Submitted:
17 September 2025
Posted:
18 September 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. RNA Profiles of Tumor-Educated Platelets in NSCLC
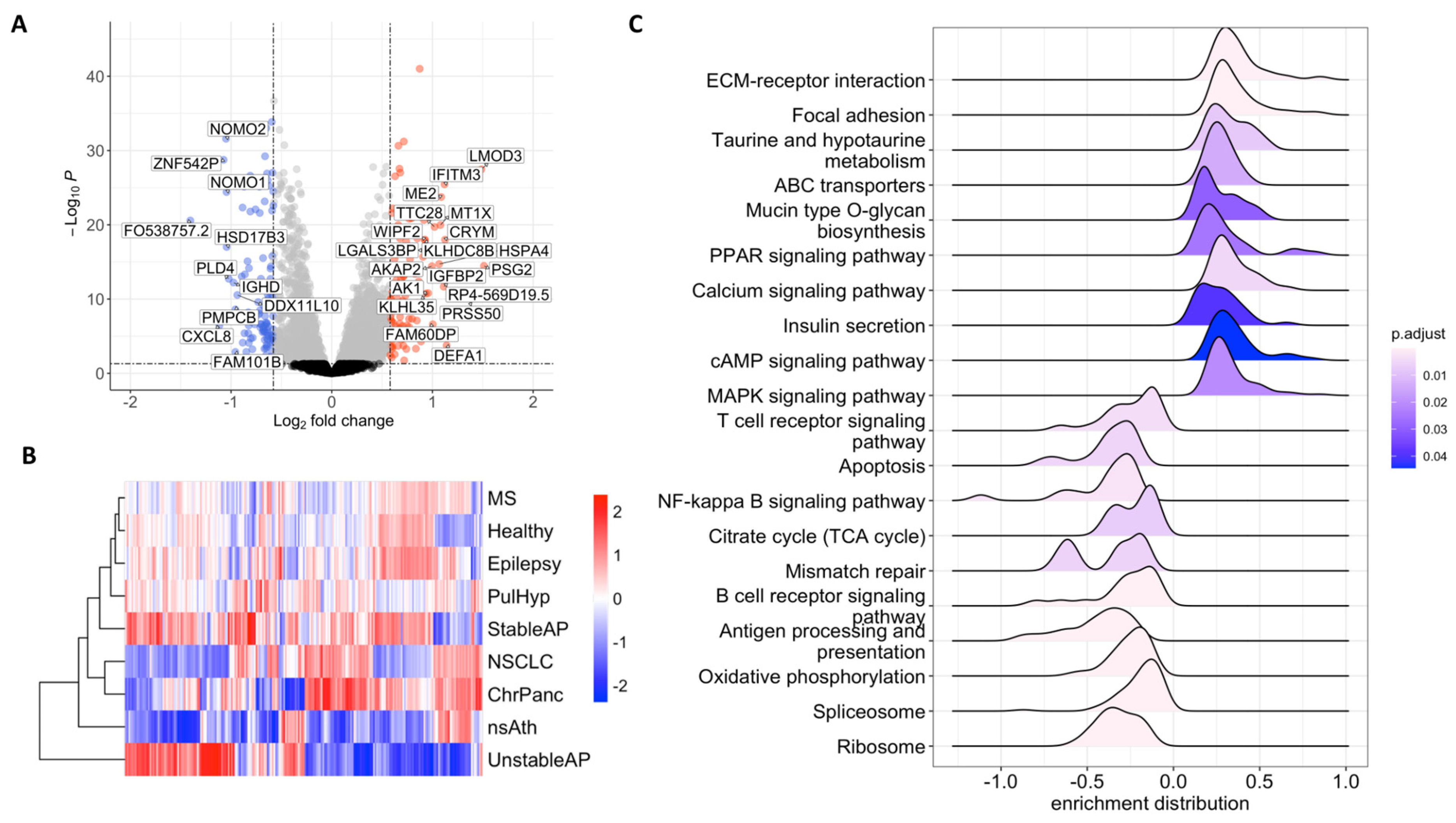
2.2. Therapeutic Target Discovery Based on Transcriptome Data
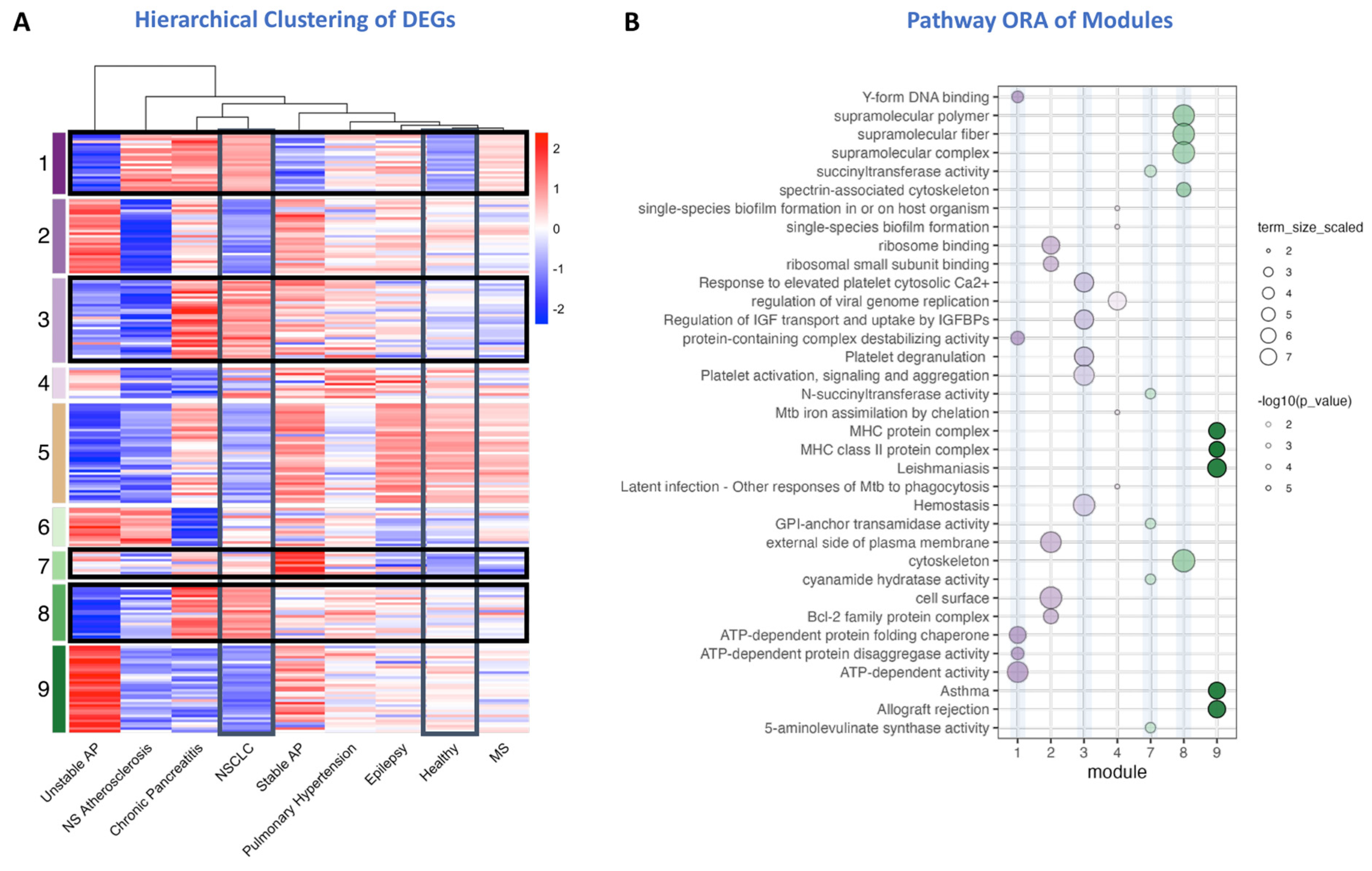
| Target Name1 | Description | Drugs2 | module |
|---|---|---|---|
| CACNA1D | calcium voltage-gated channel subunit alpha1 D | Ergocalciferol, Enflurane, Ranolazine, Phenytoin, Isradipine, Topiramate, Nimodipine, Nisoldipine, Spironolactone, Nicardipine, Magnesium sulfate, Verapamil, Levomenthol, Ethanol, Felodipine, Miconazole, Nifedipine, Amiodarone, Dronedarone, Clevidipine, Levamlodipine, Fish oil | 1 |
| PPARA | peroxisome proliferator activated receptor alpha | Valproic acid, Indomethacin, Rosiglitazone, Fenoprofen, Fenofibrate, Ibuprofen, Amiodarone, Gemfibrozil, Prasterone, Palmitic Acid, Soybean oil, Fenofibric acid, Fish oil | 1 |
| LPAR4 | lysophosphatidic acid receptor 4 | Promethazine | 1 |
| ME2 | malic enzyme 2 | NADH | 1 |
| MAOB | monoamine oxidase B | Amphetamine, Phentermine, Procaine, Tranylcypromine, Phenelzine, Zonisamide, Selegiline, Pioglitazone, Procarbazine, Isocarboxazid, Rasagiline, Metamfetamine, Flavin adenine dinucleotide, Safinamide, Viloxazine, Flortaucipir F-18 | 3 |
| FCGR2A | Fc gamma receptor IIa | Cetuximab, Etanercept, Human immunoglobulin G, Abciximab, Alemtuzumab, Bevacizumab, Sarilumab | 3 |
| SMPD1 | sphingomyelin phosphodiesterase 1 | Amlodipine, Chlorpromazine, Desipramine | 3 |
| GSTM3 | glutathione S-transferase mu 3 | Glutathione disulfide, Deoxycholic acid | 3 |
| ITGA2B | integrin subunit alpha 2b | Abciximab, Tirofiban | 3 |
| CA1 | carbonic anhydrase 1 | Topiramate, Chlorthalidone, Amlodipine, Methocarbamol, Bendroflumethiazide, Methazolamide, Hydroflumethiazide, Acetazolamide, Dorzolamide, Chlorothiazide, Zonisamide, Diclofenamide, Brinzolamide, Sodium sulfate | 7 |
| HBA1 | hemoglobin subunit alpha 1 | Iron Dextran, Nitrous acid, Copper, Sodium ferric gluconate complex, Ferric pyrophosphate citrate, Zinc acetate, Ferrous fumarate, Zinc chloride, Voxelotor, Ferric derisomaltose | 7 |
| SEC14L2 | SEC14 like lipid binding 2 | Vitamin E | 7 |
| ALAS2 | 5’-aminolevulinate synthase 2 | Glycine | 7 |
| ATOX1 | antioxidant 1 copper chaperone | Cisplatin | 8 |
| MAP1A | microtubule associated protein 1A | Estramustine | 8 |
| ID | Description | Drug Targets |
|---|---|---|
| R-HSA-445095 | Interaction between L1 and Ankyrins |
SCN1A (Sodium channel protein type 1 subunit alpha), SCN2A (Sodium channel protein type 2 subunit alpha), SCN9A (Sodium channel protein type 9 subunit alpha), SCN3A (Sodium channel protein type 3 subunit alpha), SCN11A (Sodium channel protein type 11 subunit alpha), SCN8A (Sodium channel protein type 8 subunit alpha), SCN1B (Sodium channel regulatory subunit beta-1), SCN3B (Sodium channel regulatory subunit beta-3) |
| R-HSA-3000178 | ECM proteoglycans |
APP (Amyloid-beta precursor protein), FN1 (Fibronectin), HAPLN1 (Hyaluronan and proteoglycan link protein 1), HSPG2 (Basement membrane-specific heparan sulfate proteoglycan core protein), ITGA2B (Integrin alpha-IIb) |
| R-HAS-1474228 | Degradation of the extracellular matrix |
ELN (Elastin), FBN2 (Fibrillin-2), FN1 (Fibronectin), HSPG2 (Basement membrane-specific heparan sulfate proteoglycan core protein), NID1 (Nidogen-1), PLG (Plasminogen) |
| R-HSA-446728 | Cell junction organization |
CDH11 (Cadherin-11), FLNA (Filamin-A), TESK1 (Dual specificity testis-specific protein kinase 1) |
| R-HSA-381426 | Regulation o fIGF transport and uptake by IGFBPs |
APP (Amyloid-beta precursor protein), CP (Ceruloplasmin), F5 (Coagulation factor V), FN1 (Fibronectin), ITIH2 (Inter-alpha-trypsin inhibitor heavy chain H2), PLG (Plasminogen), SERPIND1 (Heparin cofactor 2) |
| R-HSA-5173105 | O-linked glycosylation | MUC16 (Mucin-16) |
| R-HSA-1474290 | Collagen formation |
P3H2 (Prolyl 3-hydroxylase 2), P4HB (Protein disulfide-isomerase), PLOD1 (Procollagen-lysine,2-oxoglutarate 5-dioxygenase 1), PLOD2 (Procollagen-lysine,2-oxoglutarate 5-dioxygenase 2) |
| R-HSA-1592389 | Activation of Matrix Metalloproteinases | no FDA-approved drugs found. |
2.3. Therapeutic Target Discovery Based on Network Controllability
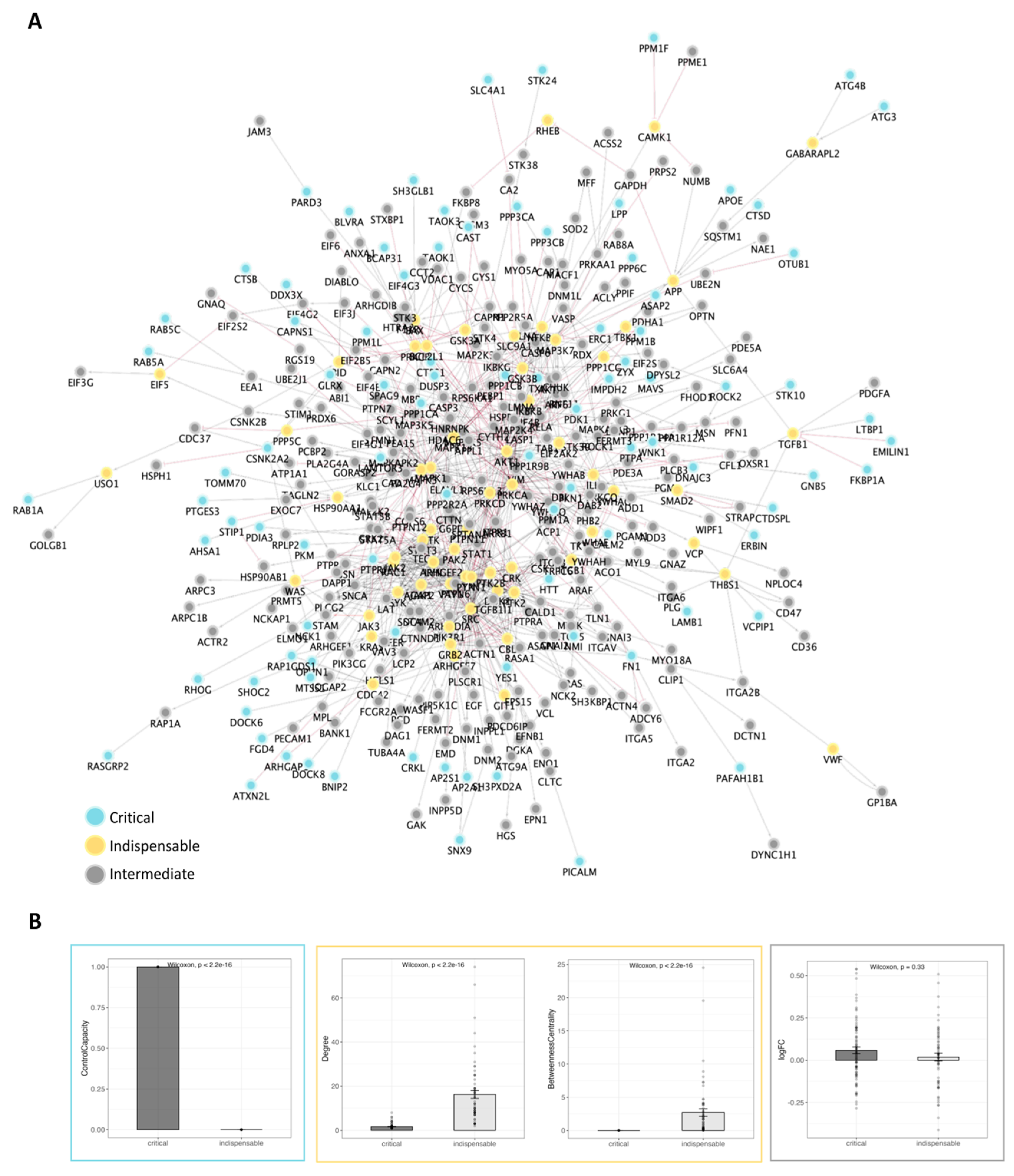
| Drug Name1 | Number of Targets | Targets2 |
| Fostamatinib | 5 | CAMK1, JAK2, MAPK14, PRKCD, PTK2 |
| Minocycline | 4 | CASP3, MAPK1, MAPK14, MAPK3 |
| Acetylsalicylic acid | 3 | CASP3, MAPK1, MAPK3 |
| Arsenic trioxide | 3 | MAPK3, MAPK1, AKT1 |
| Copper | 3 | APP, GAPDH, HSP90AA1 |
| Benzoyl peroxide | 2 | PRKCA, PRKCD |
| Dequalinium* | 2 | PRKCA, PRKCD |
| Ingenol mebutate | 2 | PRKCD, PRKCA |
| Tamoxifen | 2 | PRKCA, PRKCD |
| Abrocitinib | 1 | JAK2 |
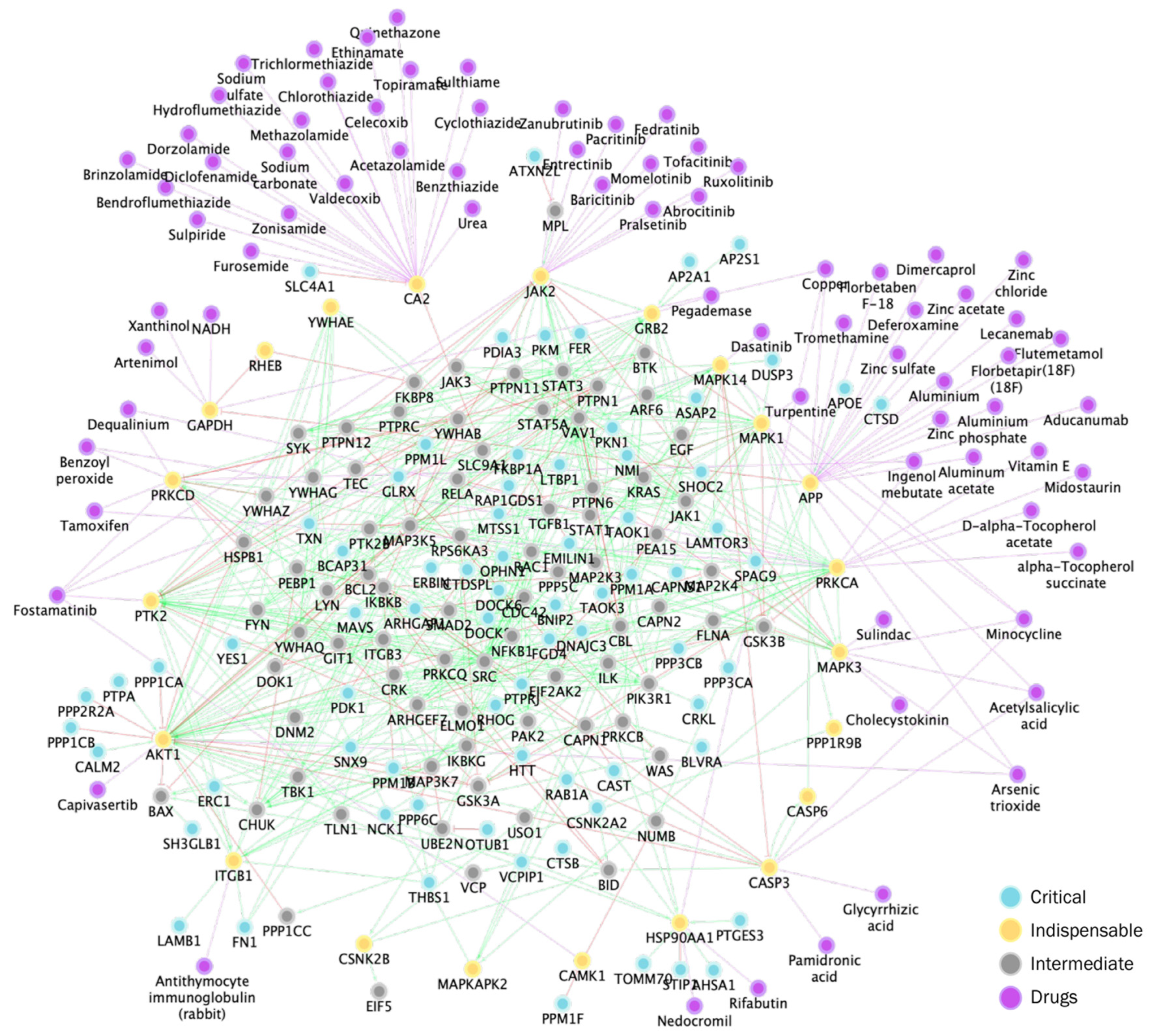
2.4. Expanded Platelet Interactome Reveals Novel Targetable Nodes
| Drug Name1 | Degree | Log2FC | Weight | GeneScore | Drugs2 |
|---|---|---|---|---|---|
| LYN | 48 | -0,24 | 11,34 | 0,48 | Dasatinib, Bosutinib, Ponatinib, Nintedanib, Fostamatinib |
| JAK1 | 21 | -- | 0 | 0,47 | Ruxolitinib, Tofacitinib, Momelotinib, Baricitinib, Fostamatinib, Fedratinib, Filgotinib, Abrocitinib, Upadacitinib, Pralsetinib |
| FCGR2A | 4 | 0,68 | 2,71 | 0,46 | Cetuximab, Etanercept, Human immunoglobulin G, Abciximab, Alemtuzumab, Bevacizumab, Catumaxomab, Sarilumab |
| TEC | 10 | 0,25 | 2,47 | 0,46 | Bosutinib, Fostamatinib, Ritlecitinib, Zanubrutinib |
| TUBA4A | 1 | -0,13 | 0,13 | 0,45 | Vincristine, Podofilox |
| PTPN6 | 29 | -0,09 | 2,73 | 0,45 | Tiludronic acid |
| SYK | 37 | -0,1 | 3,84 | 0,45 | Fostamatinib |
| ITGA5 | 5 | -- | 0 | 0,45 | Tauroursodeoxycholic acid |
| GRB2 | 37 | -- | 0 | 0,45 | Pegademase |
| JAK3 | 16 | 0,45 | 7,3 | 0,44 | Ruxolitinib, Tofacitinib, Momelotinib, Baricitinib, Fostamatinib, Ritlecitinib, Abrocitinib, Zanubrutinib |
| P2RY12 | 1 | -0,49 | 0,49 | 0,44 | Ticlopidine, Treprostinil, Clopidogrel, Promethazine, Epoprostenol, Prasugrel, Cangrelor, Ticagrelor |
| BTK | 19 | -0,271 | 5,08 | 0,44 | Dasatinib, Ibrutinib, Acalabrutinib, Fostamatinib, Ritlecitinib, Zanubrutinib, Pirtobrutinib |
| PPIA | 1 | -- | 0 | 0,44 | Cyclosporine, Copper, Artenimol |
| PIK3CB | 1 | 0,13 | 0,13 | 0,44 | Caffeine, Copanlisib |
| CSK | 18 | 0,11 | 1,95 | 0,44 | Dasatinib, Fostamatinib |
| PTK2B | 37 | 0,18 | 6,76 | 0,44 | Leflunomide, Fostamatinib |
3. Discussion
3.1. Targeting Tumor-Educated Platelet Signaling
3.2. Limitations and Outlook
4. Materials and Methods
4.1. RNAseq Data Analysis
4.2. Differential Gene Expression Analysis and Data Normalization
4.3. Volcano Plots and Heatmaps
4.4. Gene Set Enrichment Analysis
4.5. Clustering of DEGs
4.6. Construction of Platelet Signaling Network
4.7. Network Controllability and Subnetworks
4.8. Gene Scores and Subnetworks
4.9. Drug Repurposing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Metharom, P., M. Falasca, and M.C. Berndt, The History of Armand Trousseau and Cancer-Associated Thrombosis. Cancers (Basel), 2019. 11(2).
- Crissman, J.D., et al., Arrest and extravasation of B16 amelanotic melanoma in murine lungs. A light and electron microscopic study. Lab Invest, 1985. 53(4): p. 470-8.
- Gasic, G.J., et al., Aggregation of platelets and cell membrane vesiculation by rat cells transformed in vitro by Rous sarcoma virus. Cancer Res, 1978. 38(9): p. 2950-5.
- Stone, R.L., et al., Paraneoplastic thrombocytosis in ovarian cancer. N Engl J Med, 2012. 366(7): p. 610-8.
- Menter, D.G., et al., Platelets and cancer: a casual or causal relationship: revisited. Cancer Metastasis Rev, 2014. 33(1): p. 231-69.
- Haemmerle, M., et al., FAK regulates platelet extravasation and tumor growth after antiangiogenic therapy withdrawal. J Clin Invest, 2016. 126(5): p. 1885-96.
- Varki, A., Trousseau’s syndrome: multiple definitions and multiple mechanisms. Blood, 2007. 110(6): p. 1723-9.
- Khorana, A.A. and G.C. Connolly, Assessing risk of venous thromboembolism in the patient with cancer. J Clin Oncol, 2009. 27(29): p. 4839-47.
- Lyman, G.H. and A.A. Khorana, Cancer, clots and consensus: new understanding of an old problem. J Clin Oncol, 2009. 27(29): p. 4821-6.
- Bailey, S.E., et al., Clinical relevance of thrombocytosis in primary care: a prospective cohort study of cancer incidence using English electronic medical records and cancer registry data. Br J Gen Pract, 2017. 67(659): p. e405-e413.
- Abdulrahman, G.O., N. Das, and K. Lutchman Singh, The predictive role of thrombocytosis in benign, borderline and malignant ovarian tumors. Platelets, 2020. 31(6): p. 795-800.
- Skorek, P., et al., Preoperative thrombocytosis in surgically treated patients with non-small cell lung cancer. Pol Arch Intern Med, 2018. 128(9): p. 512-517.
- Li, N., Platelets in cancer metastasis: To help the “villain” to do evil. Int J Cancer, 2016. 138(9): p. 2078-87.
- Sasaki, Y., et al., Production of thrombopoietin by human carcinomas and its novel isoforms. Blood, 1999. 94(6): p. 1952-60.
- Calverley, D.C., et al., Significant downregulation of platelet gene expression in metastatic lung cancer. Clin Transl Sci, 2010. 3(5): p. 227-32.
- Sabrkhany, S., et al., Exploration of the platelet proteome in patients with early-stage cancer. J Proteomics, 2018. 177: p. 65-74.
- Denis, M.M., et al., Escaping the nuclear confines: signal-dependent pre-mRNA splicing in anucleate platelets. Cell, 2005. 122(3): p. 379-91.
- Li, S., et al., The dynamic role of platelets in cancer progression and their therapeutic implications. Nat Rev Cancer, 2024. 24(1): p. 72-87.
- Banerjee, M., et al., Cellubrevin/vesicle-associated membrane protein-3-mediated endocytosis and trafficking regulate platelet functions. Blood, 2017. 130(26): p. 2872-2883.
- Banerjee, M. and S.W. Whiteheart, The ins and outs of endocytic trafficking in platelet functions. Curr Opin Hematol, 2017. 24(5): p. 467-474.
- Kuznetsov, H.S., et al., Identification of luminal breast cancers that establish a tumor-supportive macroenvironment defined by proangiogenic platelets and bone marrow-derived cells. Cancer Discov, 2012. 2(12): p. 1150-65.
- Kerr, B.A., et al., Platelets govern pre-metastatic tumor communication to bone. Oncogene, 2013. 32(36): p. 4319-24.
- Roweth, H.G. and E.M. Battinelli, Lessons to learn from tumor-educated platelets. Blood, 2021. 137(23): p. 3174-3180.
- Weyrich, A.S., et al., Signal-dependent translation of a regulatory protein, Bcl-3, in activated human platelets. Proc Natl Acad Sci U S A, 1998. 95(10): p. 5556-61.
- Nilsson, R.J., et al., Blood platelets contain tumor-derived RNA biomarkers. Blood, 2011. 118(13): p. 3680-3.
- Ye, B., et al., A panel of platelet-associated circulating long non-coding RNAs as potential biomarkers for colorectal cancer. Genomics, 2022. 114(1): p. 31-37.
- Tabaeian, S.P., et al., Evaluation of tumor-educated platelet long non-coding RNAs (lncRNAs) as potential diagnostic biomarkers for colorectal cancer. J Cancer Res Ther, 2024. 20(5): p. 1453-1458.
- Moonmuang, S., et al., Circulating Long Non-Coding RNAs as Novel Potential Biomarkers for Osteogenic Sarcoma. Cancers (Basel), 2021. 13(16).
- Yuan, M., et al., Screening and validation of platelet activation-related lncRNAs as potential biomarkers for prognosis and immunotherapy in gastric cancer patients. Front Genet, 2022. 13: p. 965033.
- Laffont, B., et al., Activated platelets can deliver mRNA regulatory Ago2*microRNA complexes to endothelial cells via microparticles. Blood, 2013. 122(2): p. 253-61.
- Mammadova-Bach, E., et al., Platelet glycoprotein VI promotes metastasis through interaction with cancer cell-derived galectin-3. Blood, 2020. 135(14): p. 1146-1160.
- Ichikawa, J., et al., Role of Platelet C-Type Lectin-Like Receptor 2 in Promoting Lung Metastasis in Osteosarcoma. J Bone Miner Res, 2020. 35(9): p. 1738-1750.
- Mitrugno, A., et al., A novel and essential role for FcgammaRIIa in cancer cell-induced platelet activation. Blood, 2014. 123(2): p. 249-60.
- Miao, S., et al., Cancer cell-derived immunoglobulin G activates platelets by binding to platelet FcgammaRIIa. Cell Death Dis, 2019. 10(2): p. 87.
- Strasenburg, W., et al., Tumor Cell-Induced Platelet Aggregation as an Emerging Therapeutic Target for Cancer Therapy. Front Oncol, 2022. 12: p. 909767.
- Heinmoller, E., et al., Studies on tumor-cell-induced platelet aggregation in human lung cancer cell lines. J Cancer Res Clin Oncol, 1996. 122(12): p. 735-44.
- Jurasz, P., D. Alonso-Escolano, and M.W. Radomski, Platelet--cancer interactions: mechanisms and pharmacology of tumour cell-induced platelet aggregation. Br J Pharmacol, 2004. 143(7): p. 819-26.
- Cooke, N.M., et al., Increased platelet reactivity in patients with late-stage metastatic cancer. Cancer Med, 2013. 2(4): p. 564-70.
- Tesfamariam, B., Involvement of platelets in tumor cell metastasis. Pharmacol Ther, 2016. 157: p. 112-9.
- Mammadova-Bach, E., et al., Platelets in cancer. From basic research to therapeutic implications. Hamostaseologie, 2015. 35(4): p. 325-36.
- Palacios-Acedo, A.L., et al., Platelets, Thrombo-Inflammation, and Cancer: Collaborating With the Enemy. Front Immunol, 2019. 10: p. 1805.
- Filippelli, A., et al., Scoping Review on Platelets and Tumor Angiogenesis: Do We Need More Evidence or Better Analysis? Int J Mol Sci, 2022. 23(21).
- Haemmerle, M., et al., The Platelet Lifeline to Cancer: Challenges and Opportunities. Cancer Cell, 2018. 33(6): p. 965-983.
- von Hundelshausen, P., et al., Heterophilic interactions of platelet factor 4 and RANTES promote monocyte arrest on endothelium. Blood, 2005. 105(3): p. 924-30.
- Ansari, M.J., et al., Cancer combination therapies by angiogenesis inhibitors; a comprehensive review. Cell Commun Signal, 2022. 20(1): p. 49.
- Pinedo, H.M., et al., Involvement of platelets in tumour angiogenesis? Lancet, 1998. 352(9142): p. 1775-7.
- Maurer, S. and L. Ferrari de Andrade, NK Cell Interaction With Platelets and Myeloid Cells in the Tumor Milieu. Front Immunol, 2020. 11: p. 608849.
- Placke, T., et al., Platelet-derived MHC class I confers a pseudonormal phenotype to cancer cells that subverts the antitumor reactivity of natural killer immune cells. Cancer Res, 2012. 72(2): p. 440-8.
- Plantureux, L., et al., Effects of platelets on cancer progression. Thromb Res, 2018. 164 Suppl 1: p. S40-S47.
- Gay, L.J. and B. Felding-Habermann, Contribution of platelets to tumour metastasis. Nat Rev Cancer, 2011. 11(2): p. 123-34.
- Erpenbeck, L. and M.P. Schon, Deadly allies: the fatal interplay between platelets and metastasizing cancer cells. Blood, 2010. 115(17): p. 3427-36.
- Lonsdorf, A.S., et al., Engagement of alphaIIbbeta3 (GPIIb/IIIa) with alphanubeta3 integrin mediates interaction of melanoma cells with platelets: a connection to hematogenous metastasis. J Biol Chem, 2012. 287(3): p. 2168-78.
- Labelle, M., S. Begum, and R.O. Hynes, Platelets guide the formation of early metastatic niches. Proc Natl Acad Sci U S A, 2014. 111(30): p. E3053-61.
- Foss, A., et al., The contribution of platelets to intravascular arrest, extravasation, and outgrowth of disseminated tumor cells. Clin Exp Metastasis, 2020. 37(1): p. 47-67.
- Schumacher, D., et al., Platelet-derived nucleotides promote tumor-cell transendothelial migration and metastasis via P2Y2 receptor. Cancer Cell, 2013. 24(1): p. 130-7.
- Sierko, E. and M.Z. Wojtukiewicz, Inhibition of platelet function: does it offer a chance of better cancer progression control? Semin Thromb Hemost, 2007. 33(7): p. 712-21.
- Seizer, P. and A.E. May, Platelets and matrix metalloproteinases. Thromb Haemost, 2013. 110(5): p. 903-9.
- Vlodavsky, I., et al., Expression of heparanase by platelets and circulating cells of the immune system: possible involvement in diapedesis and extravasation. Invasion Metastasis, 1992. 12(2): p. 112-27.
- Giannakeas, V., et al., Analysis of Platelet Count and New Cancer Diagnosis Over a 10-Year Period. JAMA Netw Open, 2022. 5(1): p. e2141633.
- Sabrkhany, S., et al., Platelets as messengers of early-stage cancer. Cancer Metastasis Rev, 2021. 40(2): p. 563-573.
- Huong, P.T., et al., The Role of Platelets in the Tumor-Microenvironment and the Drug Resistance of Cancer Cells. Cancers (Basel), 2019. 11(2).
- Yu, L., et al., Bidirectional Interaction Between Cancer Cells and Platelets Provides Potential Strategies for Cancer Therapies. Front Oncol, 2021. 11: p. 764119.
- Franco, A.T., A. Corken, and J. Ware, Platelets at the interface of thrombosis, inflammation, and cancer. Blood, 2015. 126(5): p. 582-8.
- Battinelli, E.M., B.A. Markens, and J.E. Italiano, Jr., Release of angiogenesis regulatory proteins from platelet alpha granules: modulation of physiologic and pathologic angiogenesis. Blood, 2011. 118(5): p. 1359-69.
- Guillem-Llobat, P., et al., Aspirin prevents colorectal cancer metastasis in mice by splitting the crosstalk between platelets and tumor cells. Oncotarget, 2016. 7(22): p. 32462-77.
- Lucotti, S., et al., Aspirin blocks formation of metastatic intravascular niches by inhibiting platelet-derived COX-1/thromboxane A2. J Clin Invest, 2019. 129(5): p. 1845-1862.
- Gebremeskel, S., et al., The reversible P2Y12 inhibitor ticagrelor inhibits metastasis and improves survival in mouse models of cancer. Int J Cancer, 2015. 136(1): p. 234-40.
- Zhang, W., et al., A humanized single-chain antibody against beta 3 integrin inhibits pulmonary metastasis by preferentially fragmenting activated platelets in the tumor microenvironment. Blood, 2012. 120(14): p. 2889-98.
- Amirkhosravi, A., et al., Inhibition of tumor cell-induced platelet aggregation and lung metastasis by the oral GpIIb/IIIa antagonist XV454. Thromb Haemost, 2003. 90(3): p. 549-54.
- Khorana, A.A., et al., Thromboembolism is a leading cause of death in cancer patients receiving outpatient chemotherapy. J Thromb Haemost, 2007. 5(3): p. 632-4.
- Riedl, J., et al., Podoplanin expression in primary brain tumors induces platelet aggregation and increases risk of venous thromboembolism. Blood, 2017. 129(13): p. 1831-1839.
- Sasano, T., et al., Podoplanin promotes tumor growth, platelet aggregation, and venous thrombosis in murine models of ovarian cancer. J Thromb Haemost, 2022. 20(1): p. 104-114.
- Wojtukiewicz, M.Z., et al., Antiplatelet agents for cancer treatment: a real perspective or just an echo from the past? Cancer Metastasis Rev, 2017. 36(2): p. 305-329.
- Turei, D., T. Korcsmaros, and J. Saez-Rodriguez, OmniPath: guidelines and gateway for literature-curated signaling pathway resources. Nat Methods, 2016. 13(12): p. 966-967.
- Rolf, M.G., et al., In vitro pharmacological profiling of R406 identifies molecular targets underlying the clinical effects of fostamatinib. Pharmacol Res Perspect, 2015. 3(5): p. e00175.
- Cornish, A.J. and F. Markowetz, SANTA: quantifying the functional content of molecular networks. PLoS Comput Biol, 2014. 10(9): p. e1003808.
- Xing, S., et al., Development and Validation of Tumor-educated Blood Platelets Integrin Alpha 2b (ITGA2B) RNA for Diagnosis and Prognosis of Non-small-cell Lung Cancer through RNA-seq. Int J Biol Sci, 2019. 15(9): p. 1977-1992.
- Goswami, C., et al., Molecular signature comprising 11 platelet-genes enables accurate blood-based diagnosis of NSCLC. BMC Genomics, 2020. 21(1): p. 744.
- Best, M.G., et al., RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell, 2015. 28(5): p. 666-676.
- Ge, X., et al., Identification of seven tumor-educated platelets RNAs for cancer diagnosis. J Clin Lab Anal, 2021. 35(6): p. e23791.
- Zhang, B., et al., Prognostic value of IGFBP2 in various cancers: a systematic review and meta-analysis. Cancer Med, 2022. 11(16): p. 3035-3047.
- Moore, T. and G.S. Dveksler, Pregnancy-specific glycoproteins: complex gene families regulating maternal-fetal interactions. Int J Dev Biol, 2014. 58(2-4): p. 273-80.
- Ingman, W.V. and S.A. Robertson, The essential roles of TGFB1 in reproduction. Cytokine Growth Factor Rev, 2009. 20(3): p. 233-9.
- Zhao, M.R., et al., Dual effect of transforming growth factor beta1 on cell adhesion and invasion in human placenta trophoblast cells. Reproduction, 2006. 132(2): p. 333-41.
- Sorensen, S., J. Andersen, and T. Norgaard, Pregnancy-specific beta 1-glycoprotein (SP1) in serum and tissue from patients with benign and malignant breast tumours. Br J Cancer, 1984. 49(5): p. 663-7.
- Skinner, J.M. and R. Whitehead, Pregnancy-specific beta glycoprotein (SP1) in tumours of the human gastrointestinal tract. Br J Cancer, 1981. 44(3): p. 476-8.
- Zhao, J., R. Rabadan, and A.J. Levine, Pregnancy specific glycoproteins: a possible mediator of immune tolerance of cancers. Journal of Cellular Immunology, 2021. 3(2): p. 109-117.
- Pang, J.H., et al., Activation of tumour cell ECM degradation by thrombin-activated platelet membranes: potentially a P-selectin and GPIIb/IIIa-dependent process. Clin Exp Metastasis, 2015. 32(5): p. 495-505.
- Felding-Habermann, B., et al., Role of beta3 integrins in melanoma cell adhesion to activated platelets under flow. J Biol Chem, 1996. 271(10): p. 5892-900.
- Burdick, M.M. and K. Konstantopoulos, Platelet-induced enhancement of LS174T colon carcinoma and THP-1 monocytoid cell adhesion to vascular endothelium under flow. Am J Physiol Cell Physiol, 2004. 287(2): p. C539-47.
- Koch, A.E., et al., Interleukin-8 as a macrophage-derived mediator of angiogenesis. Science, 1992. 258(5089): p. 1798-801.
- Strieter, R.M., et al., The functional role of the ELR motif in CXC chemokine-mediated angiogenesis. J Biol Chem, 1995. 270(45): p. 27348-57.
- Cambier, S., M. Gouwy, and P. Proost, The chemokines CXCL8 and CXCL12: molecular and functional properties, role in disease and efforts towards pharmacological intervention. Cell Mol Immunol, 2023. 20(3): p. 217-251.
- Zhu, Y.M., et al., Interleukin-8/CXCL8 is a growth factor for human lung cancer cells. Br J Cancer, 2004. 91(11): p. 1970-6.
- Zhang, W., et al., Identification of cuproptosis and immune-related gene prognostic signature in lung adenocarcinoma. Front Immunol, 2023. 14: p. 1179742.
- Tyagi, T., et al., Platelet-derived TLT-1 promotes tumor progression by suppressing CD8+ T cells. J Exp Med, 2023. 220(1).
- Pandey, P., et al., New insights about the PDGF/PDGFR signaling pathway as a promising target to develop cancer therapeutic strategies. Biomed Pharmacother, 2023. 161: p. 114491.
- Hers, I., Insulin-like growth factor-1 potentiates platelet activation via the IRS/PI3Kalpha pathway. Blood, 2007. 110(13): p. 4243-52.
- Mantini, G., et al., Omics Analysis of Educated Platelets in Cancer and Benign Disease of the Pancreas. Cancers (Basel), 2020. 13(1).
- Eslami, S.Z., et al., In vitro cross-talk between metastasis-competent circulating tumor cells and platelets in colon cancer: a malicious association during the harsh journey in the blood. Front Cell Dev Biol, 2023. 11: p. 1209846.
- Li, T., et al., IGFBP2: integrative hub of developmental and oncogenic signaling network. Oncogene, 2020. 39(11): p. 2243-2257.
- Haschemi, R., et al., Insulin-like Growth Factor Binding Protein-2 (IGFBP2) Is a Key Molecule in the MACC1-Mediated Platelet Communication and Metastasis of Colorectal Cancer Cells. Int J Mol Sci, 2021. 22(22).
- Wei, L.F., et al., IGFBP2 in cancer: Pathological role and clinical significance (Review). Oncol Rep, 2021. 45(2): p. 427-438.
- Guo, C., et al., Insulin-like growth factor binding protein-2 level is increased in blood of lung cancer patients and associated with poor survival. PLoS One, 2013. 8(9): p. e74973.
- Wang, J., et al., Gene expression and prognosis of insulin-like growth factor-binding protein family members in non-small cell lung cancer. Oncol Rep, 2019. 42(5): p. 1981-1995.
- Lu, H., et al., IGFBP2/ITGA5 promotes gefitinib resistance via activating STAT3/CXCL1 axis in non-small cell lung cancer. Cell Death Dis, 2024. 15(6): p. 447.
- Saci, A., et al., Differential effect of the inhibition of Grb2-SH3 interactions in platelet activation induced by thrombin and by Fc receptor engagement. Biochem J, 2002. 363(Pt 3): p. 717-25.
- Vogtle, T., et al., Critical redundant functions of the adapters Grb2 and Gads in platelet (hem)ITAM signaling in mice. Platelets, 2020. 31(6): p. 801-811.
- Visconte, C., et al., Amyloid precursor protein is required for in vitro platelet adhesion to amyloid peptides and potentiation of thrombus formation. Cell Signal, 2018. 52: p. 95-102.
- Patel, P., et al., Platelet FcgammaRIIA in immunity and thrombosis: Adaptive immunothrombosis. J Thromb Haemost, 2021. 19(5): p. 1149-1160.
- Camerer, E., et al., Platelets, protease-activated receptors, and fibrinogen in hematogenous metastasis. Blood, 2004. 104(2): p. 397-401.
- Palumbo, J.S., et al., Platelets and fibrin(ogen) increase metastatic potential by impeding natural killer cell-mediated elimination of tumor cells. Blood, 2005. 105(1): p. 178-85.
- Trikha, M., et al., Multiple roles for platelet GPIIb/IIIa and alphavbeta3 integrins in tumor growth, angiogenesis, and metastasis. Cancer Res, 2002. 62(10): p. 2824-33.
- Jain, S., et al., Platelet glycoprotein Ib alpha supports experimental lung metastasis. Proc Natl Acad Sci U S A, 2007. 104(21): p. 9024-8.
- Osmanoglu, O., et al., Signaling network analysis reveals fostamatinib as a potential drug to control platelet hyperactivation during SARS-CoV-2 infection. Front Immunol, 2023. 14: p. 1285345.
- Spalton, J.C., et al., The novel Syk inhibitor R406 reveals mechanistic differences in the initiation of GPVI and CLEC-2 signaling in platelets. J Thromb Haemost, 2009. 7(7): p. 1192-9.
- Harbi, M.H., et al., Antithrombotic Effects of Fostamatinib in Combination with Conventional Antiplatelet Drugs. Int J Mol Sci, 2022. 23(13).
- Apostolidis, S.A., et al., Signaling Through FcgammaRIIA and the C5a-C5aR Pathway Mediate Platelet Hyperactivation in COVID-19. Front Immunol, 2022. 13: p. 834988.
- Saha, B., et al., Human tumor microenvironment chip evaluates the consequences of platelet extravasation and combinatorial antitumor-antiplatelet therapy in ovarian cancer. Sci Adv, 2021. 7(30).
- Iba, T. and J.H. Levy, The roles of platelets in COVID-19-associated coagulopathy and vaccine-induced immune thrombotic thrombocytopenia. Trends Cardiovasc Med, 2022. 32(1): p. 1-9.
- Bye, A.P., et al., Aberrant glycosylation of anti-SARS-CoV-2 spike IgG is a prothrombotic stimulus for platelets. Blood, 2021. 138(16): p. 1481-1489.
- Gonzalez-Lopez, T.J., et al., Fostamatinib effectiveness and safety for immune thrombocytopenia in clinical practice. Blood, 2024. 144(6): p. 646-656.
- Hu, M., et al., Drug repurposing of fostamatinib against cancer via potential cytotoxicity and immune checkpoint regulation. Front Immunol, 2025. 16: p. 1602189.
- Choi, Y., et al., Repurposing of the Syk inhibitor fostamatinib using a machine learning algorithm. Exp Ther Med, 2025. 29(6): p. 110.
- Park, S.R., et al., A multi-histology trial of fostamatinib in patients with advanced colorectal, non-small cell lung, head and neck, thyroid, and renal cell carcinomas, and pheochromocytomas. Cancer Chemother Pharmacol, 2013. 71(4): p. 981-90.
- Gaillard, S., et al., A phase 1 study of the SYK inhibitor fostamatinib and weekly paclitaxel for recurrent platinum-resistant ovarian cancer. 2023, American Society of Clinical Oncology.
- Schror, K., Aspirin and platelets: the antiplatelet action of aspirin and its role in thrombosis treatment and prophylaxis. Semin Thromb Hemost, 1997. 23(4): p. 349-56.
- Pulcinelli, F.M., et al., Inhibition of platelet aggregation by aspirin progressively decreases in long-term treated patients. J Am Coll Cardiol, 2004. 43(6): p. 979-84.
- van Zijverden, L.M., et al., The efficacy of aspirin to inhibit platelet aggregation in patients hospitalised with a severe infection: a multicentre, open-label, randomised controlled trial. Clin Exp Med, 2023. 23(7): p. 3501-3508.
- Florensa, D., et al., Low-dose acetylsalicylic acid for cancer prevention considering risk factors: a retrospective cohort study. Ann Epidemiol, 2023. 84: p. 60-66.
- Yang, J., et al., Aspirin prevents metastasis by limiting platelet TXA(2) suppression of T cell immunity. Nature, 2025. 640(8060): p. 1052-1061.
- Sevigny, J., et al., The antibody aducanumab reduces Abeta plaques in Alzheimer’s disease. Nature, 2016. 537(7618): p. 50-6.
- Mussbacher, M., et al., Cell Type-Specific Roles of NF-kappaB Linking Inflammation and Thrombosis. Front Immunol, 2019. 10: p. 85.
- Crimmins, E.M., Lifespan and Healthspan: Past, Present, and Promise. Gerontologist, 2015. 55(6): p. 901-11.
- Finch, C.E., Evolution in health and medicine Sackler colloquium: Evolution of the human lifespan and diseases of aging: roles of infection, inflammation, and nutrition. Proc Natl Acad Sci U S A, 2010. 107 Suppl 1(Suppl 1): p. 1718-24.
- Wendelboe, A.M. and G.E. Raskob, Global Burden of Thrombosis: Epidemiologic Aspects. Circ Res, 2016. 118(9): p. 1340-7.
- Gu, S.X. and S. Dayal, Inflammation mediated platelet hyperactivity in aging. Ann Blood, 2020. 5.
- Price, J., J.M. Lord, and P. Harrison, Inflammaging and platelet hyperreactivity: A new therapeutic target? J Thromb Haemost, 2020. 18(1): p. 3-5.
- Klavina, P.A., et al., Dysregulated haemostasis in thrombo-inflammatory disease. Clin Sci (Lond), 2022. 136(24): p. 1809-1829.
- Byun, J.S. and K. Gardner, Wounds that will not heal: pervasive cellular reprogramming in cancer. Am J Pathol, 2013. 182(4): p. 1055-64.
- Martins Castanheira, N., et al., Uptake of platelets by cancer cells and recycling of the platelet protein CD42a. J Thromb Haemost, 2022. 20(1): p. 170-181.
- Inc., A., Anaconda Software Distribution. 2020: Anaconda Documentation.
- Wilke, C., _cowplot: Streamlined Plot Theme and Plot Annotations for ‘ggplot2’_. 2024.
- Wu, L., et al., CytoCtrlAnalyser: a Cytoscape app for biomolecular network controllability analysis. Bioinformatics, 2018. 34(8): p. 1428-1430.
- Shannon, P., et al., Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res, 2003. 13(11): p. 2498-504.
- Ceccarelli, F., et al., Bringing data from curated pathway resources to Cytoscape with OmniPath. Bioinformatics, 2020. 36(8): p. 2632-2633.
- Oleś, A., DEFormats: Differential gene expression data formats converter. 2024.
- Love, M.I., W. Huber, and S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 2014. 15(12): p. 550.
- Chen Y, C.L., Lun ATL, Baldoni P, Smyth GK edgeR 4.0: powerful differential analysis of sequencing data with expanded functionality and improved support for small counts and larger datasets. bioRxiv, 2024.
- Blighe, K., S. Rana, and M. Lewis, EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling. 2023.
- Rainer, J., EnsDb.Hsapiens.v86: Ensembl based annotation package. 2017.
- Chen, S., et al., fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics, 2018. 34(17): p. i884-i890.
- Andrews, S. FastQC: a quality control tool for high throughput sequence data. 2010 [cited 2024]. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Wickham, H., ggplot2: Elegant Graphics for Data Analysis. 2016: Springer-Verlag New York.
- Kassambara, A., ggpubr: ‘ggplot2’ Based Publication Ready Plots. 2023.
- Pedersen, T.L., ggraph: An Implementation of Grammar of Graphics for Graphs and Networks. 2024.
- Ahlmann-Eltze, C. and W. Huber, glmGamPoi: fitting Gamma-Poisson generalized linear models on single cell count data. Bioinformatics, 2021. 36(24): p. 5701-5702.
- Kolberg, L., et al., gprofiler2 -- an R package for gene list functional enrichment analysis and namespace conversion toolset g:Profiler. F1000Res, 2020. 9.
- Iannone, R., et al., gt: Easily Create Presentation-Ready Display Tables. 2024.
- Csárdi, G., et al., igraph: Network Analysis and Visualization in R. 2024.
- Bray, N.L., et al., Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol, 2016. 34(5): p. 525-7.
- Ritchie, M.E., et al., limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res, 2015. 43(7): p. e47.
- Mills, B., MetBrewer: Color Palettes Inspired by Works at the Metropolitan Museum of Art. 2022.
- Ewels, P., et al., MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics, 2016. 32(19): p. 3047-8.
- Valdeolivas, A., D. Turei, and A. Gabor, OmnipathR: client for the OmniPath web service. 2019.
- Carlson, M., org.Hs.eg.db: Genome wide annotation for Human. 2023.
- Kolde, R., pheatmap: Pretty Heatmaps. 2019.
- Sievert, C., Interactive Web-Based Data Visualization with R, plotly, and shiny. 2020: Chapman and Hall/CRC Florida.
- Fischer, B.S., M.; Pau, G., rhdf5: R Interface to HDF5. R package version 2.48.0. 2024.
- Posit team, T., RStudio: Integrated Development Environment for R. 2024, Posit Software, PBC: Boston, MA.
- Risso, D., et al., Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol, 2014. 32(9): p. 896-902.
- Cornish, A.J. and F. Markowetz, SANTA: Quantifying the Functional Content of Molecular Networks. PLOS Computational Biology, 2014. 10(9): p. e1003808.
- Wickham, H.A., M,; Bryan, J.; Chang, W.; McGowan, LD.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; Kuhn, M; Pedersen, TL; Miller, E-; Bache, SM.; Müller, K.; Ooms, J.; Robinson, D.; Seidel, DP.; Spinu, V.; Takahashi, K.; Vaughan, D.; Wilke, C.; Woo, K.; Yutani, H., Welcome to the tidyverse. Journal of Open Source Software, 2019. 4(43).
- Soneson, C., M.I. Love, and M.D. Robinson, Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res, 2015. 4: p. 1521.
- Lauss, M., swamp: Visualization, Analysis and Adjustment of High-Dimensional Data in Respect to Sample Annotations. 2019.
- Wishart, D.S., et al., DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res, 2018. 46(D1): p. D1074-D1082.
- Best, M.G., et al., Swarm Intelligence-Enhanced Detection of Non-Small-Cell Lung Cancer Using Tumor-Educated Platelets. Cancer Cell, 2017. 32(2): p. 238-252 e9.
- Vinayagam, A., et al., Controllability analysis of the directed human protein interaction network identifies disease genes and drug targets. Proc Natl Acad Sci U S A, 2016. 113(18): p. 4976-81.
- Jia, T., et al., Emergence of bimodality in controlling complex networks. Nat Commun, 2013. 4: p. 2002.
- Garrido-Mesa, N., A. Zarzuelo, and J. Galvez, Minocycline: far beyond an antibiotic. Br J Pharmacol, 2013. 169(2): p. 337-52.
- Sapadin, A.N. and R. Fleischmajer, Tetracyclines: nonantibiotic properties and their clinical implications. J Am Acad Dermatol, 2006. 54(2): p. 258-65.
- Wei, X., et al., Minocycline prevents gentamicin-induced ototoxicity by inhibiting p38 MAP kinase phosphorylation and caspase 3 activation. Neuroscience, 2005. 131(2): p. 513-21.
- Chen, M., et al., Minocycline inhibits caspase-1 and caspase-3 expression and delays mortality in a transgenic mouse model of Huntington disease. Nat Med, 2000. 6(7): p. 797-801.
- Wang, X., et al., Minocycline inhibits caspase-independent and -dependent mitochondrial cell death pathways in models of Huntington’s disease. Proc Natl Acad Sci U S A, 2003. 100(18): p. 10483-7.
- Festoff, B.W., et al., Minocycline neuroprotects, reduces microgliosis, and inhibits caspase protease expression early after spinal cord injury. J Neurochem, 2006. 97(5): p. 1314-26.
- Berens, S.C., C.M. Bird, and N.A. Harrison, Minocycline differentially modulates human spatial memory systems. Neuropsychopharmacology, 2020. 45(13): p. 2162-2169.
- Murphy L, Inchauspé J, Valenzano G, Holland P, Sousos N, Belnoue-Davis HL, Li R, Jooss NJ, Benlabiod C, Murphy E, Etzioni Z, Shepherd E, Denly L, Biswas S, Chen L, O’Sullivan J, Rimmer MP, Khan AO, Karali CS, Nasreddin N, Hitchcock IS, Koupenova M, Kriaucionis S, Hughes JR, O’Neill E, Vatish M, Rees P, Leedham S, Desborough M, Mead AJ, Schuster-Böckler B, Gregory CD, Psaila B. Platelets sequester extracellular DNA, capturing tumor-derived and free fetal DNA. Science. 2025 Aug 14; 389(6761):eadp3971.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





