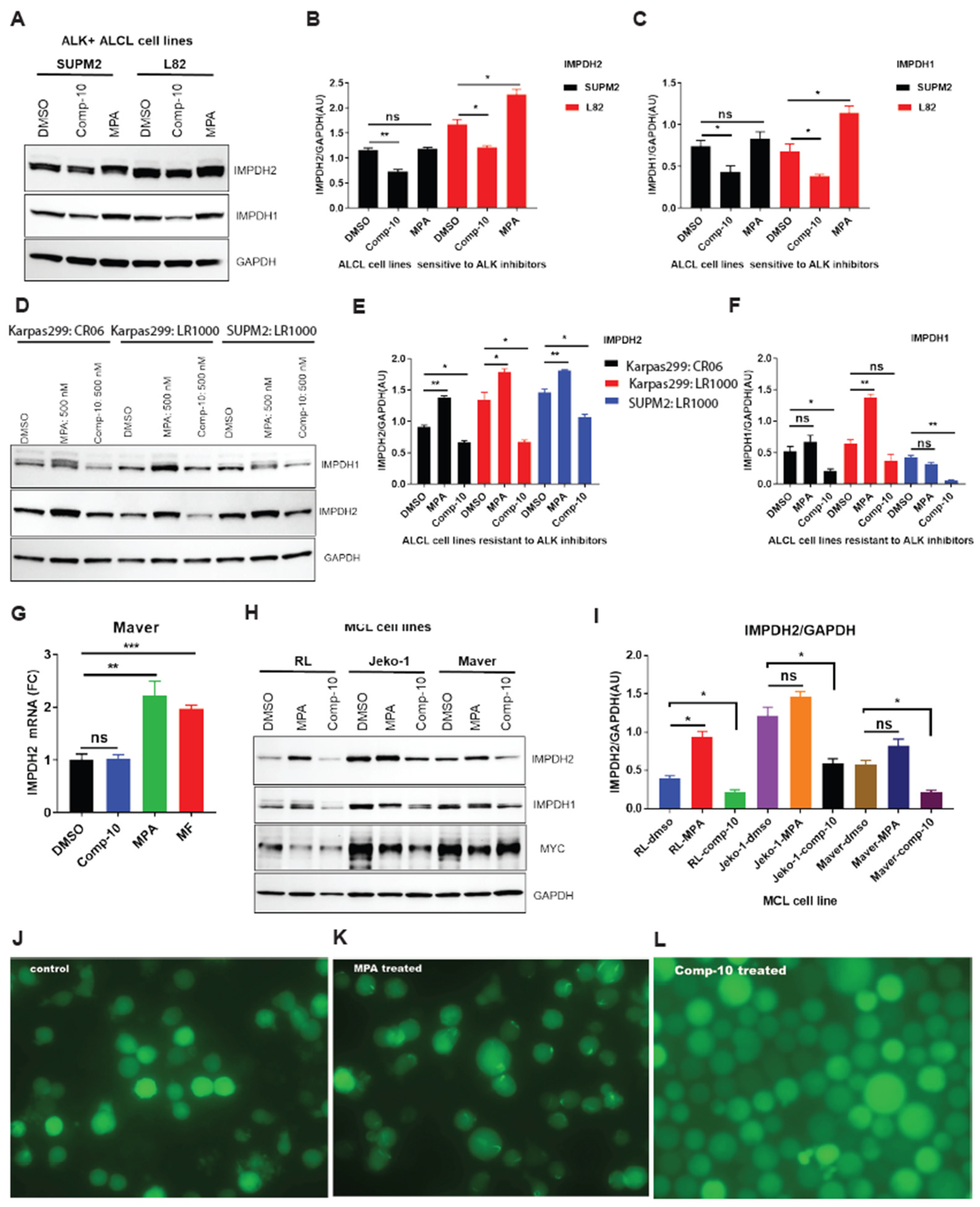
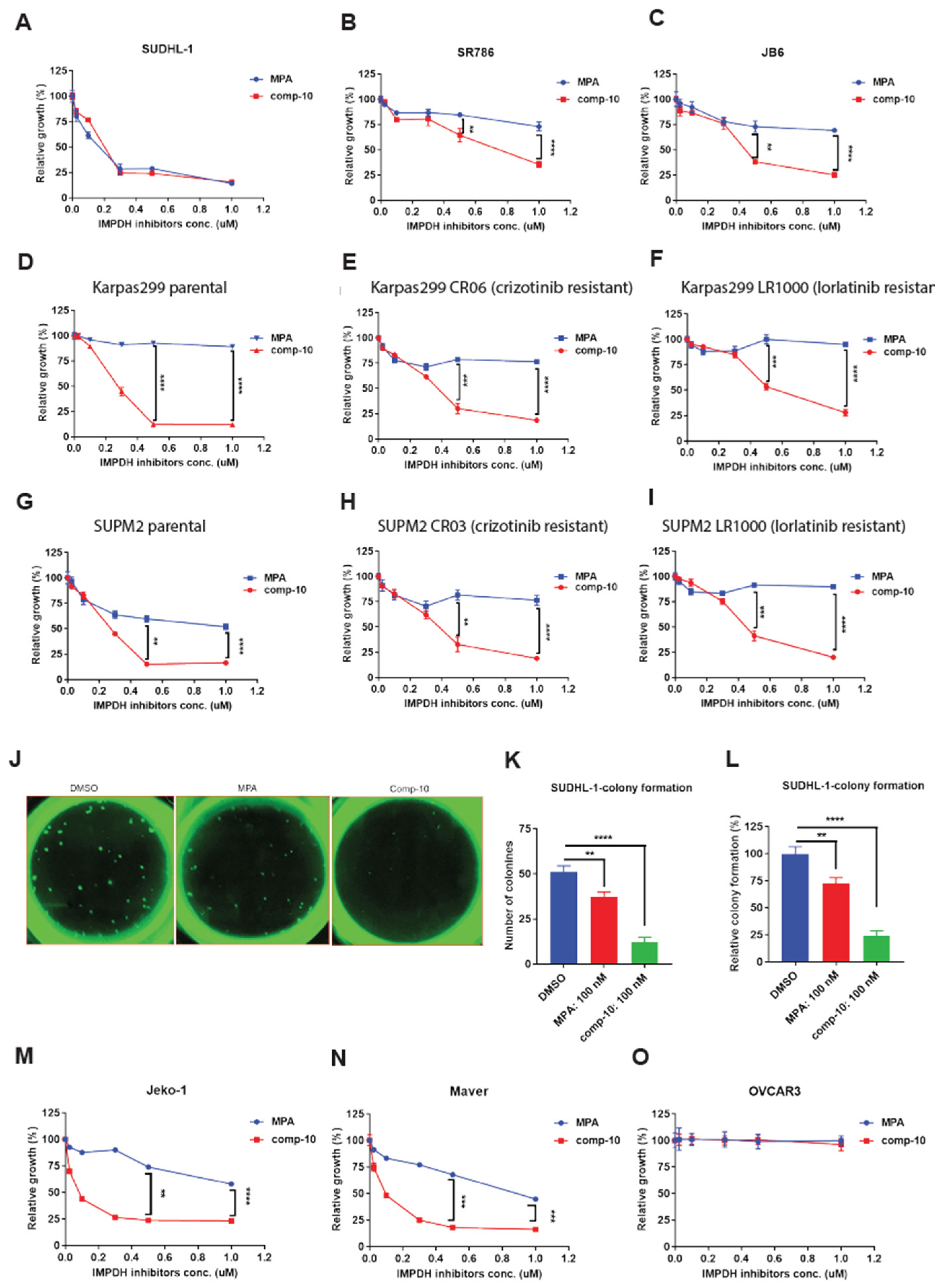
Figure 6.
Comp-10 inhibits proliferation and colony formation in ALK-positive lymphoma cells. A–C, Cell viability assays of ALCL cell lines treated with comp-10 or mycophenolic acid (MPA). While both compounds similarly inhibited growth in one line (A), comp-10 showed significantly stronger growth inhibition in two additional lines (B, C). D–F, Comp-10 potently suppressed the growth of ALK inhibitor (ALKi)-sensitive Karpas 299 cells (D) and their resistant derivatives: Crizotinib-resistant (e) and Lorlatinib-resistant (F) lines. G–I, Similar results were observed in parental SUP-M2 cells (G) and their ALKi-resistant derivatives (H, I), with comp-10 maintaining robust antiproliferative activity. J, Representative GFP fluorescence images of colonies formed by SUDHL-1 cells stably expressing IMPDH2–HA–GFP in methylcellulose after 4 weeks of treatment with DMSO, comp-10 (100 nM), or MPA (100 nM). K, Quantification of colony numbers per well. Comp-10 significantly reduced colony formation compared to both DMSO (p < 0.0001) and MPA (p < 0.01). L, Colony numbers expressed as percentage of DMSO control. Comp-10 reduced colony formation to ~12%, while MPA-treated cells retained >72% colony formation relative to control. M, N, Cell viability assays in two mantle cell lymphoma (MCL) index lines demonstrate that comp-10 effectively inhibits cell growth than MPA. O, Growth of the ovarian carcinoma cell line OVCAR3 remained unaffected by comp-10 or MPA, indicating minimal off-target toxicity in non-lymphoid malignancies. All the graphs show mean ± SD (n = 3 biological replicates), and all statistical analyses were conducted with unpaired t-tests: ∗p<0.05,∗∗p< 0.01,∗∗∗p< 0.001,∗∗∗∗p< 0.0001.
Figure 6.
Comp-10 inhibits proliferation and colony formation in ALK-positive lymphoma cells. A–C, Cell viability assays of ALCL cell lines treated with comp-10 or mycophenolic acid (MPA). While both compounds similarly inhibited growth in one line (A), comp-10 showed significantly stronger growth inhibition in two additional lines (B, C). D–F, Comp-10 potently suppressed the growth of ALK inhibitor (ALKi)-sensitive Karpas 299 cells (D) and their resistant derivatives: Crizotinib-resistant (e) and Lorlatinib-resistant (F) lines. G–I, Similar results were observed in parental SUP-M2 cells (G) and their ALKi-resistant derivatives (H, I), with comp-10 maintaining robust antiproliferative activity. J, Representative GFP fluorescence images of colonies formed by SUDHL-1 cells stably expressing IMPDH2–HA–GFP in methylcellulose after 4 weeks of treatment with DMSO, comp-10 (100 nM), or MPA (100 nM). K, Quantification of colony numbers per well. Comp-10 significantly reduced colony formation compared to both DMSO (p < 0.0001) and MPA (p < 0.01). L, Colony numbers expressed as percentage of DMSO control. Comp-10 reduced colony formation to ~12%, while MPA-treated cells retained >72% colony formation relative to control. M, N, Cell viability assays in two mantle cell lymphoma (MCL) index lines demonstrate that comp-10 effectively inhibits cell growth than MPA. O, Growth of the ovarian carcinoma cell line OVCAR3 remained unaffected by comp-10 or MPA, indicating minimal off-target toxicity in non-lymphoid malignancies. All the graphs show mean ± SD (n = 3 biological replicates), and all statistical analyses were conducted with unpaired t-tests: ∗p<0.05,∗∗p< 0.01,∗∗∗p< 0.001,∗∗∗∗p< 0.0001.