Submitted:
17 September 2025
Posted:
17 September 2025
You are already at the latest version
Abstract
The reliance on synthetic repellents such as N,N-diethyl-meta-toluamide (DEET) has raised health and environmental concerns, prompting the search for safer, plant-derived alternatives. Catnip (Nepeta cataria L.) is a rich source of iridoid monoterpenes, particularly nepetalactones, which are well known for their strong insect-repellent properties. However, the efficient extraction of nepetalactones remains challenging, and their precise mechanisms of action in insect inhibition are not yet fully understood. Thus, this study investigated the chemical composition from various methods, protein–ligand interactions, and pharmacokinetic safety profiles of catnip-derived compounds compared to DEET, with a focus on their interactions with odorant-binding proteins (OBPs) from Anopheles gambiae (AgamOBP), Culex quinquefasciatus (CquiOBP), and Aedes aegypti (AaegOBP). Gas chromatography–tandem mass spectrometry (GC–MS/MS) confirmed the presence of nepetalactone isomers as the major constituents in catnip extracts obtained through stream distillation and dried leaves extracted in olive oil fractions. Molecular docking revealed that cis,cis- and cis,trans-nepetalactones and nepetalactone exhibited high binding affinities, surpassing those of DEET. Molecular dynamics simulations demonstrated that all OBP–ligand complexes achieved stable conformations. Notably, cis,trans-nepetalactone formed a more stable complex with AgamOBP than DEET. These findings suggest that nepetalactones stabilize OBP–ligand interactions while inducing subtle conformational flexibility, potentially disrupting mosquito odorant recognition in a manner distinct from DEET. ADMET predictions indicated that nepetalactones exhibit favorable absorption, distribution, and safety profiles with reduced predicted toxicity compared to DEET. Collectively, these results establish nepetalactones as promising candidates for the development of effective, safe, and sustainable plant-based repellents.
Keywords:
1. Introduction
2. Results
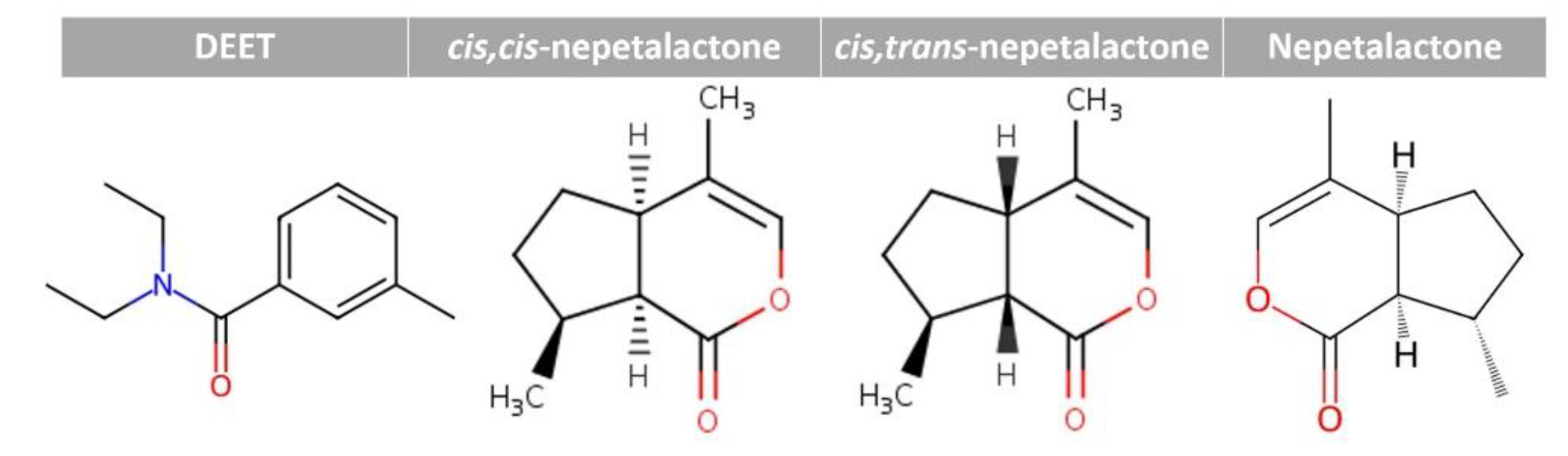
2.1. GC–MS/MS Identification of Catnip Extracts
2.2. Structural Analysis of OBP Receptors
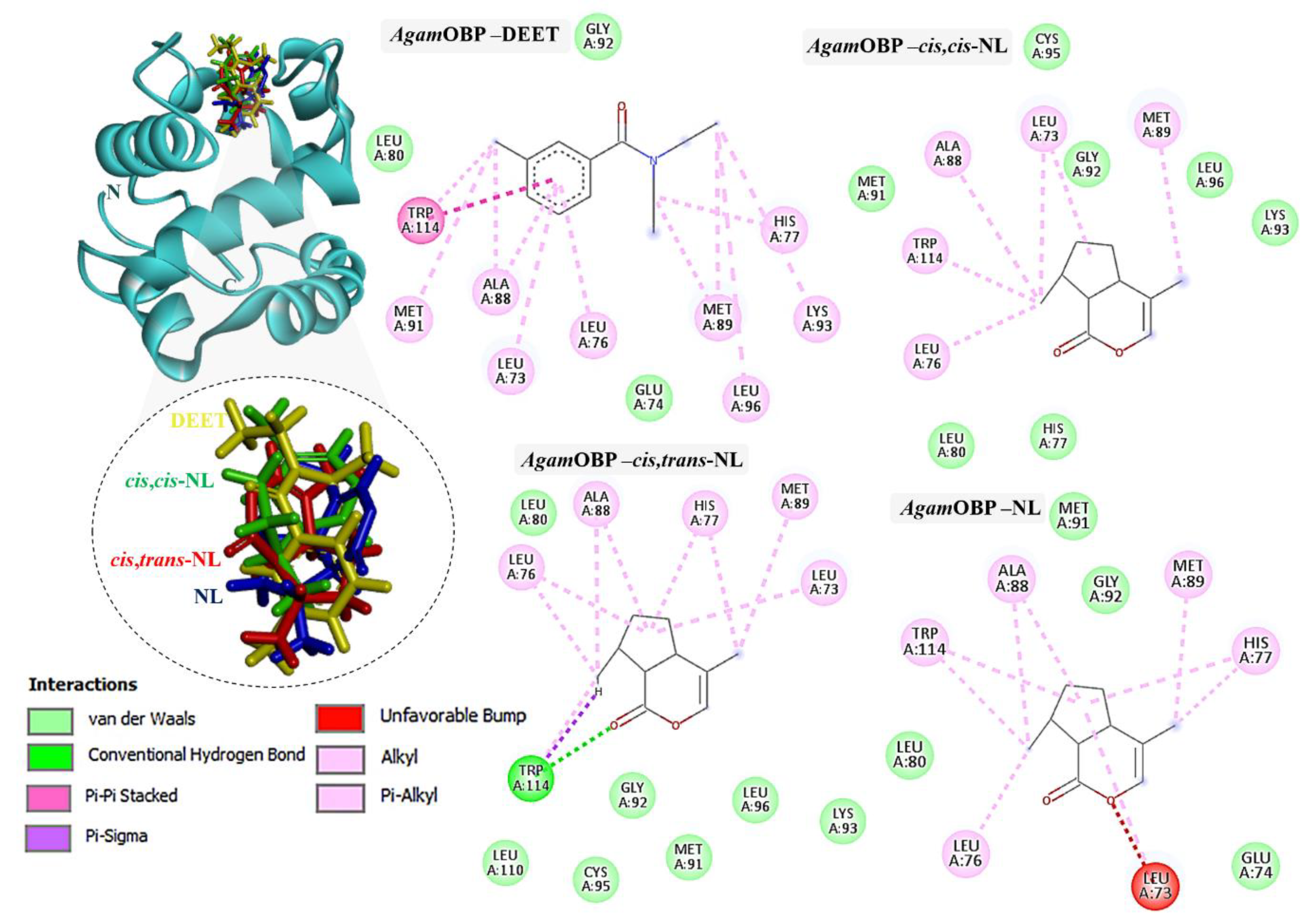
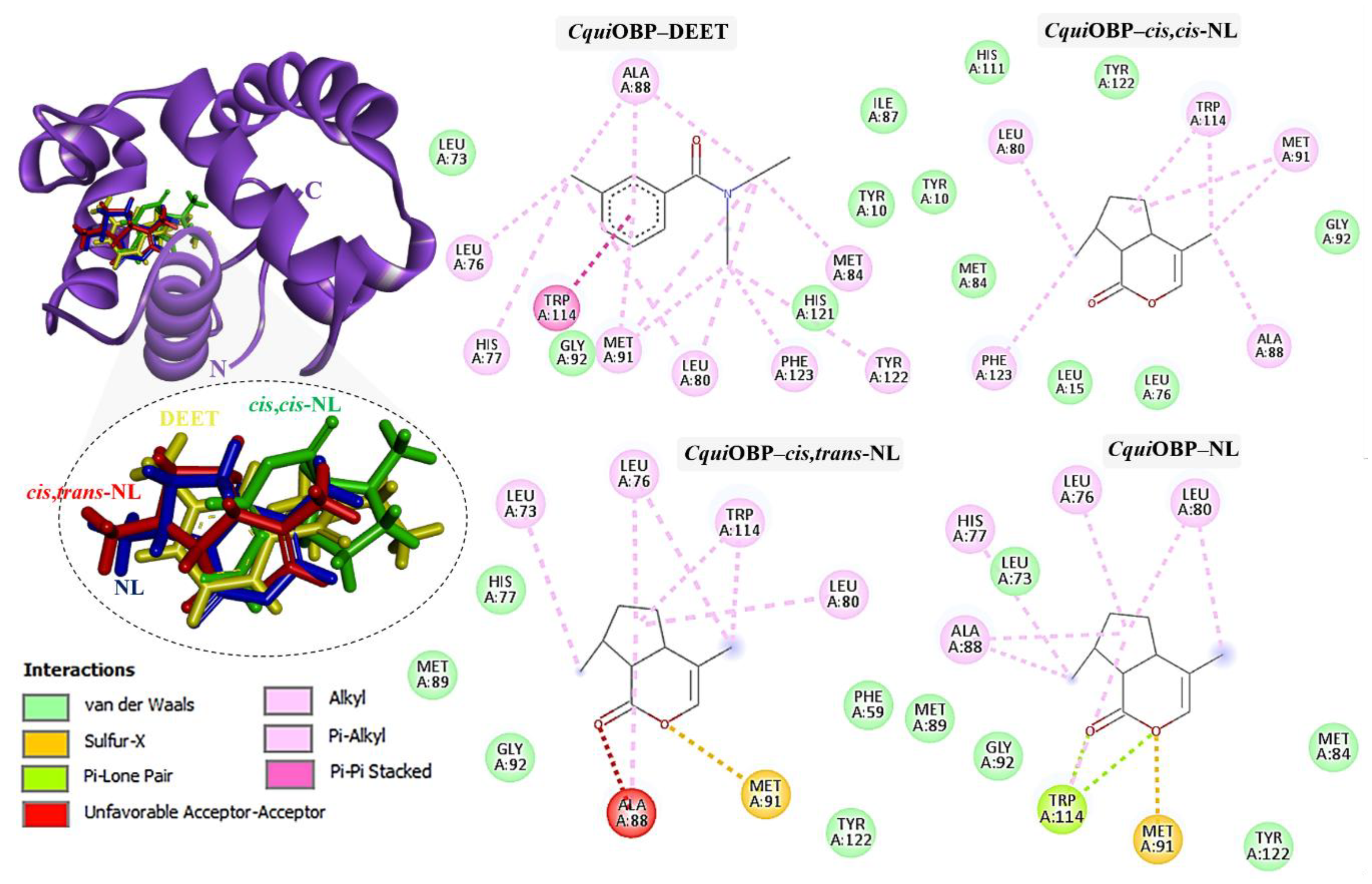
2.3. Molecular Docking of DEET and Nepetalactone Isomers with OBPs
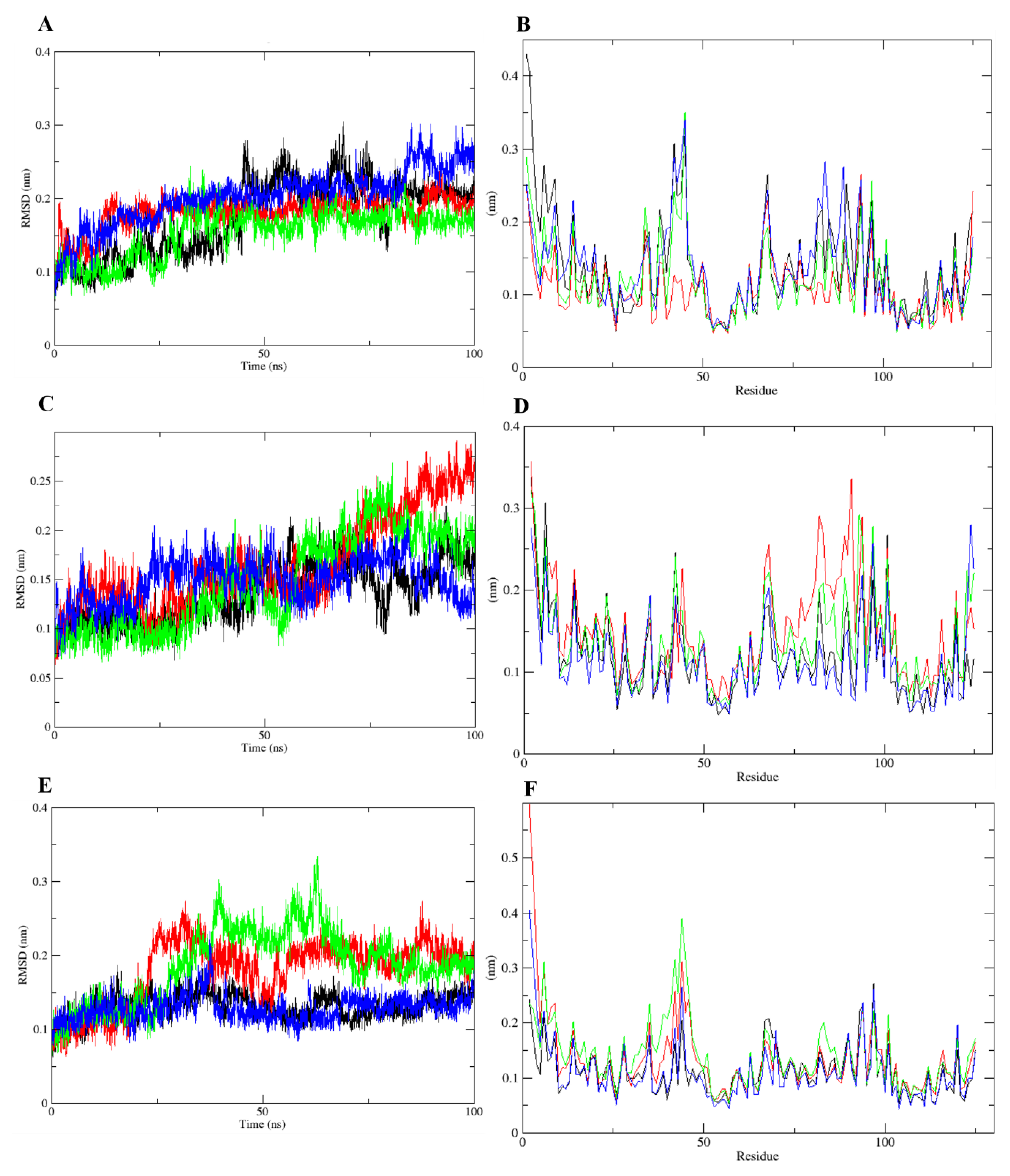
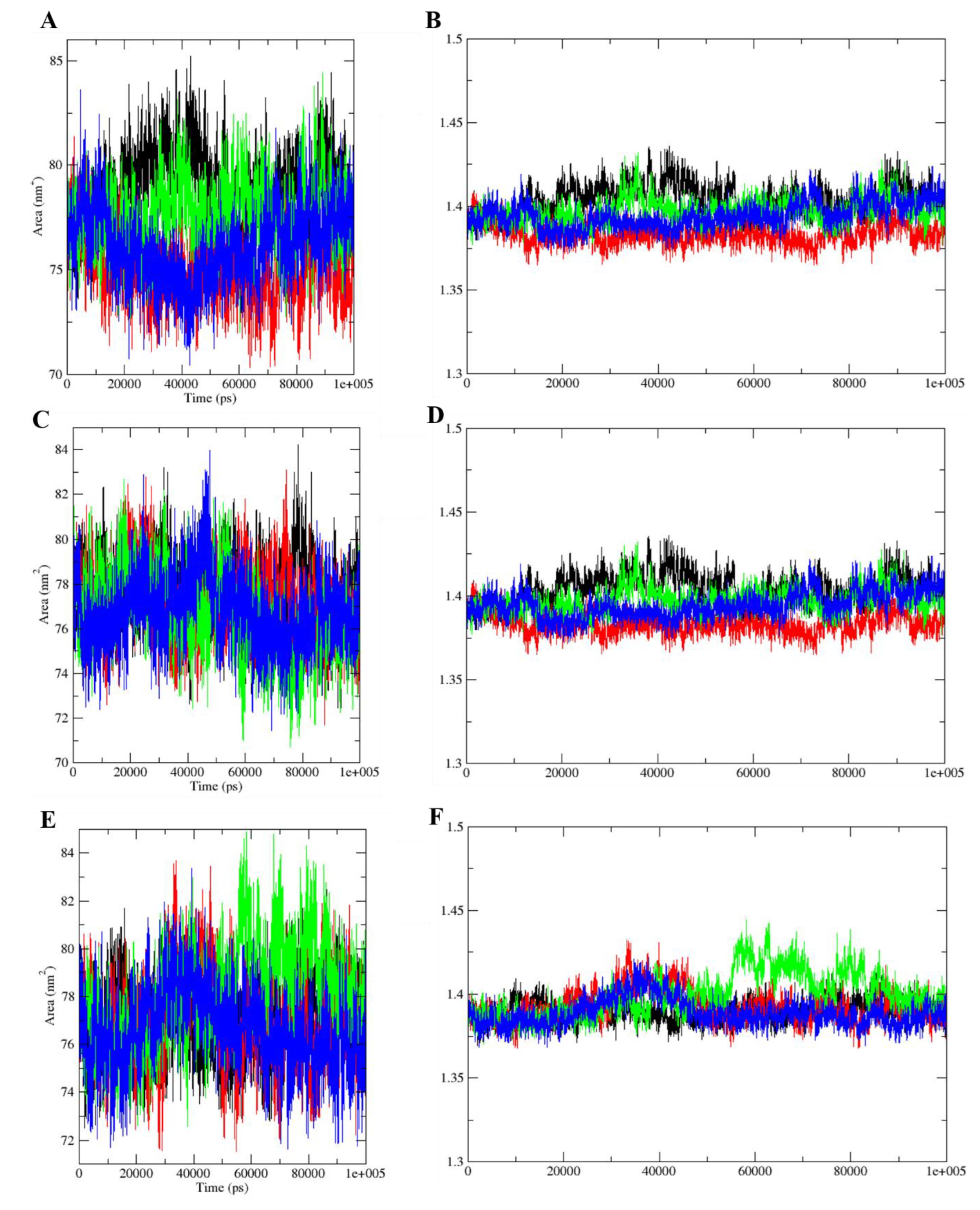
2.4. Molecular Dynamics Simulations Analysis
2.5. ADMET Analysis
3. Discussion
4. Materials and Methods
4.1. Preparation of Nepeta Cataria Leaf Extraction
4.1.1. Extraction via Steam Distillation
4.1.2. Extraction by the Maceration Method Using Olive Oil Solvent
4.2. Identification of Chemical Compositions by Using GC-MS/MS
4.3. Structural Analysis and Comparison of AgamOBP, CquiOBP, and AaegOBP
4.4. Molecular Docking of N. cataria Phytochemicals with the OBP Receptors
4.5. Molecular Dynamics Simulations and Analysis
4.6. ADMET Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DEET | N,N-diethyl-meta-toluamide |
| TRPA1 | Transient receptor potential ankyrin 1 |
| OBP | Odorant-binding protein |
| GC–MS/MS | Gas chromatography–tandem mass spectrometry |
| ADMET | Absorption, Distribution, Metabolism, Excretion, and Toxicity |
| SDE | Stream distillation extraction |
| FOE | Fresh leaf in olive oil extraction |
| DOE | Dried leaf in olive oil extraction |
| NL | Nepetalactone |
| cis,cis-NL | cis,cis-nepetalactone |
| cis,trans-NL | cis,trans-nepetalactone |
References
- Onen, H.; Luzala, M. M.; Kigozi, S.; Sikumbili, R. M.; Muanga, C. K.; Zola, E. N.; Wendji, S. N.; Buya, A. B.; Balciunaitiene, A.; Viškelis, J.; Kaddumukasa, M. A.; Memvanga, P. B. Mosquito-Borne Diseases and Their Control Strategies: An Overview Focused on Green Synthesized Plant-Based Metallic Nanoparticles. Insects 2023, 14, 221. [Google Scholar] [CrossRef]
- Bamou, R.; Mayi, M. P. A.; Djiappi-Tchamen, B.; Nana-Ndjangwo, S. M.; Nchoutpouen, E.; Cornel, A. J.; Awono-Ambene, P.; Parola, P.; Tchuinkam, T.; Antonio-Nkondjio, C. An Update on the Mosquito Fauna and Mosquito-Borne Diseases Distribution in Cameroon. Parasit. Vectors 2021, 14, 527. [Google Scholar] [CrossRef]
- Leal, W. S. The Enigmatic Reception of DEET—The Gold Standard of Insect Repellents. Curr. Opin. Insect Sci. 2014, 6, 93–98. [Google Scholar] [CrossRef]
- Rodriguez, S. D.; Drake, L. L.; Price, D. P.; Hammond, J. I.; Hansen, I. A. The Efficacy of Some Commercially Available Insect Repellents for Aedes aegypti (Diptera: Culicidae) and Aedes albopictus (Diptera: Culicidae). J. Insect Sci. 2015, 15, 140. [Google Scholar] [CrossRef] [PubMed]
- Chen-Hussey, V.; Behrens, R.; Logan, J. G. Assessment of Methods Used To Determine the Safety of the Topical Insect Repellent N,N-Diethyl-m-Toluamide (DEET). Parasit. Vectors 2014, 7, 173. [Google Scholar] [CrossRef]
- Roy, D. N.; Goswami, R.; Pal, A. The Insect Repellents: A Silent Environmental Chemical Toxicant to the Health. Environ. Toxicol. Pharmacol. 2017, 50, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Hazarika, H.; Krishnatreyya, H. Technological Advancements in Mosquito Repellents: Challenges and Opportunities in Plant-Based Repellents. Acta Parasitol. 2025, 70, 117. [Google Scholar] [CrossRef] [PubMed]
- da Costa, K. S.; Galúcio, J. M.; da Costa, C. H. S.; Santana, A. R.; Dos Santos Carvalho, V.; do Nascimento, L. D.; Lima e Lima, A. H.; Neves Cruz, J.; Alves, C. N.; Lameira, J. Exploring the Potentiality of Natural Products from Essential Oils as Inhibitors of Odorant-Binding Proteins: A Structure- and Ligand-Based Virtual Screening Approach To Find Novel Mosquito Repellents. ACS Omega 2019, 4, 22475–22486. [Google Scholar] [CrossRef]
- Melo, N.; Capek, M.; Arenas, O. M.; Afify, A.; Yilmaz, A.; Potter, C. J.; Laminette, P. J.; Para, A.; Gallio, M.; Stensmyr, M. C. The Irritant Receptor TRPA1 Mediates the Mosquito Repellent Effect of Catnip. Curr. Biol. 2021, 31, 1988–1994.e5. [Google Scholar] [CrossRef]
- Reichert, W.; Ejercito, J.; Guda, T.; Dong, X.; Wu, Q.; Ray, A.; Simon, J. E. Repellency Assessment of Nepeta cataria Essential Oils and Isolated Nepetalactones on Aedes aegypti. Sci. Rep. 2019, 9, 1524. [Google Scholar] [CrossRef]
- Birkett, M. A.; Hassanali, A.; Hoglund, S.; Pettersson, J.; Pickett, J. A. Repellent Activity of Catmint, Nepeta cataria, and Iridoid Nepetalactone Isomers against Afro-Tropical Mosquitoes, Ixodid Ticks and Red Poultry Mites. Phytochemistry 2011, 72, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Sun, J. S.; Xiao, S.; Carlson, J. R. The Diverse Small Proteins Called Odorant-Binding Proteins. Open Biol. 2018, 8, 180208. [Google Scholar] [CrossRef]
- Duarte, J. L.; Di Filippo, L. D.; Ribeiro, T. d. C.; Silva, A. C. d. J.; Hage-Melim, L. I. d. S.; Duchon, S.; Carrasco, D.; Pinto, M. C.; Corbel, V.; Chorilli, M. Effective Mosquito Repellents: Myrcene- and Cymene-Loaded Nanohydrogels against Aedes aegypti. Pharmaceutics 2024, 16, 1096. [Google Scholar] [CrossRef] [PubMed]
- Pinnelli, G. R.; Terrado, M.; Hillier, N.; Lance, D.; Plettner, E. Synthesis of Isotopically Labelled Disparlure Enantiomers and Application to the Study of Enantiomer Discrimination in Gypsy Moth Pheromone-Binding Proteins. Eur. J. Org. Chem. 2019, 2019. [Google Scholar] [CrossRef]
- Nonkhwao, S.; Plettner, E.; Daduang, S. Protein-Ligand Binding and Structural Modelling Studies of Pheromone-Binding Protein-like Sol g 2.1 from Solenopsis geminata Fire Ant Venom. Molecules 2024, 29, 1033. [Google Scholar] [CrossRef]
- Terrado, M.; Pinnelli, G. R.; Sanes, J.; Plettner, E. Binding Interactions, Structure–Activity Relationships and Blend Effects in Pheromone and Host Olfactory Detection of Herbivorous Lepidoptera. In Insect Pheromone Biochemistry and Molecular Biology; Springer, 2019; pp 1–25. [CrossRef]
- Sims, C.; Birkett, M. A.; Withall, D. M. Enantiomeric Discrimination in Insects: The Role of OBPs and ORs. Insects 2022, 13, 368. [Google Scholar] [CrossRef]
- Rao, P.; Goswami, D.; Rawal, R. M. Molecular Insights on Ar-Turmerone as a Structural, Functional and Pharmacophoric Analogue of Synthetic Mosquito Repellent DEET by Comprehensive Computational Assessment. Sci. Rep. 2022, 12, 15564. [Google Scholar] [CrossRef]
- Moustakime, Y.; Hazzoumi, Z.; Amrani Joutei, K. Aromatization of Virgin Olive Oil by Seeds of Pimpinella anisum Using Three Different Methods: Physico-Chemical Change and Thermal Stability of Flavored Oils. Grain Oil Sci. Technol. 2021, 4, 108–124. [Google Scholar] [CrossRef]
- Mohammadi, S.; Saharkhiz, M. Changes in Essential Oil Content and Composition of Catnip (Nepeta cataria L.) during Different Developmental Stages. J. Essent. Oil Bear. Plants 2013, 14, 396–400. [Google Scholar] [CrossRef]
- Gkinis, G.; Tzakou, O.; Iliopoulou, D.; Roussis, V. Chemical Composition and Biological Activity of Nepeta parnassica Oils and Isolated Nepetalactones. Z. Naturforsch., C: J. Biosci. 2003, 58, 681–686. [Google Scholar] [CrossRef] [PubMed]
- Noosidum, A.; Chareonviriyaphap, T.; Chandrapatya, A. Synergistic Repellent and Irritant Effect of Combined Essential Oils on Aedes aegypti (L.) Mosquitoes. J. Vector Ecol. 2014, 39, 298–305. [Google Scholar] [CrossRef]
- Manoharan, M.; Ng Fuk Chong, M.; Vaïtinadapoulé, A.; Frumence, E.; Sowdhamini, R.; Offmann, B. Comparative Genomics of Odorant Binding Proteins in Anopheles gambiae, Aedes aegypti, and Culex quinquefasciatus. Genome Biol. Evol. 2013, 5, 163–180. [Google Scholar] [CrossRef]
- Dassanayake, M. K.; Chong, C. H.; Khoo, T. J.; Figiel, A.; Szumny, A.; Choo, C. M. Synergistic Field Crop Pest Management Properties of Plant-Derived Essential Oils in Combination with Synthetic Pesticides and Bioactive Molecules: A Review. Foods 2021, 10, 2016. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, X.; Gan, J.; Chen, S.; Xiao, Z. X.; Cao, Y. CB-Dock2: Improved Protein–Ligand Blind Docking by Integrating Cavity Detection, Docking and Homologous Template Fitting. Nucleic Acids Res. 2022, 50, W159–W164. [Google Scholar] [CrossRef]
- Flores-Castañón, N.; Sarkar, S.; Banerjee, A. Structural, Functional, and Molecular Docking Analyses of Microbial Cutinase Enzymes against Polyurethane Monomers. J. Hazard. Mater. Lett. 2022, 3, 100063. [Google Scholar] [CrossRef]
- Ali, S.; et al. Identification and Evaluation of Inhibitors of Lipase from Malassezia restricta Using Virtual High-Throughput Screening and Molecular Dynamics Studies. Int. J. Mol. Sci. 2019, 20, 884. [Google Scholar] [CrossRef] [PubMed]
- Nonkhwao, S.; Leaokittikul, D.; Patramanon, R.; et al. Revealing the pH-Dependent Conformational Changes in Sol g 2.1 Protein and Potential Ligands Binding. Sci. Rep. 2024, 14, 21179. [Google Scholar] [CrossRef]
- Pires, D. E.; Blundell, T. L.; Ascher, D. B. pkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef] [PubMed]
- Haleem, Z. M.; Yadav, S.; Cushion, M. L.; Tanner, R. J.; Carek, P. J.; Mainous, A. G. Exposure to N,N-Diethyl-Meta-Toluamide Insect Repellent and Human Health Markers: Population-Based Estimates from the National Health and Nutrition Examination Survey. Am. J. Trop. Med. Hyg. 2020, 103, 812–814. [Google Scholar] [CrossRef]
- Lichman, B. R.; Godden, G. T.; Hamilton, J. P.; Palmer, L.; Kamileen, M. O.; Zhao, D.; Vaillancourt, B.; Wood, J. C.; Sun, M.; Kinser, T. J.; Henry, L. K.; Rodriguez-Lopez, C.; Dudareva, N.; Soltis, D. E.; Soltis, P. S.; Buell, C. R.; O’Connor, S. E. The Evolutionary Origins of the Cat Attractant Nepetalactone in Catnip. Sci. Adv. 2020, 6, eaba0721. [Google Scholar] [CrossRef]
- Bernier, U. R.; Furman, K. D.; Kline, D. L.; Allan, S. A.; Barnard, D. R. Comparison of Contact and Spatial Repellency of Catnip Oil and N,N-Diethyl-3-Methylbenzamide (DEET) against Mosquitoes. J. Med. Entomol. 2005, 42, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, K. R.; Klun, J. A.; Debboun, M.; Kramer, M. Feeding Deterrent Effects of Catnip Oil Components Compared with Two Synthetic Amides against Aedes aegypti. J. Med. Entomol. 2005, 42, 643–646. [Google Scholar] [CrossRef]
- Leal, W. S. Odorant Reception in Insects: Roles of Receptors, Binding Proteins, and Degrading Enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef] [PubMed]
- Batume, C.; Mulongo, I. M.; Ludlow, R.; Ssebaale, J.; Randerson, P.; Pickett, J. A.; Mukisa, I. M.; Scofield, S. Evaluating Repellence Properties of Catnip Essential Oil against the Mosquito Species Aedes aegypti Using a Y-Tube Olfactometer. Sci. Rep. 2024, 14, 2269. [Google Scholar] [CrossRef]
- Barnard, D. R.; Xue, R. D. Laboratory Evaluation of Mosquito Repellents against Aedes albopictus, Culex nigripalpus, and Ochlerotatus triseriatus (Diptera: Culicidae). J. Med. Entomol. 2004, 41, 726–730. [Google Scholar] [CrossRef] [PubMed]
- Souto, A. L.; Sylvestre, M.; Tölke, E. D.; Tavares, J. F.; Barbosa-Filho, J. M.; Cebrián-Torrejón, G. Plant-Derived Pesticides as an Alternative to Pest Management and Sustainable Agricultural Production: Prospects, Applications and Challenges. Molecules 2021, 26, 4835. [Google Scholar] [CrossRef]
- Angerosa, F.; d’Alessandro, N.; Konstantinou, P.; Di Giacinto, L. GC-MS Evaluation of Phenolic Compounds in Virgin Olive Oil. J. Agric. Food Chem. 1995, 43, 1802–1807. [Google Scholar] [CrossRef]
- Logan, J. G.; Stanczyk, N. M.; Hassanali, A.; Kemei, J.; Santana, A. E.; Ribeiro, K. A.; Pickett, J. A.; Mordue Luntz, A. J. Arm-in-Cage Testing of Natural Human-Derived Mosquito Repellents. Malar. J. 2010, 9, 239. [Google Scholar] [CrossRef]
- Isman, M. B. Botanical Insecticides: For Richer, for Poorer. Pest Manag. Sci. 2008, 64, 8–11. [Google Scholar] [CrossRef]
- Chen, X.; Shang, S.; Yan, F.; Jiang, H.; Zhao, G.; Tian, S.; Chen, R.; Chen, D.; Dang, Y. Antioxidant Activities of Essential Oils and Their Major Components in Scavenging Free Radicals, Inhibiting Lipid Oxidation and Reducing Cellular Oxidative Stress. Molecules 2023, 28, 4559. [Google Scholar] [CrossRef]
- Tsitsanou, K. E.; Thireou, T.; Drakou, C. E.; Koussis, K.; Keramioti, M. V.; Leonidas, D. D.; Eliopoulos, E.; Iatrou, K.; Zographos, S. E. Anopheles gambiae Odorant Binding Protein Crystal Complex with the Synthetic Repellent DEET: Implications for Structure-Based Design of Novel Mosquito Repellents. Cell. Mol. Life Sci. 2012, 69, 283–297. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Qin, Z.; Ge, X.; Xu, Y.; Chen, Z. The Odorant-Binding Proteins AspiOBP1 and AspiOBP2 in Aleurocanthus spiniferus Are Involved in the Perception of Host Volatiles. Int. J. Mol. Sci. 2025, 26, 8784. [Google Scholar] [CrossRef]
- Kröber, T.; Koussis, K.; Bourquin, M.; Tsitoura, P.; Konstantopoulou, M.; Awolola, T. S.; Dani, F. R.; Qiao, H.; Pelosi, P.; Iatrou, K.; Guerin, P. M. Odorant-Binding Protein-Based Identification of Natural Spatial Repellents for the African Malaria Mosquito Anopheles gambiae. Insect Biochem. Mol. Biol. 2018, 96, 36–50. [Google Scholar] [CrossRef]
- Ghavami, M. B.; Khoeini, S.; Djadid, N. D. Molecular Characteristics of Odorant-Binding Protein 1 in Anopheles maculipennis. Malar. J. 2020, 19, 29. [Google Scholar] [CrossRef]
- Cong, Y.; Li, M.; Feng, G.; et al. Trypsin-Ligand Binding Affinities Calculated Using an Effective Interaction Entropy Method under Polarized Force Field. Sci. Rep. 2017, 7, 17708. [Google Scholar] [CrossRef]
- Kots, E.; Shore, D. M.; Weinstein, H. Simulation of pH-Dependent Conformational Transitions in Membrane Proteins: The CLC-ec1 Cl⁻/H⁺ Antiporter. Molecules 2021, 26, 6956. [Google Scholar] [CrossRef]
- Leal, W. S.; Chen, A. M.; Erickson, M. L. Selective and pH-Dependent Binding of a Moth Pheromone to a Pheromone-Binding Protein. J. Chem. Ecol. 2005, 31, 2493–2499. [Google Scholar] [CrossRef] [PubMed]
- Sen, D.; Debnath, B.; Debnath, P.; et al. Identification of Potential Edible Mushroom as SARS-CoV-2 Main Protease Inhibitor Using Rational Drug Designing Approach. Sci. Rep. 2022, 12, 1503. [Google Scholar] [CrossRef] [PubMed]
- Sudakin, D. L.; Trevathan, W. R. DEET: A Review and Update of Safety and Risk in the General Population. J. Toxicol. Clin. Toxicol. 2003, 41, 831–839. [Google Scholar] [CrossRef]
- Osimitz, T. G.; Murphy, J. V. Neurological Effects Associated with Use of the Insect Repellent N,N-Diethyl-m-toluamide (DEET). J. Toxicol. Clin. Toxicol. 1997, 35, 435–441. [Google Scholar] [CrossRef]
- Khajeh, M.; Yamini, Y.; Shariati, S. Comparison of Essential Oils Compositions of Nepeta persica Obtained by Supercritical Carbon Dioxide Extraction and Steam Distillation Methods. Food Bioprod. Process. 2010, 88, 227–232. [Google Scholar] [CrossRef]
- Borlace, G.; Singh, R.; Seubsasana, S.; Chantaranotahi, P.; Thongkham, E.; Aiemsaard, J. Antimicrobial Effects of Catnip (Nepeta cataria L.) Essential Oil against Canine Skin Infection Pathogens. Vet. World 2024, 17, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Xu, X.; Xu, W.; Ishida, Y.; Leal, W. S.; Ames, J. B.; Clardy, J. Crystal and Solution Structures of an Odorant-Binding Protein from the Southern House Mosquito Complexed with an Oviposition Pheromone. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 19102–19107. [Google Scholar] [CrossRef]
- Leite, N. R.; Krogh, R.; Xu, W.; Ishida, Y.; Iulek, J.; Leal, W. S.; Oliva, G. Structure of an Odorant-Binding Protein from the Mosquito Aedes aegypti Suggests a Binding Pocket Covered by a pH-Sensitive “Lid”. PLoS One 2009, 4, e8006. [Google Scholar] [CrossRef]
- Yekeen, A. A.; Durojaye, O. A.; Idris, M. O.; Muritala, H. F.; Arise, R. O. CHAPERONg: A Tool for Automated GROMACS-Based Molecular Dynamics Simulations and Trajectory Analyses. Comput. Struct. Biotechnol. J. 2023, 21, 4849–4858. [Google Scholar] [CrossRef]
- Tyagi, R.; Paul, A.; Raj, V. S.; Ojha, K. K.; Kumar, S.; Panda, A. K.; Chaurasia, A.; Yadav, M. K. A Drug Repurposing Approach to Identify Therapeutics by Screening Pathogen Box Exploiting SARS-CoV-2 Main Protease. Chem. Biodivers. 2023, 20, e202200600. [Google Scholar] [CrossRef] [PubMed]
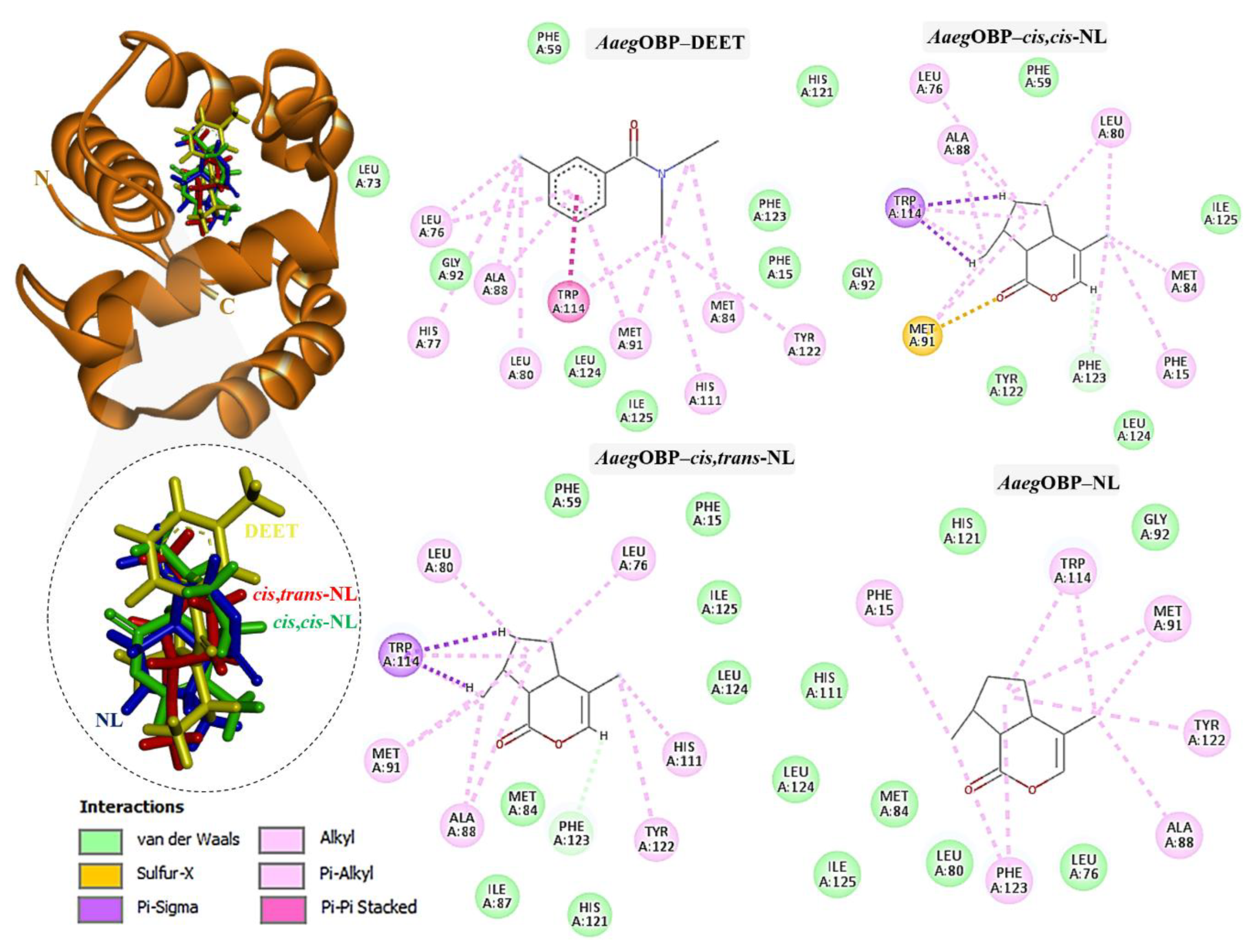
| Compound | Formula | RT (min) | % Peak area | |||
|---|---|---|---|---|---|---|
| SDE | FOE | DOE | Olive oil | |||
| Polycyclic Alkane | ||||||
| trans-4a-Methyl-decahydronaphthalene | C11H20 | 10.48 | 0 | 0.84 | 0.27 | 1.16 |
| 2-Methyldecahydronaphthalene | C11H20 | 11.18,11.08 | 0 | 1.27 | 0.69 | 1.31 |
| 1,6-Dimethyldecahydronaphthalene | C12H22 | 12.97 | 0 | 0 | 0.45 | 0 |
| Cycloalkane | ||||||
| trans-1,4-Dimethylcyclooctane | C10H20 | 9.04 | 0 | 0 | 0 | 6.86 |
| Pentylcyclohexane | C11H22 | 11.5 | 0 | 1.45 | 0.39 | 1.29 |
| 1,4-Dimethyl-2-octadecylcyclohexane | C26H54 | 46.92 | 0 | 0 | 0.59 | 0 |
| Alkene | ||||||
| 2,4-Dimethyl-1-heptene | C9H18 | 3.14 | 0 | 14.18 | 4.36 | 22.2 |
| (2Z)-7-Methyl-2-decene | C11H22 | 7.04,6.97 | 0 | 0 | 0.33 | 1.35 |
| (3E)-3-Heptadecene | C17H34 | 41.73 | 0 | 0 | 1.50 | 0 |
| 1-Tetracosene | C24H48 | 45.9 | 0 | 0 | 0.33 | 0 |
| 1-Hexacosene | C26H52 | 46.51 | 0 | 0 | 0.77 | 0 |
| Alcohol | ||||||
| 3,7-Dimethyl-1-octanol | C10H22O | 9.13 | 0 | 4.11 | 1.73 | 0 |
| 2-Butyloctanol | C12H26O | 20.58,20.5 | 0 | 3.02 | 1.78 | 3.08 |
| 2-Isopropyl-5-methyl-1-heptanol | C11H24O | 21.08,20.95 | 0 | 3.49 | 2.51 | 4.05 |
| 11-Methyldodecanol | C13H26O | 21.43 | 0 | 0 | 0 | 3.01 |
| 2-Methyl-1-decanol | C11H24O | 21.55 | 0 | 2.56 | 1.90 | 0 |
| 3,7,11-Trimethyl-1-dodecanol | C15H32O | 33.21 | 0 | 0 | 0.56 | 0 |
| 2-Hexyldecanol | C16H34O | 33.77 | 0 | 0 | 0.60 | 0.91 |
| 2-Hexyl-1-octanol | C14H30O | 34.78,33.1 | 0 | 1.49 | 0.94 | 0.87 |
| 2-Octyl-1-dodecanol | C20H42O | 35.35 | 0 | 0 | 0.71 | 0 |
| 2-Hexyldodecanol | C18H38O | 44.99 | 0 | 0 | 0.40 | 0 |
| 2-cis-9-Octadecenyloxyethanol | C20H40O2 | 55.74 | 0 | 0 | 0.65 | 0 |
| Ester | ||||||
| Tetrahydrogeranyl formate | C11H20O2 | 9.3,9.22 | 0 | 4.48 | 1.94 | 7.57 |
| Carbonic acid, eicosyl vinyl ester | C23H44O3 | 48.23 | 0 | 0 | 0.60 | 0 |
| Glycidyl (Z)-9-Heptadecenoate | C20H36O3 | 60.62 | 0 | 0 | 0.87 | 0 |
| Glycidyl palmitate | C19H36O3 | 62.54 | 0 | 0 | 10.77 | 0 |
| 9-Octadecenoic acid (Z)-, oxiranylmethyl ester | C21H38O3 | 62.72 | 0 | 0 | 5.45 | 0 |
| Sulfonamide | ||||||
| N-(2-Hydroxyethyl)-N-methyl-perfluorobutane-1-sulfonamide | C7H8F9NO3S | 15.85 | 0 | 1.87 | 0 | 0 |
| Aromatic Hydrocarbon | ||||||
| 1,3-Di-tert-butylbenzene | C14H22 | 17.63 | 0 | 0 | 4.71 | 0 |
| Lactone (Iridoid) | ||||||
| cis-trans-Nepetalactone | C10H14O2 | 23.58 | 52.80 | 0 | 4.90 | 0 |
| cis,cis-Nepetalactone | C10H14O2 | 25.21 | 47.20 | 0 | 1.80 | 0 |
| Nepetalactone | C10H14O2 | 25.58 | 0 | 0 | 0.24 | 0 |
| Sesquiterpene | ||||||
| Caryophyllene | C15H24 | 26.99 | 0 | 0 | 0.12 | 0 |
| Aromatic Ketone | ||||||
| 4-Methoxy-3-(isopenten-2-yl)acetophenone | C13H16O2 | 30.52 | 0 | 2.05 | 1.35 | 0 |
| Butylated Hydroxytoluene | C15H24O | 32.5,32.39 | 0 | 31.79 | 18.22 | 4.81 |
| Unsaturated Fatty Acid | ||||||
| 6-Octadecenoic acid | C18H34O2 | 58.47 | 0 | 0 | 0.61 | 0 |
| Oleic Acid | C18H34O2 | 59.97 | 0 | 0 | 0.75 | 0 |
| 9-Octadecenoic acid | C18H34O2 | 61 | 0 | 0 | 0.49 | 0 |
| (E)-13-Docosenoic acid | C22H42O2 | 61.48 | 0 | 0 | 0.49 | 0 |
| cis-13-Eicosenoic acid | C20H38O2 | 61.66 | 0 | 0 | 1.03 | 0 |
| cis-11-Eicosenoic acid | C20H38O2 | 62.14 | 0 | 0 | 0.83 | 0 |
| Alkane group | ||||||
| 3.43-54.91 | 0 | 26.97 | 22.80 | 41.53 | ||
| OBPs | Cavity size (ų) | DEET | cis,cis-NL | cis-trans-NL | NL |
|---|---|---|---|---|---|
| AgamOBP | 1159 | −6.4 | −6.6 | −6.6 | −6.6 |
| CquiOBP | 1643 | −6.3 | −6.6 | −6.7 | −6.5 |
| AaegOBP | 1284 | −6.2 | −7.0 | −6.7 | −7.0 |
| Compound | OBPs | ΔGMM-PBSA (kcal/mol) | RMSD (nm) | RMSF (nm) | SASA (nm²) | gyration (nm) |
|---|---|---|---|---|---|---|
| DEET | AgamOBP | −9123.22 | 0.17 | 0.43–0.05 | 78.87 | 1.40 |
| CquiOBP | −9002.52 | 0.14 | 0.34–0.05 | 77.68 | 1.39 | |
| AaegOBP | −9090.19 | 0.13 | 0.27–0.05 | 77.00 | 1.39 | |
| cis,cis-NL | AgamOBP | −9130.37 | 0.18 | 0.26–0.05 | 75.02 | 1.38 |
| CquiOBP | −8990.80 | 0.17 | 0.36–0.06 | 77.24 | 1.39 | |
| AaegOBP | −9082.88 | 0.18 | 0.60–0.06 | 76.98 | 1.39 | |
| cis-trans-NL | AgamOBP | −9120.15 | 0.15 | 0.35–0.05 | 77.76 | 1.40 |
| CquiOBP | −8996.77 | 0.15 | 0.32–0.06 | 76.62 | 1.40 | |
| AaegOBP | −9095.99 | 0.19 | 0.40–0.05 | 78.13 | 1.40 | |
| NL | AgamOBP | −9115.51 | 0.2 | 0.34–0.05 | 76.13 | 1.39 |
| CquiOBP | −8999.34 | 0.15 | 0.28–0.05 | 76.78 | 1.39 | |
| AaegOBP | −9087.15 | 0.13 | 0.41–0.05 | 76.61 | 1.39 |
| Property | Model Name | Predicted Value | Cis-cis-NL | Cis-trans-NL | NL | Unit |
|---|---|---|---|---|---|---|
| Absorption | Water solubility | −2.65 | −0.10 | −2.52 | −2.52 | Numeric (log mol/L) |
| Absorption | Skin Permeability | −2.67 | −2.28 | −2.21 | −2.21 | Numeric (log Kp) |
| Absorption | P-glycoprotein substrate | No | No | No | No | Categorical (Yes/No) |
| Absorption | P-glycoprotein I & II inhibitor | No | No | No | No | Categorical (Yes/No) |
| Distribution | VDss (human) | 0.16 | 0.28 | 0.19 | 0.19 | Numeric (log L/kg) |
| Distribution | BBB permeability | 0.36 | 0.29 | 0.70 | 0.70 | Numeric (log BB) |
| Distribution | CNS permeability | −1.93 | −2.83 | −2.08 | −2.08 | Numeric (log PS) |
| Metabolism | CYP2D6 and CYP3A4 substrate | No | No | No | No | Categorical (Yes/No) |
| Metabolism | CYP1A2 inhibitor | Yes | Yes | No | No | Categorical (Yes/No) |
| Metabolism | CYP2C9, CYP2D6 and CYP3A4 inhibitor | No | No | No | No | Categorical (Yes/No) |
| Excretion | Total Clearance | 0.59 | 0.91 | 0.11 | 0.11 | Numeric (log ml/min/kg) |
| Excretion | Renal OCT2 substrate | No | No | No | No | Categorical (Yes/No) |
| Toxicity | hERG I & II inhibitor | No | No | No | No | Categorical (Yes/No) |
| Toxicity | Oral Rat Acute Toxicity (LD50) | 2.31 | 2.48 | 1.77 | 1.77 | Numeric (mol/kg) |
| Toxicity | Oral Rat Chronic Toxicity (LOAEL) | 1.46 | 4.57 | 2.30 | 2.30 | Numeric (log mg/kg_bw/day) |
| Toxicity | Hepatotoxicity | No | No | No | No | Categorical (Yes/No) |
| Toxicity | Skin Sensitisation | Yes | No | Yes | Yes | Categorical (Yes/No) |
| Toxicity | T.Pyriformis toxicity | 0.59 | −0.96 | 0.23 | 0.23 | Numeric (log µg/L) |
| Toxicity | Minnow toxicity | 1.19 | 4.81 | 1.30 | 1.30 | Numeric (log mM) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





