1. Introduction
Archaea, first recognized as a distinct domain of life in the late 1970s by Carl Woese and George Fox [
1], have historically received far less attention than bacteria and eukaryotes in the study of human biology [
2]. For decades, archaeal species were misclassified as bacteria due to their prokaryotic morphology and inability to be readily cultured under standard laboratory conditions [
2]. Advances in molecular biology, particularly ribosomal RNA gene sequencing, have since revealed the unique evolutionary position of archaea as a separate domain [
3], yet their roles in mammalian health and disease remain underexplored [
4].
Unlike bacteria, which have yielded a long list of established pathogens in humans, no archaeal species has been definitively demonstrated to cause disease in mammals [
4]. This distinction is striking given that archaea are common members of the human microbiome [
4]. Methanogenic archaea such as
Methanobrevibacter smithii and
Methanosphaera stadtmanae are abundant in the gastrointestinal tract [
5,
6], while
Methanobrevibacter oralis is frequently detected in the oral cavity [
7,
8]. In contrast, members of the class
Nitrososphaeria, which are ammonia-oxidizing
Thaumarchaeota, are more prevalent on the skin and within the upper aerodigestive tract, where they may influence nitrogen cycling and microbial community dynamics [
9,
10]. These organisms often coexist with bacteria and may contribute to microbial ecosystem stability. However, their frequent detection in disease-associated contexts has prompted growing interest in their potential involvement in pathology [
4].
Several features differentiate archaea from bacteria in ways that may limit their pathogenic potential. Archaea generally lack the virulence factors that are hallmarks of bacterial pathogens, including pore-forming toxins, Type III and Type IV secretion systems, and specialized adhesins [
11]. Their unique membrane lipids, ether-linked rather than ester-linked, and their distinct cell wall structures further insulate them from the horizontal gene transfer events that have spread virulence determinants among bacterial pathogens [
12]. For these reasons, archaea are not considered primary pathogens in mammals [
12].
Nevertheless, evidence is mounting that archaea may exert indirect or opportunistic influences on disease processes [
4]. For example, in polymicrobial infections such as periodontal disease, methanogens can consume hydrogen, creating metabolic niches that promote the growth of pathogenic bacteria [
7,
13]. Similarly, archaeal overrepresentation has been reported in patients with inflammatory bowel disease [
14,
15] and in animal models of obesity [
16,
17], suggesting potential roles in host-microbe interactions that extend beyond simple commensalism.
The field of human archaeome research is still in its infancy. Many fundamental questions remain unresolved, including whether archaeal diversity in the human microbiome has been fully appreciated, whether some lineages are more closely linked to pathology than others, and whether the evolutionary trajectory of archaea could eventually yield pathogenic potential [
4]. With the advent of next-generation sequencing, metagenomics, and single-cell approaches, our capacity to identify and study archaea in complex microbial communities has expanded dramatically, opening new avenues for discovery [
18].
In this review, we examine current knowledge of the human archaeome, focusing on its presence in health and disease, its potential roles in pathogenesis, and the technological challenges that have limited prior investigation. We also explore possible future research directions aimed at determining whether archaea will remain categorized as harmless commensals or emerge as bona fide contributors to mammalian disease.
2. Materials and Methods
Because the study of archaea in humans is still developing, the methodologies used to investigate their presence, diversity, and potential pathogenic roles require special consideration. Traditional microbiological approaches designed for bacteria are often inadequate, necessitating tailored strategies for archaeal detection, isolation, and functional analysis.
2.1. Molecular Detection and Sequencing
The cornerstone of human archaeome research has been culture-independent molecular techniques. Ribosomal RNA gene sequencing, particularly of the 16S rRNA gene, has revealed archaeal lineages in human samples that would otherwise remain undetected [
19]. However, archaeal 16S rRNA genes are less represented in universal primer sets, leading to underestimation of archaeal abundance [
20]. To address this, archaeal-specific primers and metagenomic sequencing have been increasingly employed, allowing more accurate detection and characterization of archaeal diversity [
10,
20]. Metatranscriptomics and metaproteomics further provide insight into archaeal metabolic activity in vivo [
21].
2.2. Archaeal Culture Methods
Culturing archaea remains a significant challenge due to their slow growth rates and unique nutritional requirements. Methanogens, the dominant human-associated archaea, require strict anaerobic conditions and specific substrates such as hydrogen and carbon dioxide [
6]. Advances in anaerobic culturing techniques, along with custom-designed growth media, have enabled the isolation of Methanobrevibacter and Methanosphaera species from human samples [
6], although many archaeal taxa remain uncultured [
22]. Improving culture techniques is essential for functional studies and for testing causative roles in disease models.
2.3. In Vitro Co-Culture and Microbial Interaction Studies
Since archaea are rarely found in isolation within human microbiomes, co-culture studies with bacteria provide valuable insights [
22]. For example, co-culturing methanogens with sulfate-reducing or fermentative bacteria allows researchers to investigate metabolic cross-feeding and potential synergistic effects on host tissues [
22]. Such experiments are critical for understanding how archaea may indirectly contribute to pathogenic outcomes by enhancing bacterial virulence or altering microbial community dynamics.
2.4. Animal Models and Gnotobiotic Systems
Animal models, particularly gnotobiotic mice, offer opportunities to study archaeal colonization and host interactions under controlled conditions [
23,
24]. Colonization experiments with
M. smithii have shown increased adiposity in mice, implicating archaeal metabolism in energy harvest and obesity [
23]. These models may be expanded to test hypotheses about archaeal contributions to inflammatory diseases or co-infections. However, the lack of archaeal genetic tools limits mechanistic studies.
2.5. Bioinformatics and Systems Biology Approaches
High-throughput sequencing datasets allow integration of archaeal abundance and activity with broader microbiome analyses [
25]. Network analyses, machine learning, and systems biology frameworks can reveal archaeal-bacterial associations and identify disease-linked microbial signatures [
26]. Such approaches are particularly powerful in distinguishing correlation from potential causation, though experimental validation remains essential.
In summary, the study of the human archaeome relies on a combination of culture-independent sequencing, specialized culturing methods, co-culture experiments, and animal models. Future progress will depend on improving archaeal cultivation, developing genetic manipulation tools, and integrating multi-omics datasets to link archaeal activity with host physiology and pathology.
3. Discussion
Archaea occupy a paradoxical position in human microbiology. On one hand, they are recognized as essential members of the microbiome, especially within anaerobic environments such as the gastrointestinal tract and oral cavity [
4]. On the other hand, they have never been conclusively implicated as primary pathogens in humans or other mammals [
4]. The growing field of archaeal research has revealed intriguing associations between specific archaeal taxa and disease states, yet causality remains elusive. This discussion will explore three major themes: (1) the current evidence linking archaea to disease, (2) potential mechanisms of archaeal involvement in pathology, and (3) limitations and challenges in establishing archaea as pathogens (
Figure 1;
Table 1).
3.1. Evidence Linking Archaea to Disease
3.1.1. Inflammatory Bowel Disease (IBD)
Several studies have reported elevated levels of methanogenic archaea in patients with Crohn’s disease or ulcerative colitis compared with healthy controls [
14,
27]. Among these,
Methanosphaera stadtmanae has attracted particular attention due to its ability to elicit strong pro-inflammatory responses in vitro. For example, stimulation of human dendritic cells with
M. stadtmanae results in robust cytokine release and activation of T cell pathways, suggesting that archaea may contribute to the heightened immune activity characteristic of IBD [
14,
27,
40].
Despite these intriguing findings, the relationship between archaea and IBD remains inconsistent and far from conclusive. Not all studies observe archaeal enrichment in IBD cohorts, and many patients display no detectable increase in methanogen abundance [
14,
27]. This variability could reflect differences in geography, diet, host genetics, or even methodological limitations in archaeal detection, since many commonly used 16S rRNA primers are biased toward bacteria and miss archaeal taxa.
A further complication is that bacterial dysbiosis is a well-established hallmark of IBD [
14,
27,
39]. Shifts in
Bacteroides,
Firmicutes, and other bacterial lineages dominate the microbiome signatures of disease, raising the question of whether archaea are primary drivers of inflammation or secondary responders to an altered gut environment. One possibility is that archaeal enrichment reflects ecological “spillover” from bacterial disruptions, in which changes in available substrates (e.g., hydrogen and acetate) provide methanogens with new growth opportunities.
Alternatively, archaea may act as facilitators of disease progression rather than primary instigators. By consuming hydrogen and influencing fermentation pathways, methanogens can alter gut physiology, luminal pH, and gas dynamics, which in turn may affect bacterial community composition and mucosal interactions [
41]. This ecological role could amplify existing dysbiosis and exacerbate inflammation without making archaea independent causes of IBD.
Ultimately, more work is needed to clarify whether methanogens such as M. stadtmanae are drivers, passengers, or modulators in the IBD disease process. Longitudinal studies, archaeal-targeted metagenomics, and functional host–archaea co-culture systems will be key in disentangling cause from correlation and defining the true significance of archaea in IBD pathogenesis.
3.1.2. Periodontal Disease
The oral archaeon
Methanobrevibacter oralis has been detected at higher prevalence in individuals with periodontitis than in healthy controls [
8,
27,
28,
29]. Within the complex microbial biofilm of the oral cavity, methanogens may function as “helper” organisms, consuming hydrogen produced by bacterial fermentation and thereby creating favorable conditions for the growth of obligate anaerobic pathogens such as
Porphyromonas gingivalis and
Tannerella forsythia [
8,
27,
28,
29]. This metabolic cooperation could exacerbate dysbiosis and promote chronic inflammation in periodontal tissues.
Despite these associations, the absence of identifiable virulence factors in
M. oralis makes it unlikely to act as a primary pathogen. Instead, its role may be more ecological, indirectly influencing disease progression by stabilizing and enriching pathogenic bacterial communities [
8,
27,
28,
29]. Moreover, periodontal disease is inherently polymicrobial and influenced by host immune responses, which complicates efforts to isolate archaeal contributions [
8,
27,
28,
29]. Future work using advanced metagenomic and single-cell approaches will be necessary to clarify whether methanogens represent opportunistic bystanders, ecological facilitators, or overlooked contributors to the pathogenesis of periodontal disease.
3.1.3. Obesity and Metabolic Disorders
In gnotobiotic mouse models, colonization with
Methanobrevibacter smithii has been shown to increase adiposity [
16,
17,
30]. The mechanism is thought to involve archaeal consumption of hydrogen, which lowers hydrogen partial pressure and enables bacterial fermentation to proceed more efficiently. This enhanced fermentation increases short-chain fatty acid (SCFA) production and overall energy harvest from the diet, ultimately boosting host caloric uptake [
16,
17,
30].
In humans, the relationship between
M. smithii abundance and metabolic health appears more complex. Some studies have linked higher
M. smithii levels with obesity, consistent with the hypothesis of increased energy extraction from food, while others have reported associations with leanness, possibly reflecting improved metabolic efficiency or host-microbiome adaptation to diet [
42]. Still other studies find no significant association, suggesting that archaeal effects may be modulated by host genetics, diet composition, or co-occurring bacterial taxa.
These inconsistencies underscore the context-dependent nature of archaeal-host interactions and raise the possibility that archaea may play different roles in metabolic outcomes depending on ecological and host-specific factors [
42]. Further studies integrating metagenomics, metabolomics, and clinical phenotyping will be needed to clarify whether
M. smithii functions as a metabolic driver, adaptive symbiont, or neutral commensal in human obesity and related disorders.
3.1.4. Other Associations
Archaea have also been detected in human niches beyond the gastrointestinal and oral environments, including the skin [
9,
10], respiratory tract [
43], and urogenital tract [
44]. On the skin, members of the class
Nitrososphaeria are consistently found and are thought to participate in ammonia oxidation, potentially influencing local pH and the balance of bacterial communities. While their presence is well established, direct links to dermatological conditions such as acne, eczema, or psoriasis remain speculative.
In the lung, archaeal DNA has occasionally been identified in both healthy individuals and patients with chronic respiratory disease [
43]. Whether these findings reflect true colonization or transient presence from inhaled environmental sources is still unresolved. Similarly, in the urogenital tract, archaeal sequences have been reported in both male and female samples [
44], but functional roles and associations with reproductive or urinary pathology have yet to be defined.
Some researchers have suggested that archaeal metabolic byproducts—such as methane, which may influence motility in the gut, or ammonia, which can alter local chemical environments—could have subtle physiological consequences across these body sites [
3]. However, experimental evidence remains sparse, and the functional relevance of archaeal detection outside the gut and oral cavity is still an open question.
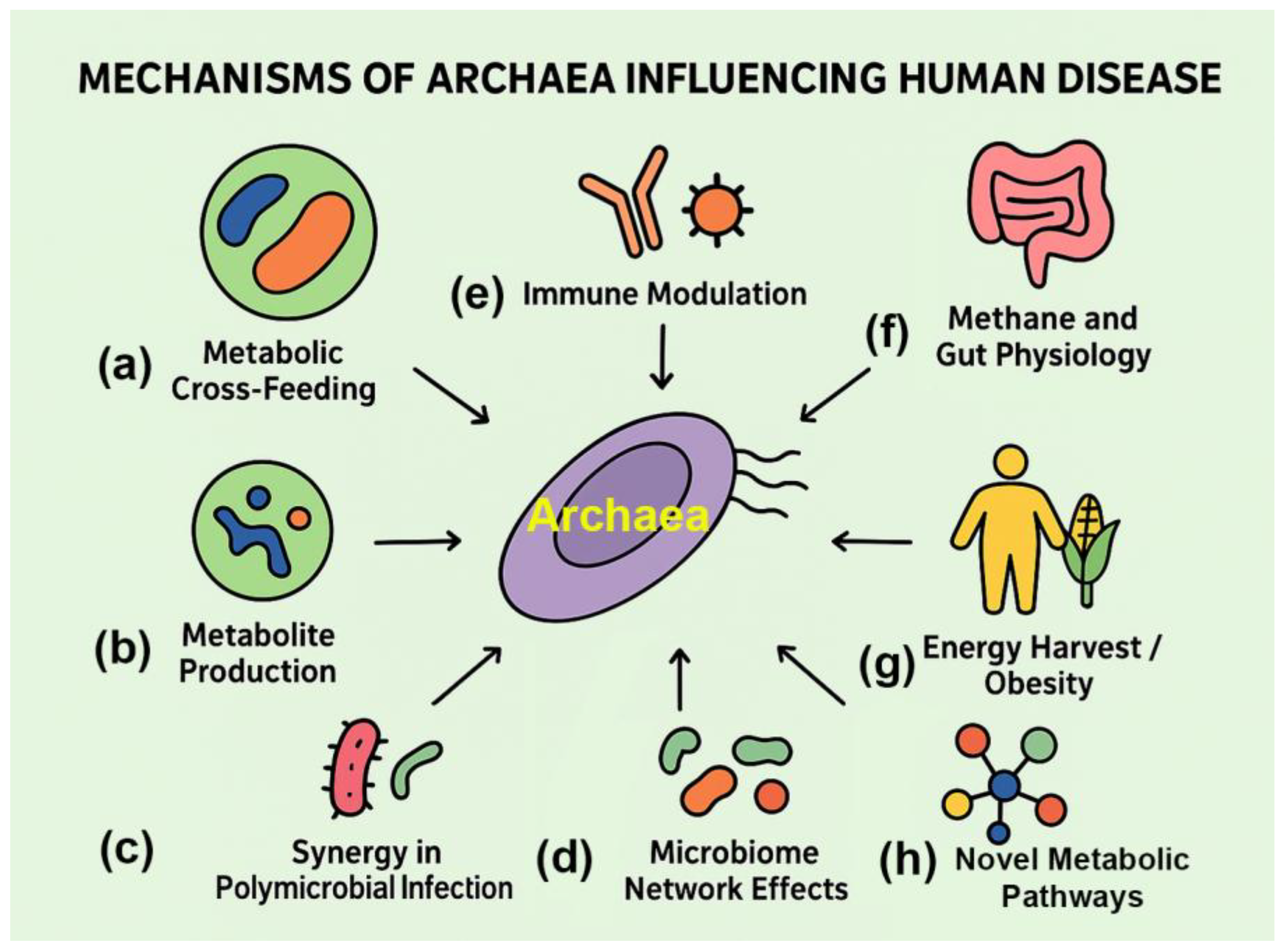
3.2. Potential Mechanisms of Archaeal Involvement in Disease
Current evidence suggests that archaea act indirectly, shaping microbial ecology, immune signaling, and host physiology, rather than functioning like classical pathogens (
Figure 2;
Table 2).
Schematic representation of potential pathways through which members of the human archaeome can impact host health. Central archaeal cells interact with multiple mechanisms: (a) Metabolic cross-feeding, where archaeal consumption of bacterial fermentation products (e.g., hydrogen) alters microbial ecosystem balance; (b) Metabolite production, involving unique archaeal metabolic pathways that yield novel small molecules influencing bacterial or host physiology; (c) Synergy in polymicrobial infection, whereby archaea enhance bacterial persistence or virulence in co-infection settings; (d) Microbiome network effects, in which archaea shape microbial community structure and inter-species interactions; (e) Immune modulation, reflecting archaeal antigens and surface molecules that may interact with or perturb host immunity; (f) Methane and gut physiology, where methane production alters gut motility and gas balance, potentially contributing to gastrointestinal disorders; (g) Energy harvest/obesity, through archaeal facilitation of caloric extraction and metabolic efficiency that can promote adiposity; and (h) Novel metabolic pathways, highlighting unexplored archaeal contributions to biochemical networks with potential implications for disease.
3.2.1. Metabolic Cross-Feeding
The best-documented role of archaea in human-associated disease contexts is through metabolic cross-feeding, a process in which archaeal metabolism indirectly shapes bacterial growth and community function. Methanogenic archaea consume molecular hydrogen generated as a byproduct of bacterial fermentation, thereby reducing hydrogen partial pressure within the local microenvironment [
3]. This thermodynamically favorable process relieves feedback inhibition on bacterial fermentative pathways, enabling bacteria to extract more energy from substrates [
3]. As a result, methanogenesis can act as a metabolic sink that enhances bacterial growth and survival, particularly under anaerobic conditions [
3].
This ecological interaction has several implications for human disease. In the gastrointestinal tract, hydrogen consumption by methanogens such as
Methanobrevibacter smithii facilitates fermentation efficiency, potentially increasing caloric harvest from polysaccharides and linking archaeal activity to obesity and metabolic disorders. In periodontal disease, methanogens such as
Methanobrevibacter oralis thrive in inflamed, anaerobic niches of the oral cavity [
8,
27,
28,
29]. By consuming hydrogen, these archaea create conditions that favor the proliferation of anaerobic bacterial pathogens, thereby exacerbating inflammation and tissue destruction. Similar dynamics may occur in polymicrobial infections elsewhere in the body, where archaeal hydrogenotrophy (i.e., attraction to hydrogen) inadvertently supports the persistence of pathogenic bacterial partners [
8,
27,
28,
29].
Beyond hydrogen metabolism, other archaeal metabolic products may also contribute to cross-feeding. For example, methane produced by methanogens can alter gut motility, indirectly influencing microbial colonization patterns, while archaeal production of unique metabolites such as methanol or methylamines may provide additional substrates for bacterial growth [
46]. Taken together, these findings suggest that archaea may act as keystone modulators of microbial community metabolism, enhancing the ecological fitness of pathogenic bacteria and contributing to disease progression without functioning as direct pathogens themselves.
3.2.2. Immune Modulation
Archaea, though not recognized as classical pathogens, appear capable of influencing host immunity through distinctive molecular features. Several studies have highlighted that
Methanosphaera stadtmanae can activate human dendritic cells, triggering the release of pro-inflammatory cytokines such as TNF-α and IL-6, suggesting potential immunopathogenic effects even in the absence of overt infection [
10,
40]. This finding underscores that archaeal recognition by the immune system may occur via mechanisms distinct from those established for bacteria.
Unlike bacteria, archaea lack canonical pathogen-associated molecular patterns (PAMPs) such as lipopolysaccharide (LPS) or peptidoglycan [
52]. Instead, their unique structural components—including pseudomurein, ether-linked membrane lipids, and heavily glycosylated S-layer proteins—may function as immunomodulatory signals [
52]. These molecules are poorly characterized in terms of host receptor interactions, but some evidence suggests they can engage pattern recognition receptors (PRRs) such as Toll-like receptors (TLRs) or C-type lectins in noncanonical ways. For example, archaeal lipids have been hypothesized to modulate TLR2 and TLR4 pathways, potentially dampening or altering inflammatory responses compared to bacterial ligands [
10,
40].
In addition to structural features, archaeal metabolic products such as methane and short-chain alcohols may indirectly influence immune tone by shaping bacterial community structure or altering epithelial barrier function [
4,
53]. Furthermore, immune responses to archaea may represent evolutionary relics of ancient host–microbe interactions, given the deep evolutionary divergence of archaea from bacteria and eukaryotes [
4,
54]. Whether these responses confer protection, promote inflammation, or contribute to immune dysregulation in disease remains unresolved.
Thus, immune modulation by archaea is emerging as a potential mechanism linking their presence in the microbiome to disease-associated states, warranting deeper investigation into archaeal-host molecular crosstalk and its consequences for inflammatory disorders.
3.2.3. Indirect Effects on Host Physiology
Beyond their direct interactions with bacteria, archaea can influence host physiology through metabolic activities that alter the biochemical environment of the gut and other body sites. The most widely studied example is methane production by methanogenic archaea such as
Methanobrevibacter smithii and
Methanosphaera stadtmanae [
55]. Methane is biologically inert in terms of host metabolism but exerts important physical effects on gut physiology. Clinical and experimental studies suggest that methane can slow intestinal transit by altering smooth muscle contractility, thereby contributing to constipation-predominant irritable bowel syndrome (IBS) [
31,
45]. The presence of methanogens has been consistently correlated with increased breath methane levels in patients with functional gastrointestinal disorders, supporting this mechanistic link.
In the context of metabolic health, archaea also play an indirect role in regulating host energy balance. By consuming hydrogen and thereby improving the efficiency of bacterial fermentation, methanogens enhance caloric extraction from dietary polysaccharides [
17,
42]. This increased energy harvest has been implicated in obesity and metabolic disorders, particularly in animal models where colonization with
M. smithii leads to greater adiposity [
17,
42]. While the effect size in humans remains debated, these findings suggest that archaeal activity can amplify the metabolic potential of the microbiome, subtly shifting host energy homeostasis.
Archaeal contributions to host physiology may extend beyond methane and energy harvest. For example, archaeal metabolites such as methylamines have been linked to cardiovascular disease risk through their role in generating trimethylamine N-oxide (TMAO), a pro-atherogenic compound [
56]. Likewise, archaeal ammonia oxidation by
Nitrososphaeria on the skin and in the upper aerodigestive tract could influence local pH, barrier integrity, and microbial community composition, with potential downstream effects on host immune responses [
57].
These examples illustrate how archaea, despite lacking canonical virulence traits, can shape host physiology indirectly through their unique metabolic outputs. Such effects underscore the need to integrate archaeal functions into broader models of host–microbe interactions, particularly in chronic diseases where small shifts in microbial metabolism can have significant clinical consequences.
3.3. Barriers to Establishing Archaeal Pathogenicity
3.3.1. Lack of Virulence Factors
A defining feature of established bacterial pathogens is their arsenal of virulence factors, which include protein toxins, adhesins, secretion systems, and specialized effector molecules that directly damage host tissues, evade immune defenses, or manipulate host cell signaling [
58]. These features are frequently encoded on mobile genetic elements such as plasmids or pathogenicity islands, enabling rapid horizontal transfer and diversification among bacterial lineages [
58]. In contrast, no archaeal species identified to date has been shown to harbor comparable systems. Comparative genomic analyses of archaeal taxa inhabiting the human body, such as
Methanobrevibacter smithii,
Methanosphaera stadtmanae, and
Methanobrevibacter oralis, consistently reveal an absence of genes encoding classical virulence determinants [
10,
40].
The structural biology of archaea also argues against traditional pathogenicity. Unlike bacteria, archaea lack peptidoglycan-based cell walls and instead possess pseudomurein, proteinaceous S-layers, or unique glycosylated cell wall structures. These distinct features not only alter their interactions with the host immune system but may also insulate archaea from acquiring bacterial virulence determinants via lateral gene transfer [
52]. The unusual lipid architecture of archaeal membranes—ether-linked isoprenoid chains rather than ester-linked fatty acids—further distinguishes them and may limit compatibility with bacterial pathogenicity machinery [
52].
The absence of classical virulence factors does not, however, preclude archaea from influencing host health. Their interactions may instead be mediated through subtler mechanisms, including metabolic cross-feeding, modulation of microbial community structure, or immune activation via unique archaeal cell envelope components [
59]. Furthermore, while no toxins or secretion systems have been identified, the possibility remains that archaeal lineages encode as-yet uncharacterized proteins or metabolites with immunomodulatory or cytotoxic potential [
4,
60]. With the expansion of archaeal genomics and metagenomic studies, previously overlooked candidates for noncanonical virulence mechanisms may yet be discovered.
Taken together, the lack of known virulence determinants strongly argues against archaea acting as primary pathogens in mammals. Rather, their role appears to be that of ecological partners, metabolic modulators, or secondary contributors in polymicrobial disease contexts. This distinction underscores the need to shift the conceptual framework from viewing archaea as putative pathogens to appreciating them as influencers of microbial community dynamics and host physiology.
3.3.2. Methodological Challenges
The study of human-associated archaea has long been constrained by methodological limitations, resulting in their consistent underrepresentation in microbiome research [
61]. Many early and even current large-scale sequencing surveys have been optimized for bacterial detection, inadvertently excluding archaea [
61]. Commonly used 16S rRNA gene primers, for example, are tailored toward bacterial conserved regions and exhibit poor coverage of archaeal taxa, particularly methanogens and members of the Nitrososphaeria [
61]. As a result, archaeal reads are often missed during amplification or discarded during bioinformatic processing pipelines, leading to a systematic bias that diminishes their apparent prevalence in human microbiomes [
61].
Culture-based studies face even greater challenges. Standard microbial growth media, incubation conditions, and atmospheric requirements are ill-suited for archaea, many of which are strict anaerobes requiring specialized substrates (e.g., hydrogen, methanol, or formate) and long doubling times [
62]. Methanogenic archaea are notoriously fastidious, often necessitating complex syntrophic relationships with bacteria in co-culture for stable growth [
62]. This makes isolation of pure archaeal strains rare and limits functional studies that could clarify their contributions to disease processes.
Even when sequencing-based approaches are employed, archaeal detection is further hindered by low abundance in mixed microbial communities and by incomplete genomic databases [
63]. Many archaeal genomes remain poorly annotated, and reference collections are far less comprehensive than those for bacteria [
63]. Consequently, archaeal reads are frequently misclassified or relegated to broad, uninformative taxonomic categories. This lack of resolution makes it difficult to link specific archaeal species or strains with disease states [
63].
Methodological hurdles also extend to functional characterization. Classical tools for assessing microbial virulence—such as cell culture infection models, mutagenesis systems, or animal challenge experiments—are not readily applicable to archaea [
64]. Genetic manipulation remains in its infancy for most archaeal lineages, restricting the ability to directly test hypotheses about immune activation, metabolite production, or ecological roles [
65]. Moreover, standard metabolomic pipelines are often not optimized for detecting unique archaeal lipids or metabolites, further obscuring their biochemical contributions [
65].
Recent advances provide reason for optimism. Archaeal-specific primers and improved shotgun metagenomic approaches now enable better detection and quantification of archaeal taxa [
66]. Expansion of archaeal genome catalogs through initiatives such as the Human Microbiome Project and large-scale metagenomic binning has begun to fill gaps in reference databases [
67]. Microfluidics, continuous-culture systems, and synthetic co-culture models also offer new avenues for maintaining and studying archaeal strains in vitro [
68,
69]. Moreover, advances in single-cell genomics [
70,
71], long-read sequencing [
72,
73], and high-resolution metabolomics [
74] may soon allow archaeal functions to be resolved with greater precision.
Until these methodological challenges are addressed, however, archaeal contributions to health and disease will remain underappreciated and potentially underestimated. The persistent biases against archaeal detection and characterization highlight a critical need for the development and adoption of archaeal-inclusive tools across microbiome research.
3.3.3. Co-Occurrence with Bacteria
A defining feature of human-associated archaea is their consistent co-occurrence with bacterial communities, particularly in disease-associated environments [
12,
59]. Rarely are archaea detected in isolation at pathological sites; instead, they appear embedded within polymicrobial consortia [
12,
59]. This observation complicates efforts to assign causality, raising the question of whether archaea act as active contributors, opportunistic colonizers, or merely incidental bystanders in disease contexts [
12,
59].
One explanation for this pattern is ecological interdependence. Many archaea, particularly methanogens, rely on bacterial partners to supply essential substrates such as hydrogen, formate, or methyl compounds [
75]. In return, archaeal consumption of these products alters the local redox balance, relieving feedback inhibition and enhancing bacterial fermentation efficiency [
75]. This metabolic interlocking creates a syntrophic partnership that not only supports archaeal survival but also indirectly boosts bacterial growth [
75]. In inflammatory niches such as the oral cavity or gut, such archaeal–bacterial synergy may amplify pathogenic processes [
8,
13,
28]. For example, in periodontal disease, methanogens co-localize with anaerobic pathogens such as
Porphyromonas gingivalis, where hydrogenotrophic activity by archaea may foster deeper tissue colonization and worsen inflammation [
8,
13,
28].
Beyond metabolism, co-occurrence may influence microbial spatial organization and biofilm dynamics [
29]. Archaea have been detected in structured biofilms on mucosal surfaces, where their presence could stabilize biofilm architecture or provide resilience against host defenses and antimicrobial treatments [
29]. Their resistance to many antibiotics further raises the possibility that archaea act as “community stabilizers,” maintaining microbial consortia under therapeutic stress and indirectly protecting co-resident bacteria [
29].
From a host perspective, archaeal–bacterial co-occurrence complicates immune recognition. Bacteria provide canonical pathogen-associated molecular patterns (PAMPs) such as lipopolysaccharide, while archaea contribute distinct cell wall structures and metabolic byproducts [
76]. Together, these signals may generate mixed or synergistic immune responses, potentially intensifying inflammation or skewing immune regulation in ways that neither group would achieve alone [
76].
Despite these intriguing possibilities, experimental proof of archaeal–bacterial synergy in disease remains limited. The technical difficulty of isolating archaeal species and recreating their interactions with bacteria in vitro has hindered mechanistic studies [
10]. Moreover, the fact that archaea are typically low-abundance members of disease-associated communities makes it challenging to separate their effects from those of dominant bacterial taxa [
10].
Nevertheless, the recurrent detection of archaea alongside bacteria in diverse disease contexts—from periodontal pockets to inflamed intestines and dysbiotic lungs—suggests that archaeal–bacterial interactions are ecologically and clinically meaningful [
35,
36]. Disentangling the directionality and functional consequences of these associations represents a critical next step in understanding the hidden role of archaea in human disease.
3.3.4. Limited Genetic Tools
One of the greatest obstacles in deciphering the role of archaea in human health and disease is the paucity of reliable genetic tools. In bacterial pathogenesis research, genetic manipulation—via knockouts, knockdowns, overexpression systems, or CRISPR-based editing—has been central to establishing causal links between candidate genes and virulence traits [
77]. By contrast, archaeal systems remain notoriously difficult to manipulate, which has severely limited mechanistic insights into their biology and potential contributions to pathology [
62].
Several factors contribute to this challenge. First, archaea possess highly distinctive cellular machinery that differs substantially from both bacteria and eukaryotes. Their DNA replication, transcription, and translation systems are a hybrid of bacterial and eukaryotic features, complicating the transfer of molecular biology techniques developed in model organisms [
78]. Second, archaeal cell envelopes—often composed of pseudomurein or heavily glycosylated S-layer proteins—pose barriers to DNA uptake, reducing the efficiency of transformation methods [
79]. Third, archaeal extremophily presents practical barriers: many archaeal species require specialized growth conditions (e.g., strict anaerobiosis, high temperatures, or unusual substrates) that are difficult to maintain in laboratory settings, further constraining experimental manipulation [
62,
80].
While genetic systems have been developed for a handful of archaeal species, such as
Haloferax volcanii [
69] and
Methanosarcina acetivorans [
81], these are not the dominant taxa found in human-associated microbiomes. The clinically relevant methanogens, such as
Methanobrevibacter smithii,
Methanosphaera stadtmanae, and
Methanobrevibacter oralis, remain refractory to routine genetic manipulation [
62]. As a result, functional hypotheses—such as whether archaeal cell wall structures act as immune activators, or whether specific metabolic pathways alter bacterial virulence—cannot yet be directly tested through targeted mutagenesis [
62].
This lack of genetic tractability creates a vicious cycle: without experimental validation, archaeal genes remain annotated largely as “hypothetical proteins,” and without functional annotation, the incentive to develop species-specific genetic tools is reduced. Consequently, the archaeal contribution to human disease is studied primarily through correlative approaches (e.g., co-occurrence networks, metagenomic association studies) rather than through rigorous functional tests of causality.
The emergence of new molecular technologies, however, offers hope. Advances in metagenomics [
82], metatranscriptomics [
83], and metaproteomics [
84] are beginning to shed light on archaeal activity in situ, while synthetic biology and cell-free expression systems may provide alternative routes to probe archaeal protein function without direct genetic manipulation. Furthermore, adaptation of CRISPR–Cas systems—many of which are natively archaeal in origin—could, with further optimization, open the door to archaeal genome editing [
85].
Until such tools are widely available, however, the molecular underpinnings of archaeal–host and archaeal–bacterial interactions will remain speculative, limiting our ability to distinguish true causal mechanisms from ecological correlations. Bridging this gap will be essential if the human archaeome is to be fully integrated into models of health and disease
3.4. Conceptual Implications
The evidence to date suggests that archaea should be considered “accessory microbes” in human disease rather than primary pathogens. Their role may resemble that of commensal organisms that can become opportunistic under certain conditions, or ecological facilitators that enhance the pathogenicity of bacteria. This does not diminish their importance; indeed, indirect contributions to disease may be clinically significant. The key challenge for future research is to shift from correlation to causation, using refined tools to test whether archaea can independently contribute to pathology.
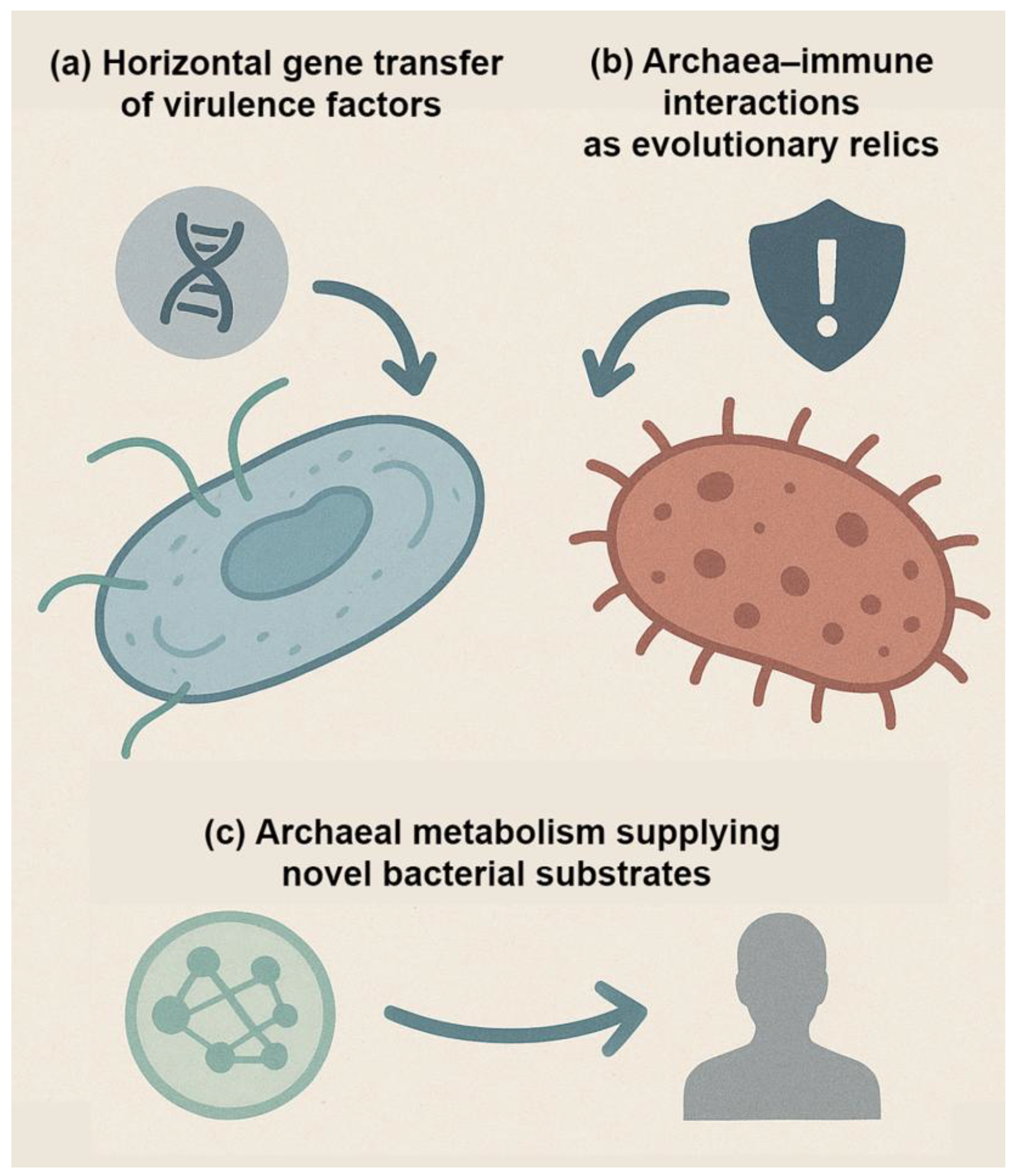
3.5. Evolutionary Perspectives
Archaea occupy a unique position in the tree of life, sharing features with both bacteria and eukaryotes yet remaining fundamentally distinct. Their unusual biology and long evolutionary history raise intriguing questions about their potential role in human disease, not only in the present but also in the future [
4]. Unlike bacteria, which have repeatedly evolved pathogenic strategies across diverse lineages, archaea have not yet produced definitive pathogens. Nevertheless, considering their deep evolutionary roots and capacity for genetic and ecological innovation, several possibilities warrant exploration (
Figure 3).
First, the potential for horizontal gene transfer (HGT) must be considered (
Figure 3a). Although archaea appear more insulated from bacterial gene exchange due to differences in cell envelope composition, plasmids and mobile elements are widespread in archaeal lineages [
86]. Experimental studies and metagenomic analyses have shown that gene flow between archaea and bacteria is possible, especially in shared niches where syntrophic interactions occur [
87]. It remains conceivable that under the right selective pressures, archaeal species could acquire bacterial virulence determinants—such as adhesins, proteases, or toxin-like proteins—and repurpose them for survival in mammalian hosts [
87]. While no such event has been documented, the evolutionary potential should not be dismissed, particularly given the dynamic nature of microbial ecosystems within the human body.
Second, archaeal interactions with the immune system may represent evolutionary relics of ancient host–microbe encounters (
Figure 3b). Archaea are thought to have coexisted with early eukaryotic ancestors and may have played a role in shaping the evolution of innate immune recognition systems [
4]. For instance, mammalian pattern recognition receptors (PRRs) evolved to detect conserved microbial molecules, many of which are absent or altered in archaea [
4]. This raises the possibility that immune responses to archaeal components, such as pseudomurein or unique glycoproteins, may reflect vestiges of interactions with extinct microbial lineages [
4]. Such immunological “fossils” could explain why archaea sometimes provoke inflammatory responses despite lacking classical bacterial PAMPs, and why their signals may synergize with bacterial stimuli to modulate disease outcomes [
4].
Third, archaea may serve as reservoirs of novel metabolic pathways that influence human disease in unanticipated ways (
Figure 3c). Their capacity for methanogenesis, ammonia oxidation, and other specialized chemistries introduces metabolic functions not typically found in bacteria [
12]. These pathways can reshape microbial community dynamics and host physiology by altering hydrogen balance, methane levels, nitrogen flux, and short-chain fatty acid availability [
12]. As new archaeal lineages are discovered in the human microbiome—including skin- and airway-associated
Thaumarchaeota (
Nitrososphaeria)—the potential for unconventional metabolic contributions to disease becomes increasingly plausible [
59]. Archaea may not act as pathogens in the classical sense, but their enzymatic repertoire could indirectly shape pathological processes by sustaining dysbiosis or fueling bacterial pathogenicity.
Addressing these evolutionary questions requires interdisciplinary collaboration. Microbiologists can uncover archaeal diversity and physiology through cultivation and -omics approaches, while evolutionary biologists can trace the origins of archaeal traits and their potential for gene exchange. Clinicians, in turn, can provide insights into how archaeal presence correlates with disease states and patient outcomes. Together, these perspectives are essential to determine whether the archaeome represents an evolutionary dead-end for pathogenesis, or a latent reservoir of microbial functions that could, under certain conditions, contribute to human disease.
3.6. Ethical and Practical Considerations
As archaeal research advances toward translational applications, ethical and practical considerations must be placed at the forefront. Unlike bacterial therapeutics, where decades of clinical experience provide a framework for safety evaluation, archaeal interventions remain largely uncharted territory. Manipulating the human archaeome—whether through archaeal probiotics, targeted antimicrobials, or engineered consortia—carries the risk of unintended ecological and physiological consequences [
59]. Archaea are deeply integrated into host-associated microbial networks, and perturbations could destabilize these ecosystems in ways that are difficult to predict.
One major challenge lies in risk assessment. Because archaea are not known pathogens, there may be a tendency to assume they are inherently benign. However, as evidence grows for their roles in metabolic cross-feeding, immune modulation, and dysbiosis, therapeutic alteration of archaeal abundance or activity could inadvertently worsen disease states [
4,
10]. Long-term safety studies, including multi-generational microbiome monitoring, will therefore be essential before archaeal-based therapies are widely adopted.
Another consideration is the ecological spillover of archaeal interventions. Engineered archaea or archaeal-targeting therapies could spread beyond the individual host into environmental reservoirs, with unknown consequences for global microbial communities [
88]. Given the resilience and adaptability of archaea, release into natural ecosystems may have profound ecological effects that warrant pre-emptive safeguards.
Ethical questions also arise around dual-use research. As tools for archaeal genetic manipulation improve, there is a theoretical risk of creating pathogenic strains, either inadvertently or deliberately. While such risks remain speculative, governance frameworks must anticipate potential misuse, just as they have for bacterial and viral genetic engineering.
From a practical standpoint, the development of archaeal diagnostics and therapeutics will require interdisciplinary oversight. Regulatory agencies, bioethicists, clinicians, and microbiologists must work together to ensure that novel interventions are evaluated not only for efficacy but also for societal and ecological safety. Public communication will also be critical: framing archaea as “hidden partners” rather than “unknown pathogens” may help prevent misperceptions and foster informed dialogue about risks and benefits [
4,
10].
In summary, as archaeal biology transitions from basic science to clinical application, the field must adopt a precautionary yet proactive stance. Ethical foresight, robust safety frameworks, and transparent communication will be essential to ensure that the integration of the archaeome into medical practice benefits human health without compromising ecological or societal stability.
4. Conclusions
The study of the human archaeome is still in its infancy, yet the evidence accumulated to date suggests that archaea are neither trivial bystanders nor classical pathogens. Instead, they occupy a gray area in which their contributions to health and disease are subtle, context-dependent, and mediated largely through interactions with bacterial partners and host physiology. This complexity makes them both fascinating and challenging to study.
Future research should prioritize the systematic expansion of archaeal diversity catalogs across multiple body sites, including underexplored niches such as the skin, respiratory tract, and urogenital tract [
4,
10]. Advances in sequencing technologies and archaeal-specific primers are beginning to uncover lineages, such as
Nitrososphaeria, that were long overlooked by bacterial-centric approaches [
4,
9]. Yet mere detection is insufficient: robust culture methods and genetic tools will be essential to bring these organisms into the laboratory, where their functions can be interrogated in controlled systems.
Equally important are functional studies in host contexts, ranging from gnotobiotic animal models to organoid co-cultures, which can provide direct evidence of how archaea modulate immune responses, alter gut motility, or interact with bacterial pathogens [
23,
24]. These approaches will help clarify whether archaeal associations with disease—such as inflammatory bowel disease, obesity, and periodontitis—reflect causality, opportunism, or ecological side effects of dysbiosis.
Another priority is deciphering archaeal–bacterial synergy, which may represent the most clinically relevant axis of archaeal influence. Metabolic cross-feeding, methane-mediated changes in gut physiology, and co-structuring of biofilms are all mechanisms by which archaea may amplify bacterial virulence or persistence [
29,
47,
68]. Parsing these relationships will require integrated ecological, genomic, and immunological perspectives.
Clinical applications may emerge even if archaea are never classified as true pathogens. Diagnostic biomarkers based on archaeal abundance or activity could provide early signals of dysbiosis, while archaeal-targeted or archaeal-modulating therapies may offer novel ways to rebalance microbial communities [
15,
55,
76]. At the same time, ethical and safety considerations must guide translation, given the unpredictability of intervening in systems that have co-evolved with humans for millennia [
88].
Ultimately, the central challenge is conceptual: to move beyond asking whether archaea are pathogens, and instead to define the spectrum of roles they play—from benign commensals and ecological stabilizers to opportunistic facilitators of disease. Recognizing archaea as integral but distinct members of the microbiome may open new frontiers in microbiology, immunology, and clinical medicine, positioning the archaeome as a vital, if still enigmatic, dimension of human health.
Funding
This research was funded by the National Institutes of Health, grant numbers UG3OD023285, P42ES030991, and P30ES036084.
Acknowledgments
During the preparation of this manuscript/study, the author used ChatGPT5 for the purposes of increasing the clarity of the text. The authors have reviewed and edited the output and takes full responsibility for the content of this publication.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| LGT |
Lower gestational tract |
| UAT |
Upper aerodigestive tract |
| IBS |
Irritable bowel syndrome |
| IBD |
Irritable bowel disease |
| SCFA |
Short chain fatty acids |
| TNF-alpha |
Tumor necrosis factor alpha |
| IL-6 |
Interleukin 6 |
| LPS |
lipopolysaccharide |
| PAMPS |
Pathology associated molecular patterns |
| TLR-2,4 |
Toll like receptor -2,4 |
| TMAO |
Trimethylamine N-oxide |
References
- Woese, C.R. and G.E. Fox, Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A, 1977. 74(11): p. 5088-90.
- Pace, N.R., J. Sapp, and N. Goldenfeld, Phylogeny and beyond: Scientific, historical, and conceptual significance of the first tree of life. Proc Natl Acad Sci U S A, 2012. 109(4): p. 1011-8.
- Lurie-Weinberger, M.N. and U. Gophna, Archaea in and on the Human Body: Health Implications and Future Directions. PLoS Pathog, 2015. 11(6): p. e1004833.
- Kuehnast, T., et al., Exploring the human archaeome: its relevance for health and disease, and its complex interplay with the human immune system. FEBS J, 2025. 292(6): p. 1316-1329.
- Gaci, N., et al., Archaea and the human gut: new beginning of an old story. World J Gastroenterol, 2014. 20(43): p. 16062-78.
- Volmer, J.G., H. McRae, and M. Morrison, The evolving role of methanogenic archaea in mammalian microbiomes. Front Microbiol, 2023. 14: p. 1268451.
- Horz, H.P. and G. Conrads, Methanogenic Archaea and oral infections - ways to unravel the black box. J Oral Microbiol, 2011. 3.
- Pilliol, V., et al., Methanobrevibacter oralis: a comprehensive review. J Oral Microbiol, 2024. 16(1): p. 2415734.
- Umbach, A.K., A.A. Stegelmeier, and J.D. Neufeld, Archaea Are Rare and Uncommon Members of the Mammalian Skin Microbiome. mSystems, 2021. 6(4): p. e0064221.
- Bang, C. and R.A. Schmitz, Archaea associated with human surfaces: not to be underestimated. FEMS Microbiol Rev, 2015. 39(5): p. 631-48.
- Gill, E.E. and F.S. Brinkman, The proportional lack of archaeal pathogens: Do viruses/phages hold the key? Bioessays, 2011. 33(4): p. 248-54.
- Eckburg, P.B., P.W. Lepp, and D.A. Relman, Archaea and their potential role in human disease. Infect Immun, 2003. 71(2): p. 591-6.
- Hajishengallis, G. and R.J. Lamont, Polymicrobial communities in periodontal disease: Their quasi-organismal nature and dialogue with the host. Periodontol 2000, 2021. 86(1): p. 210-230.
- Cisek, A.A., et al., The Role of Methanogenic Archaea in Inflammatory Bowel Disease-A Review. J Pers Med, 2024. 14(2).
- Santana, P.T., et al., Dysbiosis in Inflammatory Bowel Disease: Pathogenic Role and Potential Therapeutic Targets. Int J Mol Sci, 2022. 23(7).
- Clarke, S.F., et al., The gut microbiota and its relationship to diet and obesity: new insights. Gut Microbes, 2012. 3(3): p. 186-202.
- Murphy, E.F., et al., Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut, 2010. 59(12): p. 1635-42.
- Kim, N., et al., Genome-resolved metagenomics: a game changer for microbiome medicine. Exp Mol Med, 2024. 56(7): p. 1501-1512.
- Clarridge, J.E., 3rd, Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev, 2004. 17(4): p. 840-62, table of contents.
- Koskinen, K., et al., First Insights into the Diverse Human Archaeome: Specific Detection of Archaea in the Gastrointestinal Tract, Lung, and Nose and on Skin. mBio, 2017. 8(6).
- Fernandez-Carro, E., et al., Alternatives Integrating Omics Approaches for the Advancement of Human Skin Models: A Focus on Metagenomics, Metatranscriptomics, and Metaproteomics. Microorganisms, 2025. 13(8).
- Hofer, U., The majority is uncultured. Nat Rev Microbiol, 2018. 16(12): p. 716-717.
- Samuel, B.S. and J.I. Gordon, A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci U S A, 2006. 103(26): p. 10011-6.
- Samuel, B.S., et al., Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc Natl Acad Sci U S A, 2007. 104(25): p. 10643-8.
- Franzosa, E.A., et al., Sequencing and beyond: integrating molecular ‘omics’ for microbial community profiling. Nat Rev Microbiol, 2015. 13(6): p. 360-72.
- Sabih Ur Rehman, S., et al., Integrative systems biology approaches for analyzing microbiome dysbiosis and species interactions. Brief Bioinform, 2025. 26(4).
- Blais Lecours, P., et al., Increased prevalence of Methanosphaera stadtmanae in inflammatory bowel diseases. PLoS One, 2014. 9(2): p. e87734.
- Lepp, P.W., et al., Methanogenic Archaea and human periodontal disease. Proc Natl Acad Sci U S A, 2004. 101(16): p. 6176-81.
- Vianna, M.E., et al., Quantitative analysis of three hydrogenotrophic microbial groups, methanogenic archaea, sulfate-reducing bacteria, and acetogenic bacteria, within plaque biofilms associated with human periodontal disease. J Bacteriol, 2008. 190(10): p. 3779-85.
- Mbakwa, C.A., et al., Gut colonization with methanobrevibacter smithii is associated with childhood weight development. Obesity (Silver Spring), 2015. 23(12): p. 2508-16.
- Triantafyllou, K., C. Chang, and M. Pimentel, Methanogens, methane and gastrointestinal motility. J Neurogastroenterol Motil, 2014. 20(1): p. 31-40.
- Ghoshal, U., et al., Irritable Bowel Syndrome, Particularly the Constipation-Predominant Form, Involves an Increase in Methanobrevibacter smithii, Which Is Associated with Higher Methane Production. Gut Liver, 2016. 10(6): p. 932-938.
- Li, T., et al., Multi-Cohort Analysis Reveals Altered Archaea in Colorectal Cancer Fecal Samples Across Populations. Gastroenterology, 2025. 168(3): p. 525-538 e2.
- Mathlouthi, N.E.H., et al., The Archaeome’s Role in Colorectal Cancer: Unveiling the DPANN Group and Investigating Archaeal Functional Signatures. Microorganisms, 2023. 11(11).
- Malat, I., M. Drancourt, and G. Grine, Methanobrevibacter smithii cell variants in human physiology and pathology: A review. Heliyon, 2024. 10(18): p. e36742.
- Hassani, Y., et al., Detection of Methanobrevobacter smithii and Methanobrevibacter oralis in Lower Respiratory Tract Microbiota. Microorganisms, 2020. 8(12).
- Culp, E.J. and A.L. Goodman, Cross-feeding in the gut microbiome: Ecology and mechanisms. Cell Host Microbe, 2023. 31(4): p. 485-499.
- Goyal, A., et al., Ecology-guided prediction of cross-feeding interactions in the human gut microbiome. Nat Commun, 2021. 12(1): p. 1335.
- Houshyar, Y., et al., Going Beyond Bacteria: Uncovering the Role of Archaeome and Mycobiome in Inflammatory Bowel Disease. Front Physiol, 2021. 12: p. 783295.
- Vierbuchen, T., et al., The Human-Associated Archaeon Methanosphaera stadtmanae Is Recognized through Its RNA and Induces TLR8-Dependent NLRP3 Inflammasome Activation. Front Immunol, 2017. 8: p. 1535.
- Campbell, A., et al., H(2) generated by fermentation in the human gut microbiome influences metabolism and competitive fitness of gut butyrate producers. Microbiome, 2023. 11(1): p. 133.
- Duranti, S., et al., Obesity and microbiota: an example of an intricate relationship. Genes Nutr, 2017. 12: p. 18.
- Li, R., J. Li, and X. Zhou, Lung microbiome: new insights into the pathogenesis of respiratory diseases. Signal Transduct Target Ther, 2024. 9(1): p. 19.
- Kim, Y.B., et al., In-depth metataxonomic investigation reveals low richness, high intervariability, and diverse phylotype candidates of archaea in the human urogenital tract. Sci Rep, 2023. 13(1): p. 11746.
- Pimentel, M., et al., Methane, a gas produced by enteric bacteria, slows intestinal transit and augments small intestinal contractile activity. Am J Physiol Gastrointest Liver Physiol, 2006. 290(6): p. G1089-95.
- van de Pol, J.A., et al., Gut Colonization by Methanogenic Archaea Is Associated with Organic Dairy Consumption in Children. Front Microbiol, 2017. 8: p. 355.
- Radaic, A. and Y.L. Kapila, The oralome and its dysbiosis: New insights into oral microbiome-host interactions. Comput Struct Biotechnol J, 2021. 19: p. 1335-1360.
- Wang, P., et al., Gut microbiome-derived ammonia modulates stress vulnerability in the host. Nat Metab, 2023. 5(11): p. 1986-2001.
- Matchado, M.S., et al., Network analysis methods for studying microbial communities: A mini review. Comput Struct Biotechnol J, 2021. 19: p. 2687-2698.
- Liu, Z., et al., Network analyses in microbiome based on high-throughput multi-omics data. Brief Bioinform, 2021. 22(2): p. 1639-1655.
- Abram, F., Systems-based approaches to unravel multi-species microbial community functioning. Comput Struct Biotechnol J, 2015. 13: p. 24-32.
- Papageorgiou, A.C. and P.S. Adam, Archaea appropriate weaponry to breach the bacterial cell wall. PLoS Biol, 2025. 23(8): p. e3003323.
- Takiishi, T., C.I.M. Fenero, and N.O.S. Camara, Intestinal barrier and gut microbiota: Shaping our immune responses throughout life. Tissue Barriers, 2017. 5(4): p. e1373208.
- van Wolferen, M., et al., The cell biology of archaea. Nat Microbiol, 2022. 7(11): p. 1744-1755.
- Kerr, M., et al., Beyond the Gut: Unveiling Methane’s Role in Broader Physiological Systems. FASEB Bioadv, 2025. 7(8): p. e70048.
- Brugere, J.F., et al., Archaebiotics: proposed therapeutic use of archaea to prevent trimethylaminuria and cardiovascular disease. Gut Microbes, 2014. 5(1): p. 5-10.
- Jung, M.Y., et al., Ammonia-oxidizing archaea possess a wide range of cellular ammonia affinities. ISME J, 2022. 16(1): p. 272-283.
- Green, E.R. and J. Mecsas, Bacterial Secretion Systems: An Overview. Microbiol Spectr, 2016. 4(1).
- Duller, S. and C. Moissl-Eichinger, Archaea in the Human Microbiome and Potential Effects on Human Infectious Disease. Emerg Infect Dis, 2024. 30(8): p. 1505-13.
- Karavaeva, V. and F.L. Sousa, Navigating the archaeal frontier: insights and projections from bioinformatic pipelines. Front Microbiol, 2024. 15: p. 1433224.
- Guerra, A., Human associated Archaea: a neglected microbiome worth investigating. World J Microbiol Biotechnol, 2024. 40(2): p. 60.
- Rafiq, M., et al., Challenges and Approaches of Culturing the Unculturable Archaea. Biology (Basel), 2023. 12(12).
- Kioukis, A., et al., Global Archaeal Diversity Revealed Through Massive Data Integration: Uncovering Just Tip of Iceberg. Microorganisms, 2025. 13(3).
- Atomi, H., T. Imanaka, and T. Fukui, Overview of the genetic tools in the Archaea. Front Microbiol, 2012. 3: p. 337.
- Eren, A.M. and J.F. Banfield, Modern microbiology: Embracing complexity through integration across scales. Cell, 2024. 187(19): p. 5151-5170.
- Bahram, M., et al., Newly designed 16S rRNA metabarcoding primers amplify diverse and novel archaeal taxa from the environment. Environ Microbiol Rep, 2019. 11(4): p. 487-494.
- Chibani, C.M., et al., A catalogue of 1,167 genomes from the human gut archaeome. Nat Microbiol, 2022. 7(1): p. 48-61.
- Kapoore, R.V., et al., Co-culturing microbial consortia: approaches for applications in biomanufacturing and bioprocessing. Crit Rev Biotechnol, 2022. 42(1): p. 46-72.
- Pohlschroder, M., et al., Haloferax volcanii: a versatile model for studying archaeal biology. J Bacteriol, 2025. 207(6): p. e0006225.
- Bai, D., J. Peng, and C. Yi, Advances in single-cell multi-omics profiling. RSC Chem Biol, 2021. 2(2): p. 441-449.
- Lim, J., et al., Advances in single-cell omics and multiomics for high-resolution molecular profiling. Exp Mol Med, 2024. 56(3): p. 515-526.
- Gonzalez, A., A. Fullaondo, and A. Odriozola, Why Are Long-Read Sequencing Methods Revolutionizing Microbiome Analysis? Microorganisms, 2025. 13(8).
- Wen, L. and F. Tang, Single-cell omics sequencing technologies: the long-read generation. Trends Genet, 2025.
- Tronel, A., et al., Untargeted and semi-targeted metabolomics approach for profiling small intestinal and fecal metabolome using high-resolution mass spectrometry. Metabolomics, 2025. 21(4): p. 84.
- Buan, N.R., Methanogens: pushing the boundaries of biology. Emerg Top Life Sci, 2018. 2(4): p. 629-646.
- Aggarwal, N., et al., Microbiome and Human Health: Current Understanding, Engineering, and Enabling Technologies. Chem Rev, 2023. 123(1): p. 31-72.
- Vento, J.M., N. Crook, and C.L. Beisel, Barriers to genome editing with CRISPR in bacteria. J Ind Microbiol Biotechnol, 2019. 46(9-10): p. 1327-1341.
- De Lise, F., et al., Archaea as a Model System for Molecular Biology and Biotechnology. Biomolecules, 2023. 13(1).
- Sleytr, U.B., et al., S-layers: principles and applications. FEMS Microbiol Rev, 2014. 38(5): p. 823-64.
- Cowan, D.A., et al., Extremophiles in a changing world. Extremophiles, 2024. 28(2): p. 26.
- Zhu, P., et al., CRISPR/Cas12a toolbox for genome editing in Methanosarcina acetivorans. Front Microbiol, 2023. 14: p. 1235616.
- Wani, A.K., et al., Discovering untapped microbial communities through metagenomics for microplastic remediation: recent advances, challenges, and way forward. Environ Sci Pollut Res Int, 2023. 30(34): p. 81450-81473.
- Ojala, T., et al., Current concepts, advances, and challenges in deciphering the human microbiota with metatranscriptomics. Trends Genet, 2023. 39(9): p. 686-702.
- Wolf, M., et al., Advances in the clinical use of metaproteomics. Expert Rev Proteomics, 2023. 20(4-6): p. 71-86.
- Ruden, D.M., TIGR-Tas and the Expanding Universe of RNA-Guided Genome Editing Systems: A New Era Beyond CRISPR-Cas. Genes (Basel), 2025. 16(8).
- Pfeifer, E. and E.P.C. Rocha, Phage-plasmids promote recombination and emergence of phages and plasmids. Nat Commun, 2024. 15(1): p. 1545.
- Yin, Q., et al., Potential interactions between syntrophic bacteria and methanogens via type IV pili and quorum-sensing systems. Environ Int, 2020. 138: p. 105650.
- Ghaly, T.M., et al., Discovery of integrons in Archaea: Platforms for cross-domain gene transfer. Sci Adv, 2022. 8(46): p. eabq6376.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).