1. Introduction
Plants of section Chrysantha Chang belongs to the genus Camellia of the family Theaceae. This group of plants is named for its “pure yellow flowers like gold” and is the only camellia that has golden yellow petals. Their buds of these plants are round, delicate, and beautiful, and they are known as the “Queen of the Tea Family” and the “Giant Panda of the Plant World” (Zhang and Ren 1998). There are 42 species and 5 varieties belonging to sect. Chrysantha that are known to be native to southern China and Vietnam, most of which are distributed in Guangxi Province, and a few are distributed in Guizhou, Yunnan, and Sichuan Provinces (Liang 2007). Sect. Chrysantha is a group of subtropical plants that are found in warm and humid climates. They utilize fertilizer and have strong waterlogging resistance. Their soil requirements are not high, and they can grow in slightly acidic to neutral soils (Wei et al. 2007). The plants in sect. Chrysantha are evergreen shrubs or small trees with yellow‒brown, nearly smooth bark. Their leaves are leathery, oblong, lanceolate, or rarely oblanceolate. The upper surface of the leaf blade is dark green, the reticulate veins of the leaf blade are inconspicuous, and the margin is serrulate. The flowers in the leaf axils are yellow and solitary. Petals 10–13 are fleshy and glabrous; the outer whorl is suborbicular; and the inner whorl is obovate or elliptic. The ovaries are superior, 3-loculed, glabrous; styles 3–4, glabrous. They flower from December to March. The capsule was depressed and globose, 3.5 cm long and 4.5 cm wide, with 2-3 seeds per locule, and the apex was concave. The seed coat is brown, glossy, hemispherical, and 1.5–2 cm in diameter (Zhang 2023). Section Chrysantha plants have important ornamental, medicinal, and economic value (Huang 1994; Xia et al. 2013; Huang et al. 2009; Wang et al. 2021).
Camellia tianeensis S. Yun Liang et Y. T. Luo is a member of the sect. Chrysantha. In 1995, Liang Shengye et al. recognized this plant as a new species of the genus Camellia characterized by its characteristics of elliptic leaves, 6-7 pairs of lateral veins, solitary flowers, a purplish-red or light-red surface during the bud stage, a yellowish color after opening, and brown seeds (Liang and Luo 1995).In contrast, it was treated as a synonym of Camellia huana in Flora of China (Min and Bruce, 2007). To address the taxonomic issues of C. tianeensis, we conducted research involving the examination of type specimens, field observations of wild populations, micro-morphological analyses of leaves and pollen, and multi-year introduction and cultivation studies, ultimately confirming that C. tianeensis is an independent species and C. liberofilamenta is a synonym of C. huana (Jiang et al., 2024). Currently, relevant studies have been conducted on the distribution status, conservation strategies, cultivation, and population characterization of this species (Xie et al. 2013, 2014; Su et al. 2017; Luo et al. 2021; Xu et al. 2022; Yang et al. 2023). However, there are no molecular data available for exploring the phylogenetic position of C. tianeensis. Therefore, the present study reports the whole chloroplast genome of C. tianeensis, which not only includes the genetic information of the species but also provides important theoretical references for further classification, evolution, and exploitation of the species.
2. Materials and Methods
2.1. Material Collection, DNA Extraction, and Sequencing
Samples of the
C. tianeensis plants in this study were collected from the Forestry Bureau of Ceheng County, Guizhou Province, China (N 24.98465303°, E 105.81570840°) (
Figure 1). The specimens were preserved in the Tree Specimen Laboratory, Guizhou Academy of Forestry (GZAF, LH-20221101). Fresh and tender leaves were collected, and chloroplast DNA was extracted in the laboratory using an optimized CTAB method (Pahlich and Gerliz 1980). The integrity of the DNA was determined by 1% glucose agar gel electrophoresis, and the purity and content of the DNA were determined using a nucleic acid proteometer. The DNA was then subjected to interruption, end repair, and splice junction detection to construct a sequencing library. Sequencing was performed using the NovaSeq 6000 high-throughput sequencing platform on the libraries that passed the quality test.
2.2. Assembly and Annotation of the Chloroplast Genome
Using GetOrganelle 1.7.5.3 (Jin et al. 2020) software, the screened data were spliced from scratch to obtain a circular chloroplast gene map. After that, online annotation, BLAST comparison, and manual correction were performed using CPGAVAS2 (Shi et al. 2019), and the sequence was finally uploaded to NCBI (GenBank ID: PP187689). The chloroplast genome map was constructed online via OGDRAW (
https://chlorobox.mpimp-golm.mpg.de/OGDraw.html) (Lohse et al. 2007).
2.3. Repeat Sequence Analysis and Codon Preference
In this study, we searched for SSRs in the chloroplast genome sequence of
C. tianeensis using MISA 2.1 software (Beier et al. 2017) and set the minimum number of repeat units and the number of repetitions as follows: at least 10 for single-nucleotide units, at least 5 for dinucleotide units, at least 4 for trinucleotide units, at least 3 for tetranucleotide units, at least 3 for pentanucleotide units, and at least 3 for hexanucleotide units (Zheng et al. 2020). The REPuter (
https://bibiserv.cebitec.uni-bielefeld.de/reputer) (Kurtz et al. 2001) online tool was used to search for larger repeat sequences with a Hamming distance of 3 and a minimum repeat size of 30 bp. The four types were forward (F), reverse (R), complement (C), and palindromic (P). The codon preference relative synonymous codon usage (RSCU) was statistically analyzed using CodonW 1.4.2 software (Shield and Sharp 1987) and plotted using R4.0.5.
2.4. Phylogenetic Analysis
To understand the phylogenetic relationships of
C. tianeensis, the chloroplast genome sequences of 22 species of sect.
Chrysantha were queried and downloaded from the NCBI database (
https://www.ncbi.nlm.nih.gov/) in this study. The phylogenetic analysis was performed using
Camellia pyxidiacea (GenBank ID: OP058659) as an outgroup. After comparison via MAFFT7 (Katoh and Standiey 2014), the phylogenetic tree was constructed (GTR+I+G model) via manual correction in MEGAX (Kumar et al. 2018), after which the best maximum likelihood (ML) method was selected. The phylogenetic tree was subsequently constructed in IQ-TREE 2.2.0, and the self-expansion support was set to 1000 (Trifinopoulos et al. 2016). The optimal model (HKY+G+I) was identified using MrModeltest v2.3, and a Bayesian inference (BI) phylogenetic tree was subsequently constructed using MrBayes v3.2.7 (Huelsenbeck and Ronquist 2001). Finally, the phylogenetic tree was visualized using the online tool iTOL V4 (
https://itol.embl.de/) (Letunic and Bork 2021).
3. Results
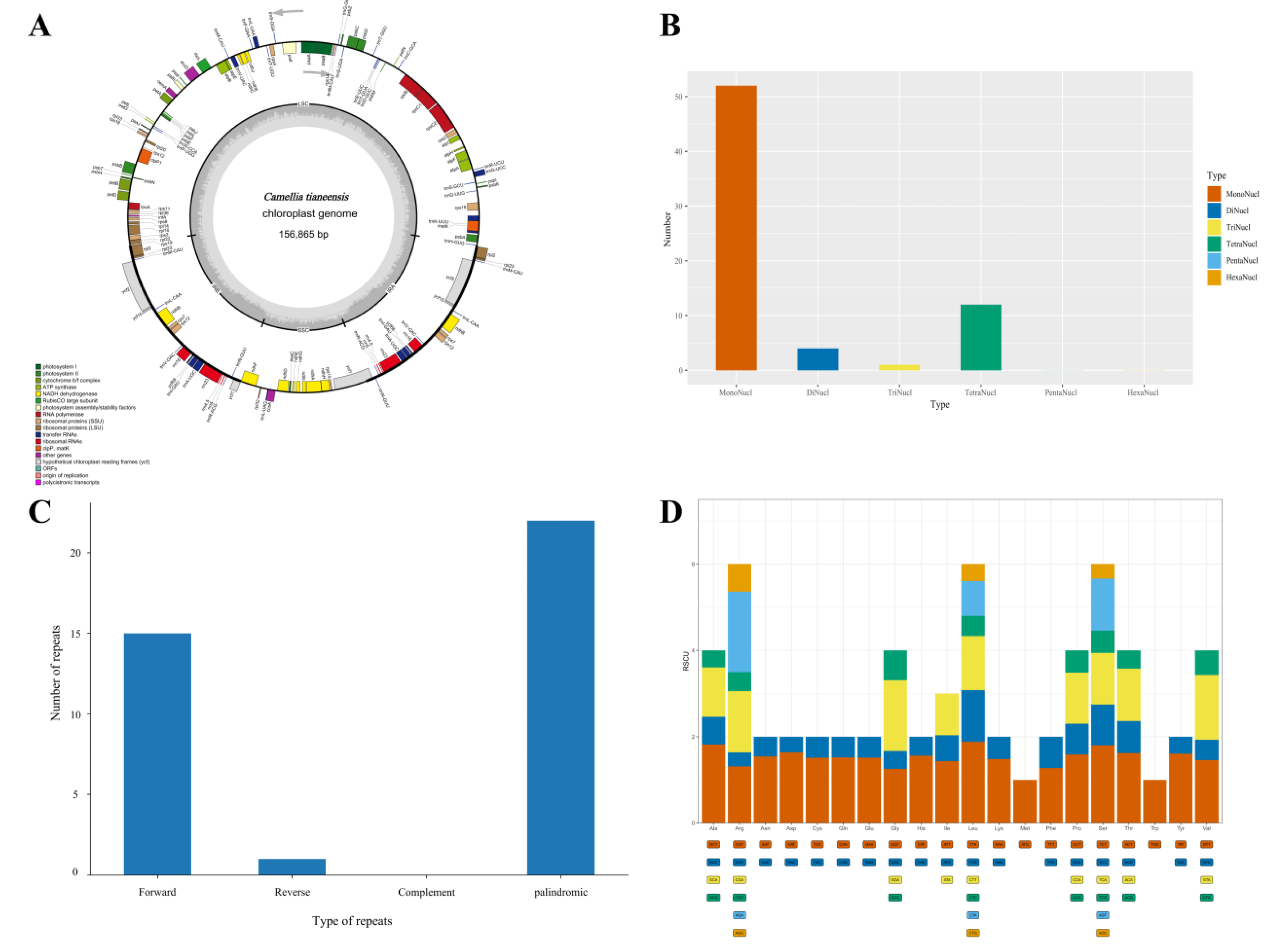
This study yielded a total chloroplast whole genome (
Figure 2A) for
C. tianeensis, with a length of 156,865 bp and exhibiting the typical tetrameric structural features of one large-copy region (LSC), one small-copy region (SSC), and two inverted-repeat regions (IRs). The length of each region was 86,579 bp (LSC), 18,236 bp (SSC), and 26,025 bp (IRs). The total GC content was 37.32%, including 35.33% of the LSC, 30.59% of the SSC, and 42.99% of the IRs. A total of 164 genes, including 111 protein-coding genes, 45 tRNA genes, and 8 rRNA genes, were identified by chloroplast-wide genome annotation of
C. tianeensis. A total of 69 simple repeat sequences(SSRs), including 52 single nucleotides (A-21, T-31), 4 dinucleotides (AT-3, TA-1), 1 trinucleotide (TTC-1), and 12 tetranucleotides (AGAT-1, AAAT-1, AATA-1, ATAG-1, GTCT-1, GAAA-1, GAGG-1, CCCT-1, TCTT-1, TTTC-1, TCTA-1), were identified from four repetitive modes across the whole genome of chloroplasts. The single nucleotide repeats were the most abundant and were significantly more abundant than the other repeat types (
Figure 2B). A total of 38 dispersed repeats were found and categorized into three types: 15 forward (F) repeats, 1 reverse (R) repeat, and 22 palindromic (P) repeats. No complement (C) repeat sequences were found (
Figure 2C). Codon preferences were also analyzed in this study, and a total of 61 codons encoding 20 amino acids were detected, in addition to the termination codons UAA, UAG, and UGA. A total of 27,001 codons were observed, of which Tle was the most abundant with 1118 codons, accounting for 4.14%, and Cys was the least abundant with 72, accounting for 0.27%. Thirty-one codons had an RSCU greater than 1, 12 had an A in the final position, 16 had a T in the final position, and 3 had a G in the final position. Species with A/T as the final position were the most common, indicating that the whole-genome codon preference of
C. tianeensis chloroplasts ended in A/T (
Figure 2D).
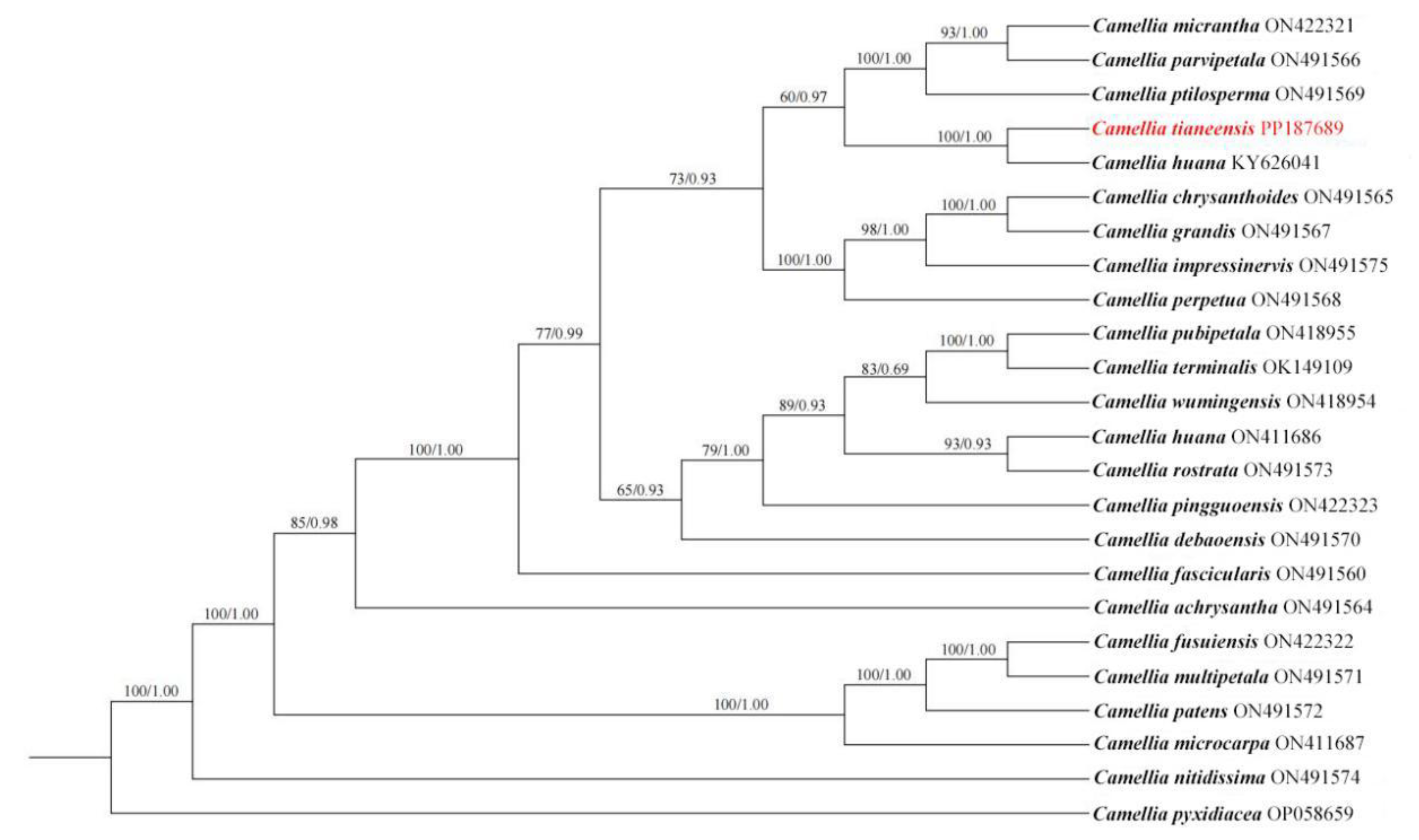
A phylogenetic tree constructed based on 22 published complete whole chloroplast genomes of from sect.
Chrysantha showed that
C. tianeensis is a constituent of sect.
Chrysantha (
Figure 3), and clustered on the same small branch as
Camellia liberofilamenta (BS/PP = 100/1.00). Most of the tree nodes were highly supported, and
Camellia pyxidiacea (outgroup) was completely separated from the goldenrod group of plants.
4. Discussion
Chloroplasts, the main organelles involved in photosynthesis, have independent and complete genomes and are uniparental in most species (Zou et al. 2021). The chloroplast genome has been widely used in plant taxonomic revision, population genetic structure, genetic diversity, population dynamic history, and kinship relationship studies (Zou et al. 2021; Chen et al. 2010). As soon as the chloroplast whole-genome sequence was reported, it was studied by many scholars and applied to plants, thus solving many important problems, such as ambiguous taxonomic identification. By analyzing the NCBI database, we obtained genome-wide information on the chloroplasts of more than 100 species of the genus Camellia. These chloroplast DNAs exhibit very small differences, ranging from 150 to 160 kb, and all of them have a typical tetrameric structure with relatively conserved structural variation. The degree of conservatism of the chloroplast DNA increases with increasing GC content. In this study, we submitted whole-genome sequence information for the chloroplast of C. tianeensis to the NCBI database to enrich the available chloroplast genome data for the genus Camellia.
SSRs refer to a segment of the chloroplast genome with a length of 1-6 nucleotides and are often applied to plant species identification, genetic map construction, population systematic evolution, and germplasm resource genetic diversity research because of their rich content and high polymorphism (Du et al. 2019; Liu et al. 2019; Duan et al. 2019). The results of this study showed that there are 69 SSRs in the chloroplast genome of C. tianeensis, including 52 mononucleotides, 4 dinucleotides, 1 trinucleotide (TTC-1), and 12 tetranucleotides. Among the cpSSR categories, mononucleotide SSRs were the most abundant and significantly more common than the other repeat types. There were 38 dispersed repeats, and no complement (C) repeats were found. The sequence composition of the SSR loci in this study is consistent with the results of existing studies, thus proving that the SSR loci are dominated by polyA and polyT (Kuang et al. 2011); in particular, the SSR gene loci obtained will lay the foundation for further molecular genetic analysis of sect. Chrysantha.
Codon preference is the phenomenon of unequal use of synonymous codons coding for the same amino acid in an organism that has developed a set of commonly used codons that are compatible during evolution. Factors such as mutation, selection, gene length, gene function, and genetic drift can have some effect on this population. The preference for codon usage varies among species, so the closeness of kinship can be judged based on codon preference (Zhang 2007). The RSCU represents the ratio between the actual usage value and the theoretical usage value of the codon. An RSCU less than 1 indicates that the codon is used less frequently than other synonymous codons; an RSCU greater than 1 indicates that the codon is used more frequently than the synonymous codon; and an RSCU equal to 1 indicates that there is no preference for the codon (Zhao et al. 2008). Of the two amino acids with an RSCU equal to 1, one is tryptophan (Trp), and the other is methionine (Met). Both have only one codon encoding an amino acid; there is no codon preference, and the C. tianeensis codons are more inclined to end in A/U, which is in agreement with the results of the study by Zhang Xiaoyu (Zhang 2023) on the group of plants of the sect. Chrysantha.
Phylogenetic analysis can provide an effective basis for discriminating affinities between species. Wei et al. (2022) reconstructed the phylogenetic relationship of yellow-flowered Camellia species based on multi-molecular data, and they proposed a new taxonomic treatment for the sect. Chrysantha recognized 20 species inclued in this section. Although they provided a comprehensive analysis for the sect. Chrysantha, this result lacked C. tianeensis because it was considered a synonym of C. huana at the time. Moreover, among the molecular data used in this study by Wei et al. (2022), the chloroplast genome was only partially represented—limited to the small single-copy (SSC) region—rather than the complete genome sequence. The phylogenetic tree based on single-copy homologous genes showed that golden camellia species with shorter geographical distances were closer phylogenetically (Xie et al., 2025). Based on whole chloroplast genome sequences, the present phylogenetic analysis demonstrates that C. tianeensis is sister to C. huana with strong support, providing valuable insights into the evolutionary relationships within the sect. Chrysantha at the genomic level, This molecular assessment of C. tianeensis not only clarifies its phylogenetic position but also lays a foundation for future studies on the molecular systematics and population evolution of this section.
5. Conclusions
In this study, the chloroplast genome of C. tianeensis was characterized for the first time by high-throughput sequencing technology, and its repeat sequence and codon preference were analyzed. The phylogenetic position of the species was initially explored through the construction of a phylogenetic tree with other plants in sect. Chrysantha. of the chloroplast genome contained 164 genes with a length of 156,865 bp and four SSR types scattered among the repeat sequence types; no complement (C) repeats were found; the codon preference of the whole-genome codon of the chloroplasts of C. tianeensis ended in A/T, and C. tianeensis was clustered in the same branch as the other plants in sect. Chrysantha. Therefore, the chloroplast genomic data of this study may aid in exploring the phylogenetic relationships of sect. Chrysantha, and additionally, this study provides a theoretical basis and technical support for the rational exploitation and effective conservation of this species in the future.
Author Contributions
Ju-Yan CHEN designed, wrote and revised this study. Lun-Xiu Deng and He Li participated in the collection and identification of plant material. All the authors read and approved the final manuscript and agreed to be accountable for all the aspects of the work.
Funding
This project was supported by the Basic Research of Guizhou Science and Technology - ZK [2024] General 620, Guizhou Province”Hundred Levels’Talent Project”[Qiankehe Platform Talent (2020) 6017-2] and The research project of the director of the Forestry Science Institute of Guizhou Province(Guilin Kehe [2024]09); Key Laboratory of National Forestry and Grassland Administration on Biodiversity Conservation in Karst Mountainous Areas of Southwestern China.
Ethical statement
All the materials used in this study were in compliance with international and national legal standards. The collected species material does not pose a threat to other species, and the collection of the species is recognized by the relevant authorities.
Disclosure statement
The authors declare that there are no conflicts of interest regarding the publication of this article. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The genome data that supported the findings of this study are openly available in GenBank of NCBI at [
https://www.ncbi.nlm.nih.gov] under accession no. PP187689.
References
- Beier S, Thiel T, Munch T, Scholz U, Mascher M. 2017. MISA-web: a web server for microsatellite prediction. Bioinformatics. 33(16): 2583-2585. [CrossRef]
- Chen SL, Yao H, Han JP, Liu C, Song JY, Shi LC, Zhu YG, Ma XY, Gao T, Pang XH, Yao H, Luo K, Li Y, Li XW, Jia XC, Lin YL, Leon C. 2010. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One. 5(1): e8613. [CrossRef]
- Du QZ, Wang BW, Wei ZZ, Zhang ZZ, Zhang DQ, Li BL. 2012. Genetic diversity and population structure of Chinese white poplar (Populus tomentosa) revealed by SSR markers. Journal of Heredity. 103(6): 853-862. [CrossRef]
- Duan YZ, Du ZY, Wang HT. 2019. Chloroplast genome characteristics of endangered relict plant tetraena mongolica in the arid region of northwest China. Bulletin of Botanical Research. 39(5): 653-663.
- Huang, XC. 1994. Overview of the development and utilization of Camellia nitidssima and prospect prediction. Chinese Journal of Information on Traditional Chinese Medicine. (06): 10-11.
- Huang YL, Chen YY, Wen YX, Li DP, Liang RG, Wei X. 2009. Effects of the Extracts from Camellia nitidssima Leaves on Blood Lipids. Lishizhen Medicine and Materia Medica Research. 20(04): 776-777.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17 (8): 754-755. [CrossRef]
- Jiang GB, Li H, Yang YB, Xu CR, Guo ZX, Deng LX. 2024. Taxonomic Notes on Camellia huana (Theaceae). Bulletin of Botanical Research. 44(06): 901–913.
- Jin JJ, Yu WB, Yang JB, Song Y, Depamphilis CW, Yi TS, Li DZ. 2020. Get organelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biology. 21(1): 1-31. [CrossRef]
- Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. 2001. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic acids research. 29(22): 4633-4642. doi.org/10.1093/nar/29.22.4633.
- Kuang DY, Wu H, Wang YL, Gao LM, Zhang ZZ, Lu L. 2011. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): implication for DNA barcoding and population genetic. Genome. 54(8): 663-673. [CrossRef]
- Katoh K, Standley DM. 2014. MAFFT: iterative refinement and additional methods. Methods Mol Biol. 1079: 131-46. [CrossRef]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution. 35(6): 1547-1549. [CrossRef]
- Liang, SY. 2007. The World List of Camellia. Guangxi Forestry Science. (04): 221-223. [CrossRef]
- Liang SY, Luo YT. 1995. A new species of Camellia sinensis from China. Guangxi Forestry Science. 24(02): 83-85. [CrossRef]
- Luo QG, Shu FB, Feng LX, Huang HH, Su BC, Li S, Mai KL. 2021. Study on the effect of fertilization on the growth of a high crown and the total flavonoid content of Camellia tianeensis. Journal of Green Science and Technology. 23(07): 52-54. [CrossRef]
- Lohse M, Drechsel O, Bock R. 2007. Organellar Genome DRAW (OGDRAW): a tool for the easy generation of highquality custom graphical maps of plastid and mitochondrial genome. Current Genetics. 52(5): 267-274. [CrossRef]
- Letunic I, Bork P. 2021. Interactive Tree Of Life (iTOL)v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Research. 49 (W1): 293-296. [CrossRef]
- Liu LY, Geng YP, Song ML, Zhang PF, Hou JL, Wang WQ. 2019. Genetic structure and diversity of glycyrrhiza populations based on transcriptome SSR markers. Plant Molecular Biology Reporter. 37:401-412. [CrossRef]
- Min T, Bruce B. 2007. Flora of China. Beijing: Science Press and Missouri Botanical Garden Press. 367-372.
- Pahlich E, Gerliz C. 1980. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemistry. 19(1): 11-13.
- Shi LC, Chen H, Jiang M, Wang LQ, Wu X, Huang LF, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Research. 47(W1): 65-73. [CrossRef]
- Shield DC, Sharp PM. 1987. Synonymous codon usage in Bacillus subtilis reflects both translational selection and mutational biases. Nucleic Acids Research. 15(19): 8023-8040.
- Su FB, Feng LX, Huang HH, Luo QG, Xu RR, Lin H, Li RZ, Li S. 2017. Preliminary Study of the Selection of Superior Individuals from Late Blooming Camellia nitidssima Chi. in Tian’e. Journal of Hechi University. 37(05): 12-16.
- Trifinopoulos J, Nguyen LT, Von HA, Minh BQ. 2016. W-IQTREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research. 44(W1): 232-235. [CrossRef]
- Wei SJ, Liufu, YQ, Zheng, HW, Chen, HL, Lai, YC, Liu, Y, Ye, QQ, Tang, SQ. Using phylogenomics to untangle the taxonomic incongruence of yellow-flowered Camellia species (Theaceae) in China. 2022. Journal of Systematics and Evolution. 61(5): 748-763. [CrossRef]
- Wei X, Jiang YS, Wei JQ, Chen ZY, Wang ML, Zhao RF. 2007. Investigation on the geographical distribution and habitat of Camellia nitidissma. Ecology and Environmental Sciences. (03): 895-899. [CrossRef]
- Wang ZL, Guo YJ, Zhu YY, Chen L, Wu T, Liu DH, Huang BS, Du HZ. 2021. Active fractions of Camellia nitidissima inhibit non-small cell lung cancer via suppressing epidermal growth factor receptor. China Journal of Chinese Materia Medica. 46(20): 5362-5371. [CrossRef]
- Xia X, Huang JX, Wang ZP, Wang Q, Pan LG. 2013. Studies on the hypoglycemic effect and acute toxicity of Camellia nitidssima leaves. Lishizhen Medicine and Materia Medica Research. 24(05): 1281-1282.
- Xie DZ, Qin LX, Qin GL, Wei L, Fu YJ, Ya ZG. 2013. The Community Characteristics of Accompanying Plants of Camellia tianeensis in Guangxi. Guizhou Agricultural Sciences. 41(05): 40-43.
- Xie DZ, Ya ZG, Han JY, Wei Y, Wei L. 2014. Study of Distribution and Protection Stategies of Camellia tianeensis. Journal of Green Science and Technology. (04): 89-91.
- Xie YJ, Bai YY, Gao H, Li YY, Su MX, Li SS, Chen JM, Li T, Yan GY. Phylotranscriptomics resolved phylogenetic relationships and divergence time between 20 golden camellia species. Scientific Reports, 2025. 15(1). [CrossRef]
- Xu YT, Su FB, Feng LX, Yang CS, Huang ZX, He HJ, Su BC. 2022. High-yield Cultivation Techniques of Camellia tianeensis in Forests. Journal of Smart Agriculture. 2(11): 49-51. [CrossRef]
- Yang CS, Luo QG, Su FB, Feng LX, Mai KL, Su BC. 2023. Selection of superior individual of early-flowering and late-flowering Camellia tianeensis. South China Agriculture. 17(01):13-17+22. [CrossRef]
- Zhang, WJ. 2007. Codon Analysis and Its Application in Bioinformatics and Evolutionary Studies, Dissertation for Ph.D., Fudan University in Shanghai, Supervisors: Zhong Y., pp.10-23.
- Zhao Y, Liu HM, Gu Y, Huang YB. 2008. Analysis of Characteris tic of Codon Usage in waxy Gene of Zea mays. Journal of Maize Sciences. 16(2): 16-21.
- Zhang HD, Ren SX. 1998. Flora of China. Beijing: Science Press. pp 101-112.
- Zhang, XY. 2023. Study on the evolution of chloroplast genome of Sect. Chrysantha Chang. ShanXi University.
- Zheng W, Zhang H, Wang QM, Gao Y, Zhang ZH, Sun YX. 2020. Complete Chloroplast Genome Sequence of Clivia miniata and Its Characteristics. Acta Horticulturae Sinica. 47(12): 2439-2450. [CrossRef]
- Zou WJ, Liu K, Gao XP, Yu CJ, Wang XF, Shi JJ, Chao YR, Yu Q, Zhou GK, Ge L. 2021. Diurnal variation of transitory starch metabolism is regulated by plastid proteins WXR1/WXR3 in Arabidopsis young seedlings. Journal of Experimental Botany. 72(8): 3074–3090. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).