1. Introduction
Freshwater fungi are a group of microorganisms that play a crucial role in the ecological dynamics of freshwater ecosystems such as rivers, streams, and lakes. They function as key decomposers of phytoplankton and other organic matter in these environments. To date, approximately 3,870 freshwater fungal species have been identified [
1]. These fungi have developed unique metabolic pathways to adapt to various environmental stresses, resulting in the production of diverse secondary metabolites [
2]. These metabolic products are not only essential for their survival and growth but also exhibit various biological activities [
2]. Research has shown that freshwater fungi can produce compounds with physiological activities, such as antibacterial, antifungal, antiviral, and anticancer activities, rendering them potential candidates for drug development. Although fungi are widely acknowledged as a prolific source of bioactive secondary metabolites, freshwater fungi have been relatively underexplored in this regard [
2,
3]. Research on the metabolic products of freshwater fungi remains in its early stages and is significantly less advanced compared to studies on marine and terrestrial fungi. However, interest in the metabolites produced by freshwater fungi has grown in recent years, and their potential medicinal applications are being actively investigated [
3,
4].
The emergence and spread of microorganisms have become a significant public health concern in contemporary society. Pathogenic microorganisms, in particular, pose a significant threat to human health by causing infectious diseases [
5]. Consequently, the effective suppression and management of these microorganisms have become crucial areas of research and development. Although traditional antibiotics and antifungal agents are widely used, the growing prevalence of resistance to these treatments highlights the urgent need for novel alternative compounds [
6].
Trichoderma flavipes is a species within the phylum Ascomycota, class Sordariomycetes, order Hypocreales, and genus
Trichoderma. The genus
Trichoderma is recognized for its notable biological and ecological significance [
7,
8]. Species of this genus produce antibiotics that inhibit the growth of other microorganisms, including fungi, bacteria, and viruses, with a particular efficacy for suppressing plant pathogens [
9]. Additionally,
Trichoderma species are known to secrete plant growth hormones, such as auxins and cytokinins, and produce enzymes, such as cellulase, chitinase, and protease [
9,
10,
11]. Owing to these beneficial properties, the genus
Trichoderma has become an important focus of ongoing research in life sciences and industrial applications. However,
T. flavipes has not been extensively studied.
This study aimed to explore and analyze the chemical properties of metabolites produced by the freshwater fungus Trichoderma flavipes FBCC-F1632 and assess its antibacterial activities.
2. Materials and Methods
2.1. Isolation and Identification of FBCC-F1632
The fungal strain FBCC-F1632 was isolated from a freshwater sample collected from Kimyong-ri (N36°44′26.1″, E128°13′19″), Sanbuk-myeon, Mungyeong-si, South Korea, during the summer of July 2021. At the sampling site, a 50 mL sample of freshwater was filtered directly using a hand pump and an MCE membrane filter (HAWP04700, MF-Millipore™, Tullagreen, Ireland). Subsequently, the filter paper was placed on water agar plates containing 20 g/L agar (Difco; BD, Franklin Lakes, NJ, USA), 1 L distilled water, and 100 ppm streptomycin (Sigma, Darmstadt, Germany) to inhibit bacterial growth. The samples were transported to the laboratory and incubated at 15 °C to promote spore isolation. Single spores were isolated, and purified fungal cultures were transferred to potato dextrose agar (PDA; Difco) and incubated at 20 °C to establish pure cultures. All fungal strains used in this study were maintained on PDA at 25 °C. Morphological characteristics were observed using an Eclipse Ni-U microscope (Nikon, Tokyo, Japan).
2.2. DNA Extraction and Phylogenetic Analysis
Fungal genomic DNA was extracted using the NucleoSpin Plant II DNA extraction kit (Macherey-Nagel, Duren, Germany) according to the manufacturer’s instructions. To identify the fungal species, the RNA polymerase II (RPB2) gene was targeted and amplified using specific primers 5f (5′-GAYGAYMGWGATCAYTTYGG-3′) and 7cr (5′-CCCATRGCTTGYTTRCCCAT-3′) [
12]. DNA sequence homology was assessed by performing BLAST searches on the National Center for Biotechnology Information (NCBI) database to compare the sequences with existing records. For phylogenetic analysis, MEGA11 software [
13] was used to construct and analyze the evolutionary relationships among the fungal strains. A phylogenetic tree was generated using the neighbor-joining method with 1,000 bootstrap replications. This analysis included 36 nucleotide sequences. All positions with less than 95% site coverage were eliminated, indicating that fewer than 5% alignment gaps, missing data, or ambiguous bases were allowed at any position (partial deletion option). The final dataset comprised 889 positions. Reference sequences for other fungi were obtained from GenBank at NCBI.
2.3. Antimicrobial Activity
A disk diffusion assay was conducted to evaluate the antibacterial activity against Staphylococcus aureus CCARM3089 and Bacillus cereus CCARM0120. The overlay medium was prepared by pouring 20 mL of R2A agar (Difco) containing a 5% suspension of S. aureus and B. cereus (adjusted to OD600 = 0.5) into 9 cm petri dishes and allowing it to solidify. Next, 50 µL of the extract from T. flavipes FBCC-F1632 was carefully applied to an 8 mm sterile paper disc, which was subsequently placed at the center of the solidified overlay medium. The plates were incubated at 30 °C for 1–2 days to enable the extract to diffuse into the medium and interact with the pathogen for the determination of MIC and MBC values.
2.4. Material Separation and Purification
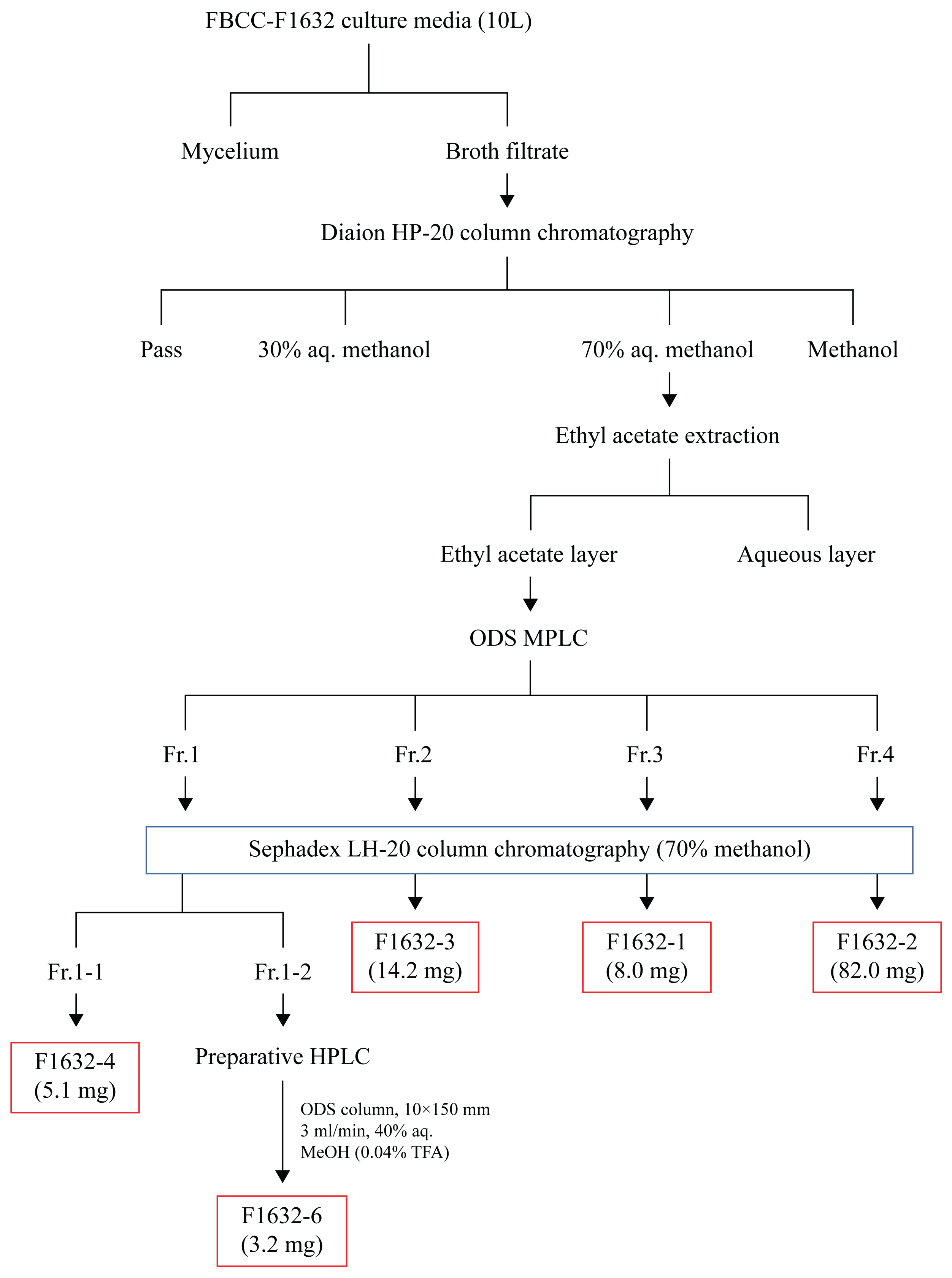
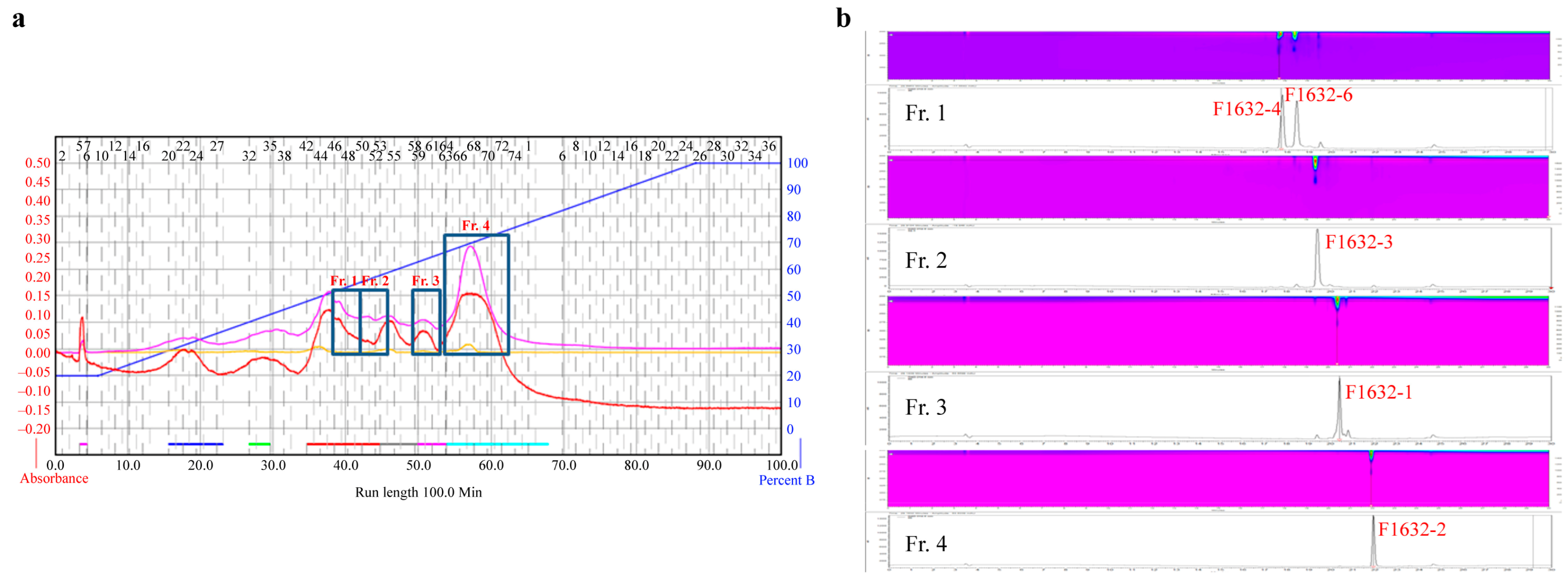
FBCC-F1632 culture broth (10 L) was centrifuged at 3000 rpm for 30 min at 4 ℃, separating the supernatant and pellet. Next, 1 L Diaion HP-20 resin (Supelco, Darmstadt, Germany) was added to the supernatant to adsorb the active compounds. The adsorbed resin was filled into a column and eluted using 30%, 70%, and 100% methanol aqueous solutions (Thermo Fisher Scientific, Waltham, MA, USA). The fraction eluted with the 70% methanol aqueous solution, which exhibited activity, was concentrated under reduced pressure to remove the solvent. Ethyl acetate (EA, Thermo Fisher Scientific) was subsequently added for distribution extraction. The dispensed EA layer was concentrated under reduced pressure and fractionated using octadecylsilane (ODS; Sigma, Darmstadt, Germany) medium-pressure liquid chromatography (MPLC). ODS MPLC was conducted using Combiflash RF+ (Teledyne ISCO, Nebraska, USA) equipment, employing deionized water-methanol as a mobile phase and a C18 column (86 g, Teledyne ISCO, Nebraska, USA). Based on the ODS MPLC analysis, the compound was separated and purified by dividing it into four groups (fraction [Fr.] 1, Fr. 2, Fr. 3, and Fr. 4), followed by Sephadex LH-20 column chromatography (conditions added). The entire separation process is schematically shown in
Figure 1.
2.5. Structure Analysis
Electrospray ionization (ESI)-mass spectra were obtained using ESI-QTRAP-3200 Mass Spectrometer (Applied Biosystems, Thermo Fisher Scientific), whereas NMR spectra were obtained using a JEOL JNM-ECZ500R 500 MHz FT-NMR Spectrometer (JEOL, Ltd., Tokyo, Japan) at 500 MHz for 1H NMR and at 125 MHz for 13C NMR, using CD3OD (deuterated methanol, Thermo Fisher Scientific) as the solvent. Chemical shifts are presented in parts per million (ppm, δ) with tetramethylsilane as the internal standard. For NMR analysis, both two-dimensional NMR, such as 1H-1H correlation spectroscopy, heteronuclear single quantum coherence (HMQC), and heteronuclear multiple bond correlation (HMBC), and one-dimensional NMR, such as 1H NMR and 13C NMR, were employed.
3. Results
3.1. Isolation and Identification of FBCC-F1632
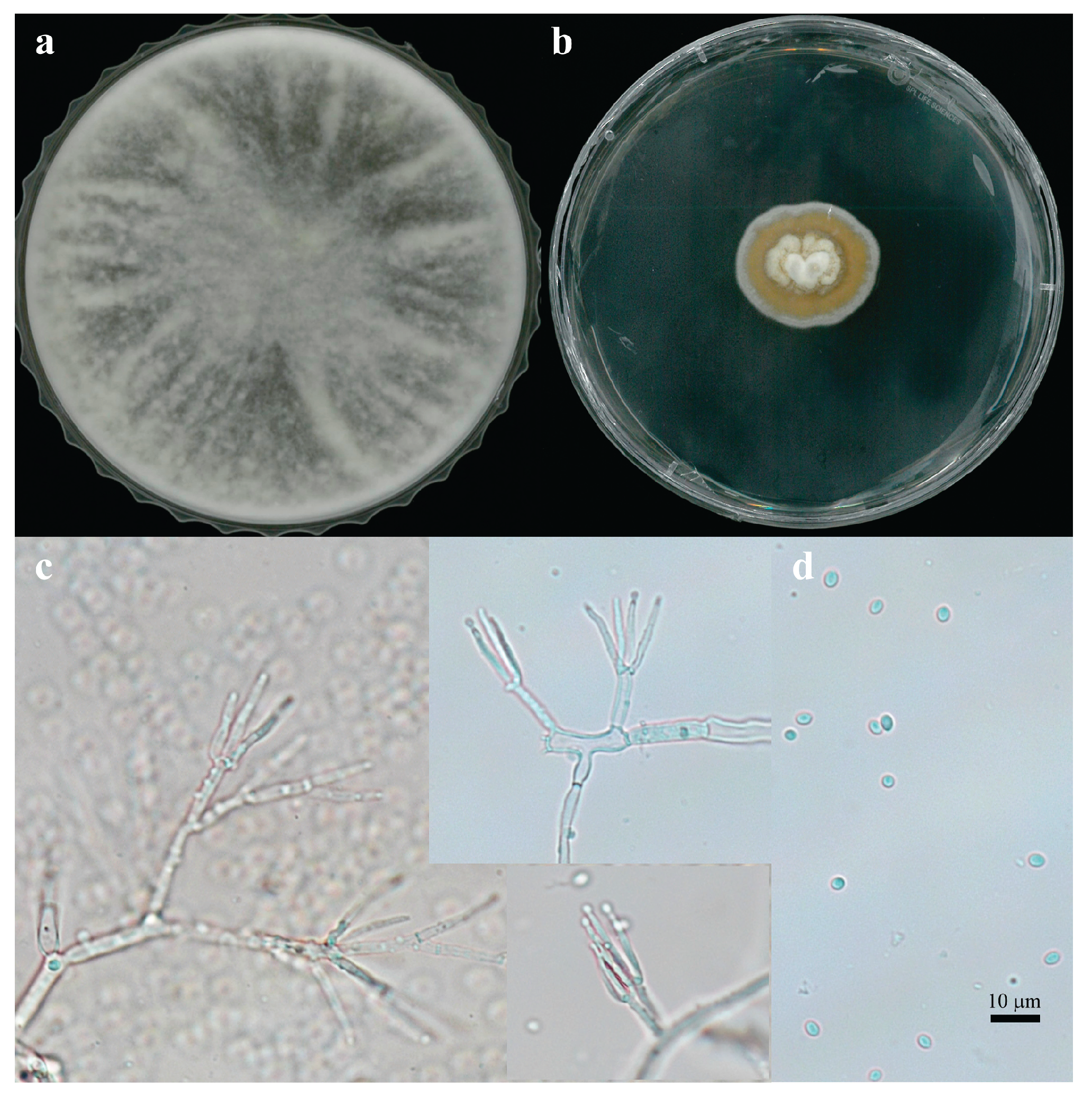
The unique morphological and genetic characteristics of FBCC-F1632 provided valuable insights into its classification and ecological adaptability. The dual-colony morphology observed in FBCC-F1632, with growth patterns resembling those of both
Trichoderma and
Stilbella species, suggests a possible adaptive mechanism that enables this isolate to thrive under varying environmental conditions. This adaptability is particularly noteworthy, given that
Trichoderma species are generally known for their rapid growth, whereas
Stilbella species exhibit slower, more conservative growth patterns [
14]. Such morphological variability could be a response to differing nutrient availability or environmental pressures in freshwater ecosystems (
Figure 2a, b).
The conidial morphology of FBCC-F1632, characterized by globose conidia and penicillate, slender phialides, further supports its close relationship with
T. flavipes (
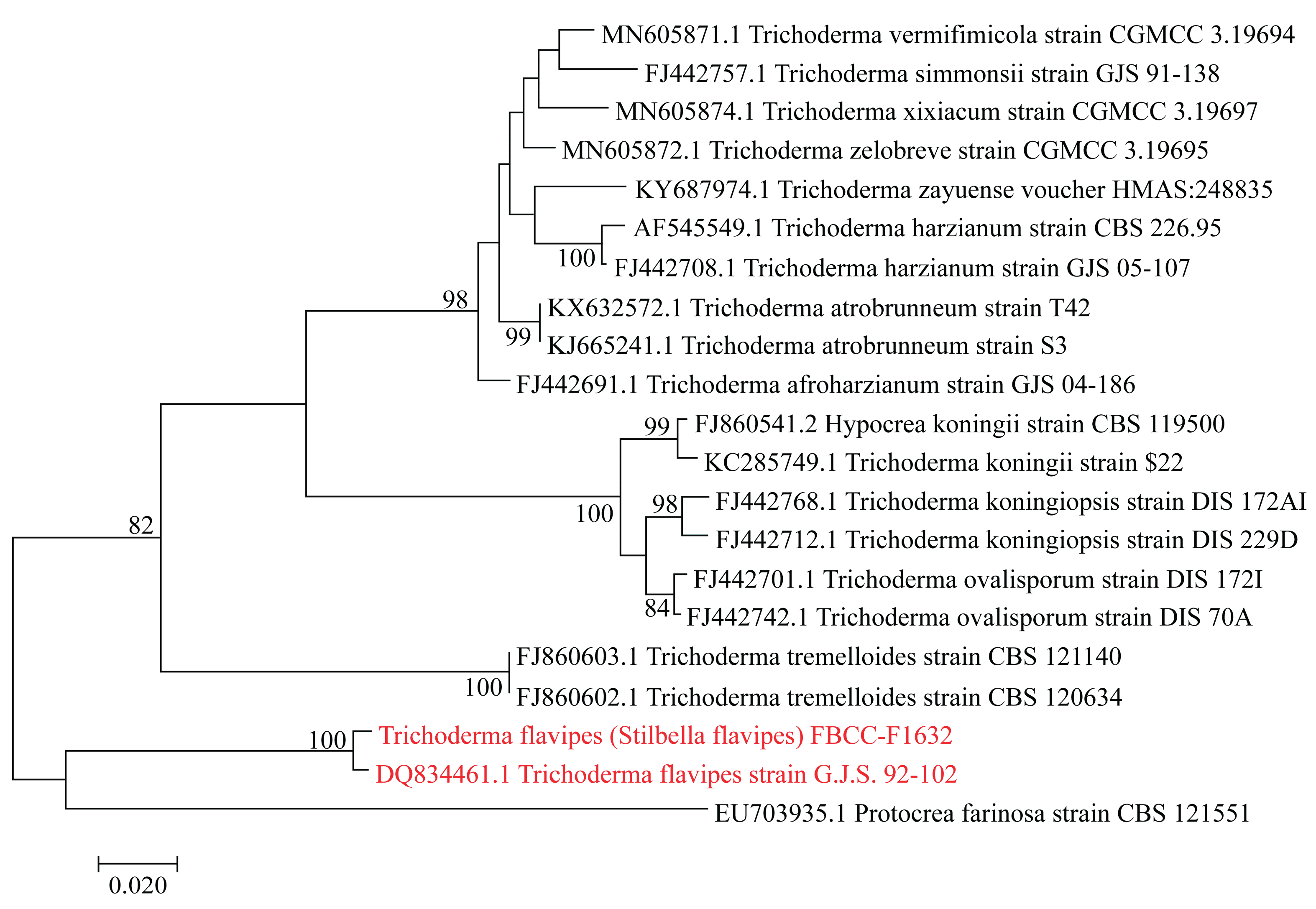
Figure 2c). These morphological features, coupled with the high sequence similarity (99.10%) to
T. flavipes based on the RPB2 gene, firmly place FBCC-F1632 within the
Trichoderma clade (
Figure 3). The genetic similarity also suggests that FBCC-F1632 may share biological or ecological functions with
T. flavipes, such as antifungal or antibacterial activities, which are commonly observed in many
Trichoderma species known for their biocontrol properties [
15].
Phylogenetic note: phylogenetic analysis revealed a consistency index of 0.663194 (0.630476), a retention index of 0.819535 (0.819535), and a composite index of 0.543511 (0.516697) for all sites and parsimony-informative sites (in parentheses). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is indicated next to the branches [
16]. The maximum parsimony tree was constructed using the tree-bisection-regrafting algorithm [
17] with a search level of 2, where initial trees were generated by the random addition of sequences (10 replicates). This analysis involved 20 nucleotide sequences, with a total of 858 positions in the final dataset. Evolutionary analyses were conducted using MEGA11 [
13].
3.2. Antimicrobial Activity of FBCC-F1632
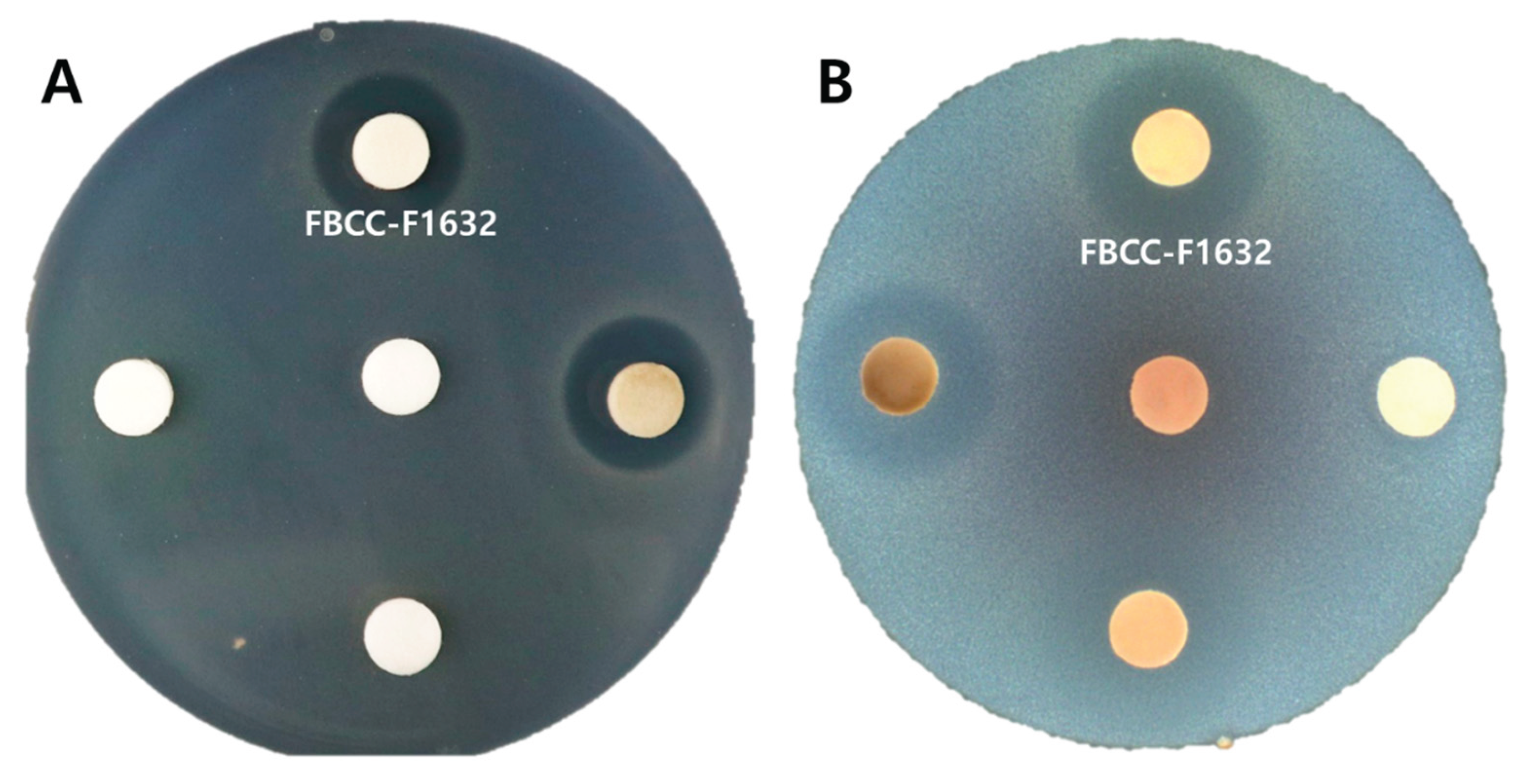
This study investigates the antimicrobial efficacy of FBCC-F1632 against two clinically relevant bacterial strains:
Staphylococcus aureus CCARM3089 and
Bacillus cereus CCARM0120. The observed inhibition zone of 17.67 ± 0.58 mm and 22.33 ± 0.31 indicates that
T. flavipes produces compounds capable of targeting resistant strains (
Figure 4), which is significant given the increasing concern over antibiotic resistance in pathogenic bacteria. These results suggest that the culture filtrate of
T. flavipes may contain metabolites with unique antibacterial mechanisms that could overcome common resistance pathways in
S. aureus and
B. cereus.
The ability of T. flavipes to inhibit antibiotic-resistant S. aureus expands its potential applications, extending beyond agriculture to areas such as food safety and potentially clinical settings, where resistant infections pose substantial health risks. Identifying and characterizing the specific compounds responsible for this activity could pave the way for developing alternative antimicrobial agents or supplements to existing antibiotics. Additionally, investigating whether these compounds are effective against other resistant strains could provide a broader understanding of the antibacterial spectrum of T. flavipes.
3.3. Compound Extraction from FBCC-F1632
Bioactive compounds were successfully extracted from FBCC-F1632 and fractionated, highlighting the potential of this isolate as a source of novel metabolites. A combination of EA extraction, ODS MPLC, and Sephadex LH-20 column chromatography facilitated the effective separation and isolation of distinct compounds, representing essential steps in identifying and characterizing bioactive molecules. Specific compounds were isolated in varying quantities, such as F1632-2 (58.0 mg), the most abundant compound, and F1632-6 (3.2 mg), which was further purified using preparative high-performance liquid chromatography (HPLC). These results suggest a wide range of metabolite diversity within FBCC-F1632. The successful isolation of F1632-6 in pure form is particularly significant, as it can facilitate its subjection to further structural and bioactivity studies to assess its potential applications (
Figure 5). These methodologies, including fractionation by ODS MPLC and final purification through HPLC, highlight the importance of multistep chromatographic processes for accessing pure forms of bioactive metabolites from complex fungal extracts.
3.4. Structure Analysis
3.4.1. Structure of F1632-1
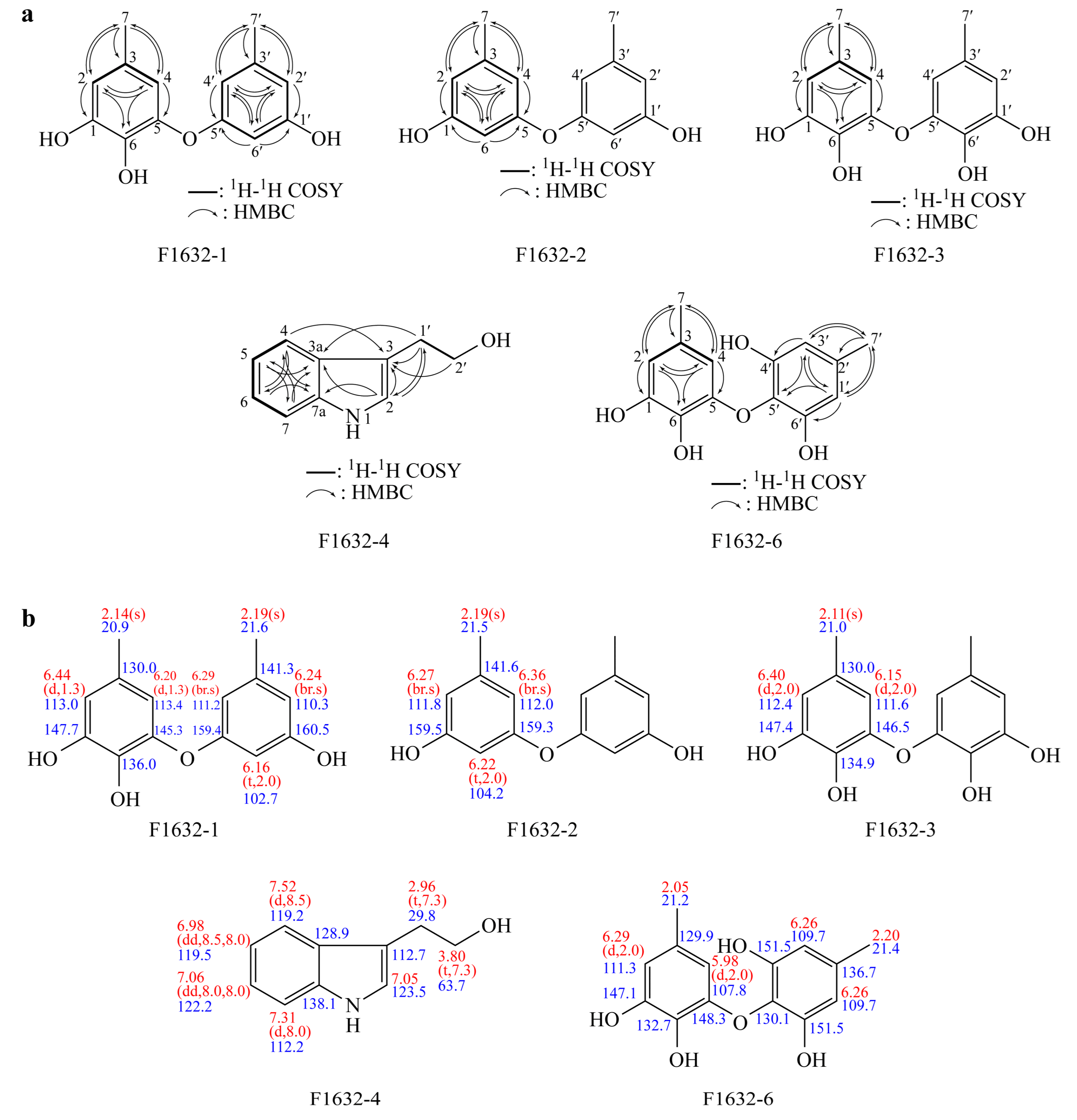
The comprehensive spectral analysis of F1632-1 provided detailed information on its molecular structure and potential biological activities. ESI-MS analysis determined a molecular weight of 246, consistent with that of known metabolites in the cordycepin family. This molecular characterization is crucial for understanding the properties of the compound and its possible applications in pharmaceuticals and natural product development. The
1H NMR spectrum revealed distinctive peaks corresponding to aromatic methine protons and methyl groups, indicating a complex structure typical of phenolic compounds. Five aromatic methine protons were identified, suggesting a highly substituted aromatic ring, which is characteristic of compounds with potential antioxidant or antimicrobial properties. Chemical shifts and splitting patterns were observed, providing a strong basis for deducing the connectivity of the protons and the overall arrangement of the molecule. The
13C NMR spectrum revealed multiple oxygenated sp² carbons, indicating the presence of functional groups known for their biological activities, such as hydroxyl or carbonyl groups, which may contribute to the reactivity of the compound and its interactions with biological targets (
Table 1). Long-range correlations from HMQC and HMBC spectra further elucidated the structural framework, indicating a well-defined spatial arrangement, which is essential for understanding the mechanism of action of the compound (
Figure 6).
3.4.2. Structure of F1632-2
The spectral analysis of F1632-2 provided crucial information on its molecular structure and potential biological properties. ESI-mass analysis determined a molecular weight of 230, consistent with that of known polyphenolic metabolites, which are often associated with significant bioactivity. The
1H NMR spectrum revealed multiple aromatic methine protons, suggesting a symmetrical aromatic system typical of polyphenols. Splitting patterns and chemical shifts were observed, indicating the electronic environment of the protons and offering insights into the connectivity within the molecule. The methyl proton peak at 2.19 ppm, with a relative intensity of 6H, indicates the presence of two equivalent methyl groups, which is a hallmark of compounds such as diorcinol. In the
13C NMR spectrum, oxygenated sp² carbon peaks at 159.5 and 159.3 ppm were identified, indicating the presence of functional groups often implicated in antioxidant and antimicrobial activities. The distribution of sp² methine and methyl carbon peaks further supports the notion of a complex molecular framework, which is critical for understanding the interactions between the compound and biological systems (
Table 1). Long-range correlations were observed from the HMQC and HMBC spectra, confirming the structural integrity of F1632-2. The correlations observed between the methyl protons and various carbons within the aromatic system highlight the importance of these groups in stabilizing the overall structure through intramolecular interactions (
Figure 6). This connectivity is vital for elucidating the potential mechanisms by which diorcinol exerts its biological effects.
3.4.3. Structure of F1632-3
The characterization of F1632-3 as violaceol I involved a comprehensive analytical approach, highlighting the importance of employing a multi-faceted analytical approach to identify and understand natural products. ESI-mass analysis determined the molecular weight of the compound, providing a solid foundation for subsequent structural elucidation and confirming the identity of the compound [
18]. The
1H NMR spectrum exhibited distinct patterns, indicating a well-defined structure with symmetrical features, likely contributing to the stability and bioactivity of the compound. The methyl proton peak revealed multiple methyl groups, which may influence the solubility and interactions of the compound with biological targets. In the
13C NMR spectrum, oxygenated sp² carbons were identified, which are often associated with enhanced biological activities, including antioxidant and antimicrobial effects (
Table 1) [
19]. Long-range correlations from the HMQC and HMBC spectra further validated the structural assignments, providing further insights into the connectivity of the molecule and elucidating the relationships between various functional groups (
Figure 6).
3.4.4. Structure of F1632-4
The characterization of F1632-4 as tryptophol involved comprehensive spectroscopic analysis, highlighting the importance of employing detailed spectroscopic analysis to elucidate the structures of natural products. ESI-mass analysis determined the molecular weight of the compound, providing a solid foundation for subsequent structural elucidation and confirming the identity of the compound. The
1H NMR spectrum revealed multiple aromatic methine protons, suggesting a well-defined aromatic system crucial for the biological activity typical of tryptophols. Moreover, the presence of methylene protons indicated a complex molecular framework that may facilitate interactions with biological targets. In the
13C NMR spectrum, sp² carbon peaks were identified, indicating functional groups that may contribute to the potential bioactivity of the compound (
Table 1). Long-range correlations were observed in the HMQC and HMBC spectra, providing further insights into the connectivity of the compound and supporting the structural assignments made based on the proton and carbon chemical shifts (
Figure 6).
3.4.5. Structure of F1632-6
The characterization of F1632-6 as violaceol II involved detailed spectroscopic analysis, highlighting the importance of comprehensive spectroscopic analysis for the structural elucidation of natural products. ESI-mass analysis determined the molecular weight of the compound, providing a strong basis for subsequent structural interpretations. The
1H NMR spectrum revealed several aromatic methine protons, indicating a robust aromatic system critical for biological activity typical of compounds such as violaceol II [
18]. The presence of multiple methyl groups suggests that these structural elements may enhance the lipophilicity of the compound, potentially influencing its bioavailability and interaction with biological targets. In the
13C NMR spectrum, sp² carbon peaks were detected, confirming the presence of functional groups relevant to the known properties of violaceol II (
Table 1) [
19]. Long-range correlations were observed in the HMQC and HMBC spectra, providing further insights into the connectivity of the compound and corroborating the assignments made based on the chemical shifts (
Figure 6).
3.5. Antimicrobial Activity of Compounds Isolated from FBCC-F1632
The antimicrobial activities of five isolated compounds (F1632-1 to F1632-5) were evaluated against Staphylococcus aureus and Bacillus cereus using minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) assays (
Table 2). The selection of these bacterial strains is particularly relevant, as both strains are known to cause serious infections in humans and are associated with various foodborne illnesses [
20,
21].
Among the tested compounds, Violaceol I (F1632-3) exhibited the lowest MIC value against S. aureus (12.5 µg/mL), indicating the highest inhibitory potency toward this strain. For B. cereus, Cordyol C (F1632-1) demonstrated the lowest MIC (12.5 µg/mL), followed by Violaceol II (F1632-5) and Violaceol I (F1632-3) with MIC values of 25 µg/mL.
MBC values were generally higher or equal to the MICs, with most compounds showing an MBC of 50 µg/mL against both bacterial species. Notably, Cordyol C displayed a fourfold difference between MIC and MBC against B. cereus (12.5 µg/mL vs. 50 µg/mL), suggesting a primarily bacteriostatic effect at lower concentrations.
4. Discussion
This study investigated the antimicrobial potential of compounds isolated from T. flavipes FBCC-F1632, including cordyol C, diorcinol, violaceol I, tryptophol, and violaceol II. Although this strain demonstrated effectiveness against fungal pathogens under co-culture conditions, the extracted compounds did not exhibit antifungal activity in isolation. This discrepancy suggests that certain synergistic effects or environmental conditions present during co-culture may be necessary to activate or enhance the antifungal properties of these compounds.
The identification of F1632-1 as cordyol C through database searches underscores its importance in natural product chemistry. Cordyol C, initially isolated from
Cordyceps sp. BCC1861, exhibits diverse bioactivities, including antimicrobial, antimalarial, and cytotoxic effects, indicating its potential as a multifunctional therapeutic agent. Similarly,
Aspergillus sydowii ZSDS1-F6, associated with marine sponges, produces various antimicrobial and antiviral sesquiterpenoids [
22]. Among related compounds, cordyol E from
Aspergillus sp. XS-20090066 has demonstrated significant antibacterial effects against pathogens, such as
Staphylococcus epidermidis, S. aureus, Vibrio anguillarum, Vibrio parahemolyticus, and Pseudomonas putida [
22,
23,
24]. F1632-2 was identified as diorcinol, a compound initially derived
from Aspergillus tabacinus and
Emericella falconensis, underscoring its relevance in natural product research [
25,
26]. Diorcinol has been demonstrated to exhibit antimicrobial properties against plant pathogenic fungi and bacteria, further supporting its potential in therapeutic applications [
25,
26]. Furthermore, F1632-3 and F1632-6 were identified as violaceol I and violaceol II, respectively, both of which were originally isolated from
A. tabacinus, Trichoderma polyalthiae, and
Emericella violacea [
18,
19,
25]. These compounds exhibit a broad range of bioactivities, including antibacterial, antimalarial, anti-tuberculous, anti-herpes simplex virus type I, and cytotoxic effects [
18,
19,
25]. The extensive bioactivities associated with these compounds highlight their potential for various medical applications, although further studies are required to elucidate their specific mechanisms of action.
Violaceol I demonstrated the strongest inhibitory effect against S. aureus, while Cordyol C was most effective against B. cereus. The relatively small gap between MIC and MBC values for most compounds suggests that bactericidal activity is achieved at concentrations close to the inhibitory threshold. Interestingly, Diorcinol (F1632-2) and Tryprophol (F1632-4) exhibited identical MIC and MBC values (50 µg/mL) for both bacterial strains, indicating moderate and non-selective antibacterial activity. In contrast, the violaceol derivatives (F1632-3 and F1632-5) showed more variable MICs, implying that subtle structural modifications may enhance or reduce potency against specific Gram-positive bacteria.
In conclusion, freshwater fungi represent a promising source of antimicrobial metabolites. Future research should focus on the bioprospecting of these organisms, focusing on isolating and characterizing novel compounds that can address current challenges in health and agriculture. By doing so, this study not only provides a better understanding of the complexities of microbial interactions within freshwater ecosystems but also paves the way for practical applications across various fields.
Author Contributions
Conceptualization, H.Y.M.; methodology, W.S.C., S.L., and S.J.; formal analysis, H.Y.M.; investigation, J.G., W.S.C., S.L., and S.J.; writing—original draft preparation, J.T.K. and H.Y.M.; writing—review and editing, J.G. and C.S.L.; visualization, J.T.K. and H.Y.M.; project administration, C.S.L. and H.Y.M.; funding acquisition, C.S.L. and H.Y.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Nakdonggang National Institute of Biological Resources (NNIBR) (Grant number NNIBR20251105) and the Korea Environment Industry & Technology Institute (KEITI) through a project to make multi-ministerial national biological research resources a more advanced program funded by the Korean Ministry of Environment (MOE) (Grant number 2021003420002).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
We gratefully acknowledge NP ChemPharma Co., Ltd. for their valuable assistance in the structural analysis.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Calabon, M.S.; Hyde, K.D.; Jones, E.B.G.; Luo, Z.-L.; Dong, W.; Hurdeal, V.G.; Gentekaki, E.; Rossi, W.; Leonardi, M.; Thiyagaraja, V.; Lestari, A.S.; Shen, H.-W.; Bao, D.-F.; Boonyuen, N.; Zeng, M. Freshwater fungal numbers. Fungal Divers. 2022, 114, 3–235. [Google Scholar] [CrossRef]
- El-Elimat, T.; Raja, H.A.; Figueroa, M.; Al Sharie, A.H.; Bunch, R.L.; Oberlies, N.H. Freshwater fungi as a source of chemical diversity: A review. J. Nat. Prod. 2021, 84, 898–916. [Google Scholar] [CrossRef]
- Calabon, M.S.; Hyde, K.D.; Jones, E.B.G.; Bao, D.; Bhunjun, C.; Phukhamsakda, C.; Shen, H.; Gentekaki, E.; Al Sharie, A.; Barros, J.; Chandrasiri, K.; Hu, D.; Hurdeal, V.; Rossi, W.; Valle, L.; Zhang, H.; Figueroa, M.; Raja, H.; Seena, S.; Song, H.; Dong, W.; El-Elimat, T.; Leonardi, M.; Li, Y.; Li, Y.; Luo, Z.; Ritter, C.; Strongman, D.; Wei, M.; Balasuriya, A. Freshwater fungal biology. Mycosphere 2023, 14, 195–413. [Google Scholar] [CrossRef]
- Ślusarczyk, J.; Adamska, E.; Czerwik-Marcinkowska, J. Fungi and algae as sources of medicinal and other biologically active compounds: A review. Nutrients 2021, 13, 3178. [Google Scholar] [CrossRef] [PubMed]
- Hammerschmidt, S.; Hacker, J.; Klenk, H.-D. Threat of infection: Microbes of high pathogenic potential – strategies for detection, control and eradication. Int. J. Med. Microbiol. 2005, 295, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Huemer, M.; Mairpady Shambat, S.; Brugger, S.D.; Zinkernagel, A.S. Antibiotic resistance and persistence-Implications for human health and treatment perspectives. E.M.B.O. Rep. 2020, 21, e51034. [Google Scholar] [CrossRef] [PubMed]
- Jaklitsch, W.M.; Voglmayr, H. New combinations in Trichoderma (Hypocreaceae, Hypocreales). Mycotaxon 2013, 126, 143–156. [Google Scholar] [CrossRef]
- Zin, N.A.; Badaluddin, N.A. Biological functions of Trichoderma spp for agriculture applications. Ann. Agric. Sci. 2020, 65, 168–178. [Google Scholar] [CrossRef]
- Guo, R.; Li, G.; Zhang, Z.; Peng, X. Structures and biological activities of secondary metabolites from Trichoderma harzianum. Mar. Drugs 2022, 20, 701. [Google Scholar] [CrossRef]
- Siscar-Lewin, S.; Hube, B.; Brunke, S. Emergence and evolution of virulence in human pathogenic fungi. Trends Microbiol. 2022, 30, 693–704. [Google Scholar] [CrossRef]
- Contreras-Cornejo, H.A.; Schmoll, M.; Esquivel-Ayala, B.A.; González-Esquivel, C.E.; Rocha-Ramírez, V.; Larsen, J. Mechanisms for plant growth promotion activated by Trichoderma in natural and managed terrestrial ecosystems. Microbiol. Res. 2024, 281, 127621. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Samuels, G.J. Trichoderma: Systematics, the sexual state, and ecology. Phytopathology 2006, 96, 195–206. [Google Scholar] [CrossRef]
- Kubiak, A.; Wolna-Maruwka, A.; Pilarska, A.A.; Niewiadomska, A.; Piotrowska-Cyplik, A. Fungi of the Trichoderma genus: Future perspectives of benefits in sustainable agriculture. Appl. Sci. 2023, 13, 6434. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, 2000. [Google Scholar]
- Yamazaki, M.; Maebayashi, Y. Structure determination of violaceol-I and -II, new fungal metabolites from a strain of Emericella violacea. Chem. Pharm. Bull. 1982, 30, 514–518. [Google Scholar] [CrossRef]
- Nuankeaw, K.; Chaiyosang, B.; Suebrasri, T.; Kanokmedhakul, S.; Lumyong, S.; Boonlue, S. First report of secondary metabolites, Violaceol I and Violaceol II produced by endophytic fungus, Trichoderma polyalthiae and their antimicrobial activity. Mycoscience 2020, 61, 16–21. [Google Scholar] [CrossRef]
- Bottone, E.J. Bacillus cereus, a volatile human pathogen. Clin. Microbiol. Rev. 2010, 23, 382–398. [Google Scholar] [CrossRef]
- Liu, Q.; Meng, X.; Li, Y.; Zhao, C.-N.; Tang, G.-Y.; Li, H.-B. Antibacterial and antifungal activities of spices. Int. J. Mol. Sci. 2017, 18, 1283. [Google Scholar] [CrossRef]
- Wang, J.-F.; Lin, X.-P.; Qin, C.; Liao, S.-R.; Wan, J.-T.; Zhang, T.-Y.; Liu, J.; Fredimoses, M.; Chen, H.; Yang, B.; Zhou, X.-F.; Yang, X.-W.; Tu, Z.-C.; Liu, Y.-H. Antimicrobial and antiviral sesquiterpenoids from sponge-associated fungus, Aspergillus sydowii ZSDS1-F6. J. Antibiot. (Tokyo) 2014, 67, 581–583. [Google Scholar] [CrossRef]
- Bunyapaiboonsri, T.; Yoiprommarat, S.; Intereya, K.; Kocharin, K. New diphenyl ethers from the insect pathogenic fungus Cordyceps sp BCC 1861. Chem. Pharm. Bull. (Tokyo) 2007, 55, 304–307. [Google Scholar] [CrossRef]
- Xu, L.; Meng, W.; Cao, C.; Wang, J.; Shan, W.; Wang, Q. Antibacterial and antifungal compounds from marine fungi. Mar. Drugs 2015, 13, 3479–3513. [Google Scholar] [CrossRef] [PubMed]
- Itabashi, T.; Nozawa, K.; Nakajima, S.; Kawai, K. A new azaphilone, falconensin H, from Emericella falconensis. Chem. Pharm. Bull. 1993, 41, 2040–2041. [Google Scholar] [CrossRef]
- Van Nguyen, M.V.; Han, J.W.; Kim, H.; Choi, G.J. Phenyl ethers from the marine-derived fungus Aspergillus tabacinus and their antimicrobial activity against plant pathogenic fungi and bacteria. A.C.S. Omega 2022, 7, 33273–33279. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).