Submitted:
03 January 2024
Posted:
04 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
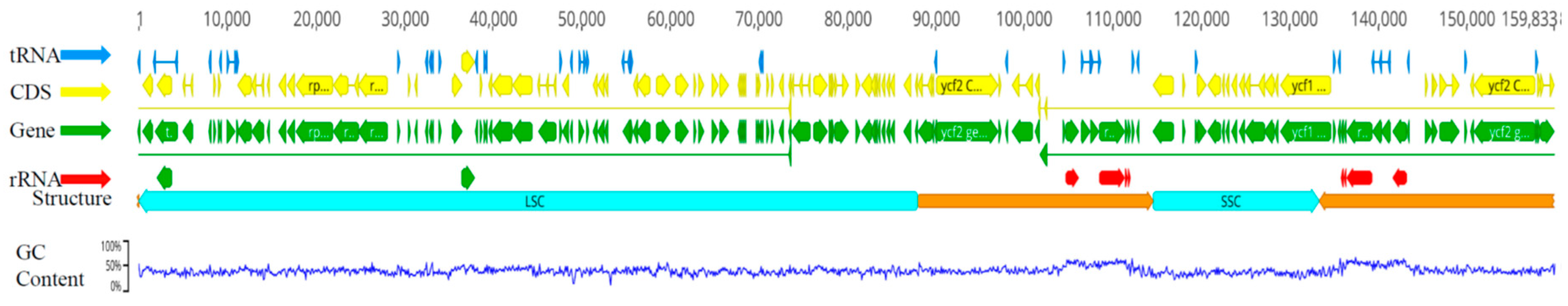
2.1. General characteristics of eight chloroplast genome of Lirianthe Species
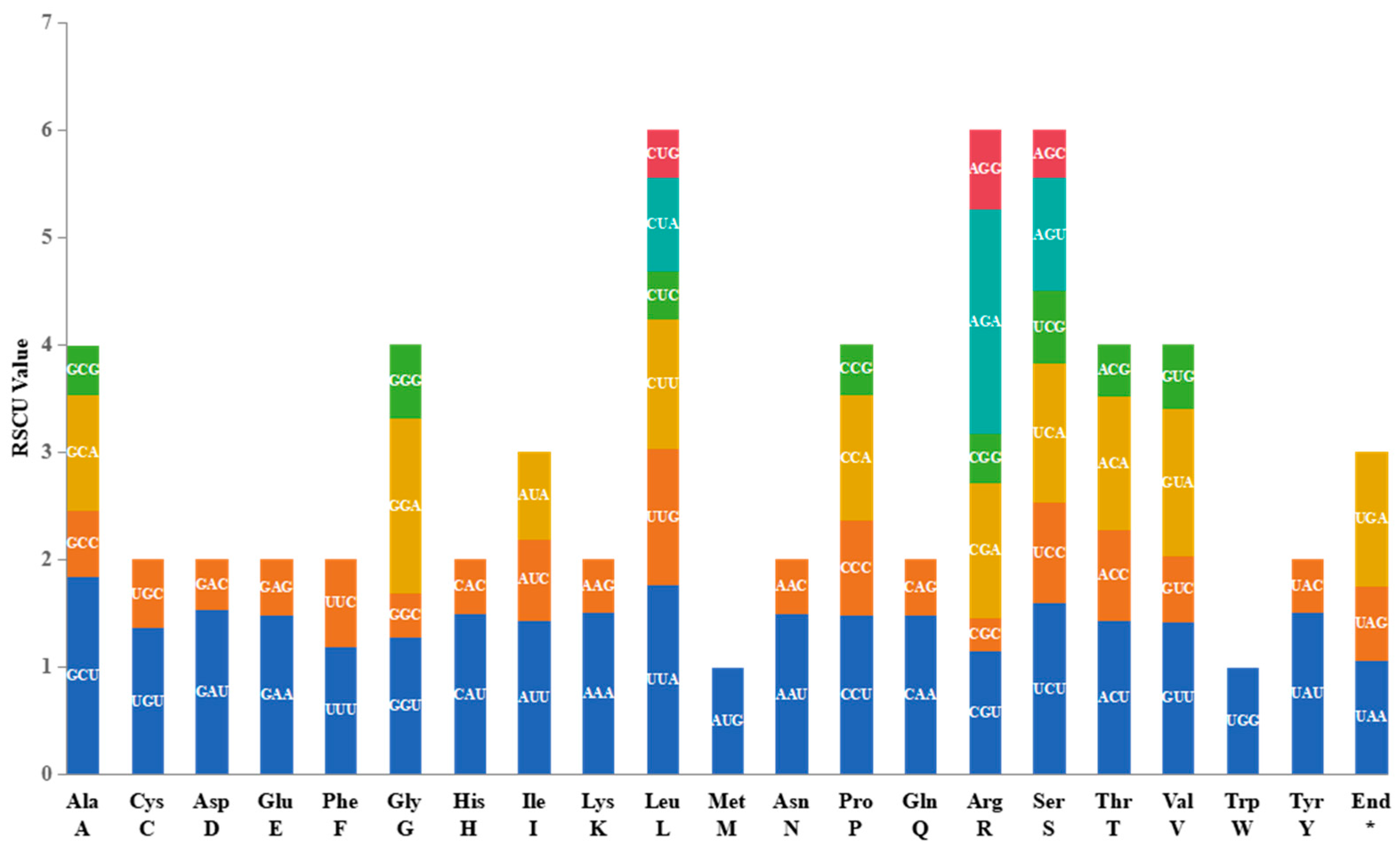
2.2. Codon usage of protein coding genes
2.3. Repeat identification
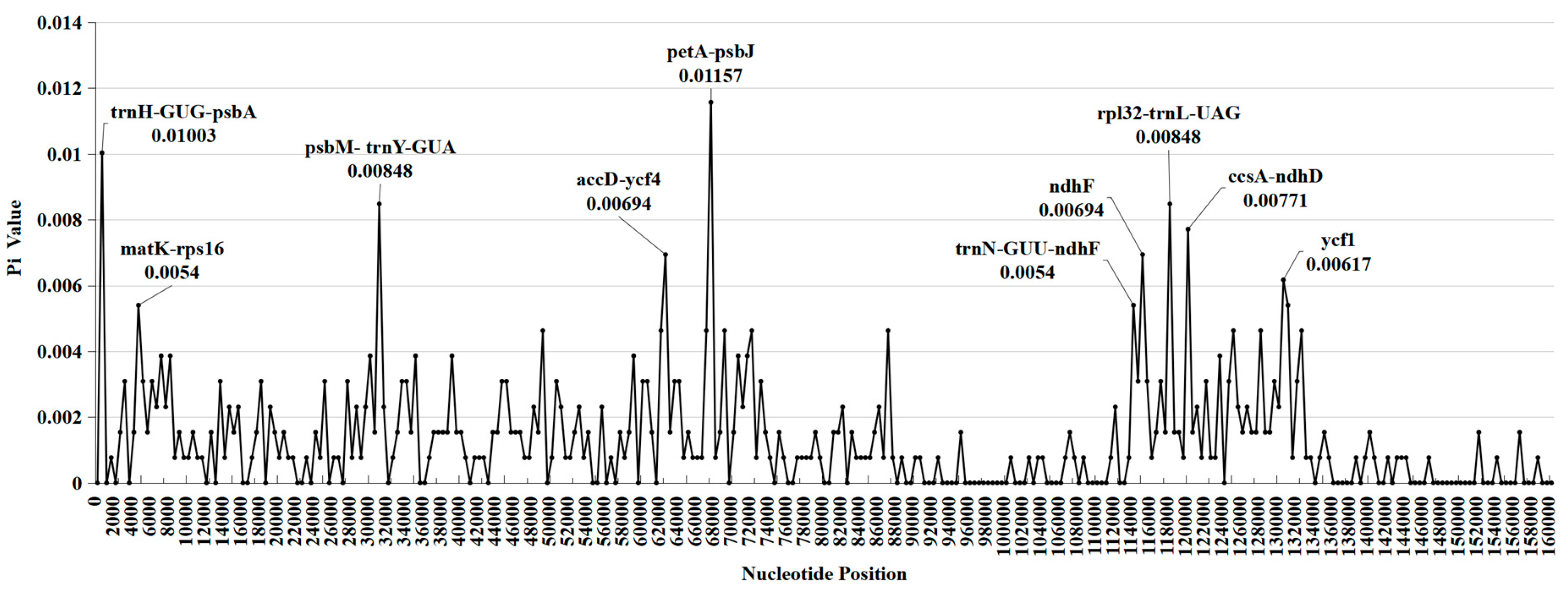
2.4. DNA polymorphism
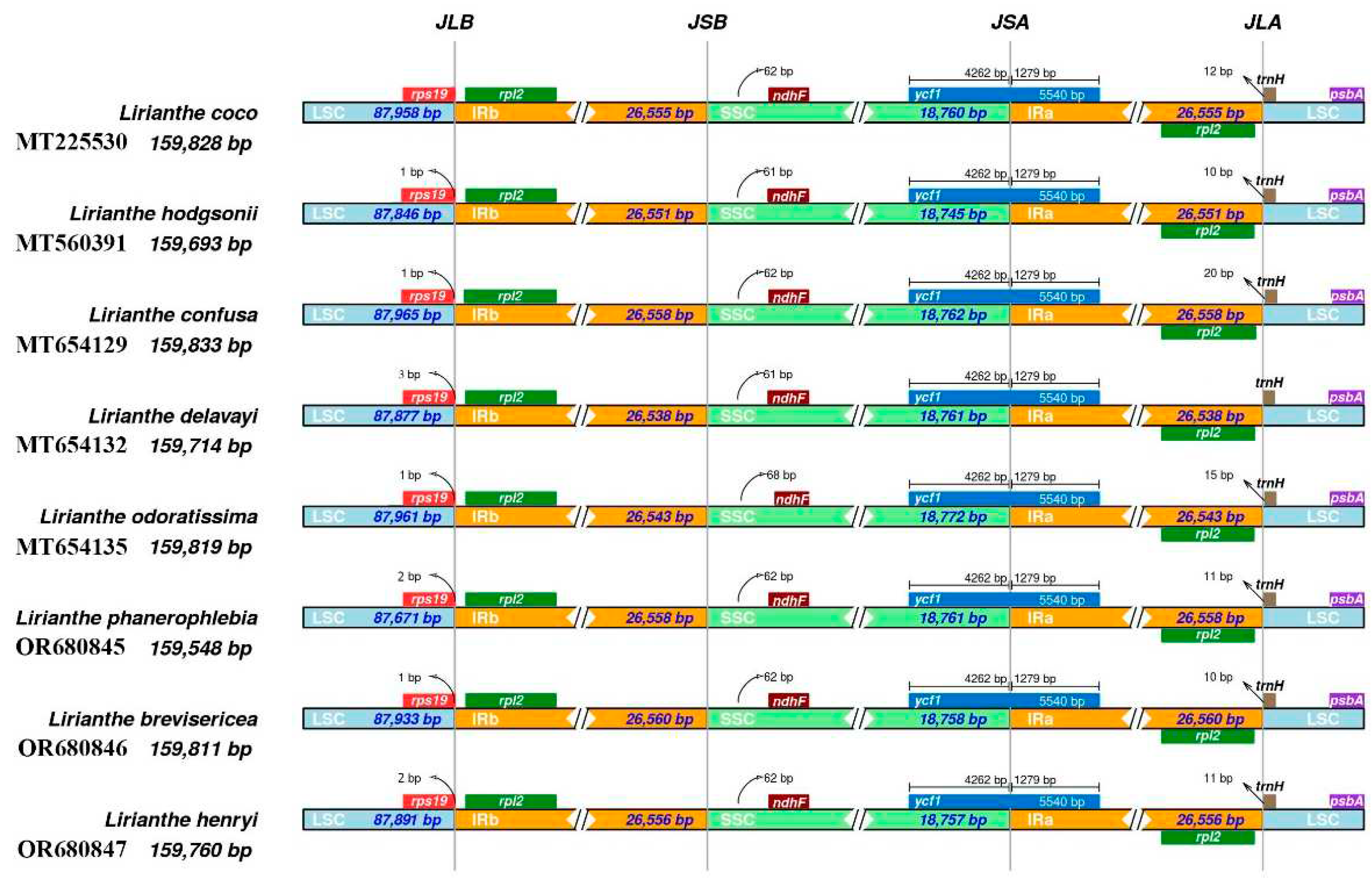
2.5. Contraction and expansion of IR region
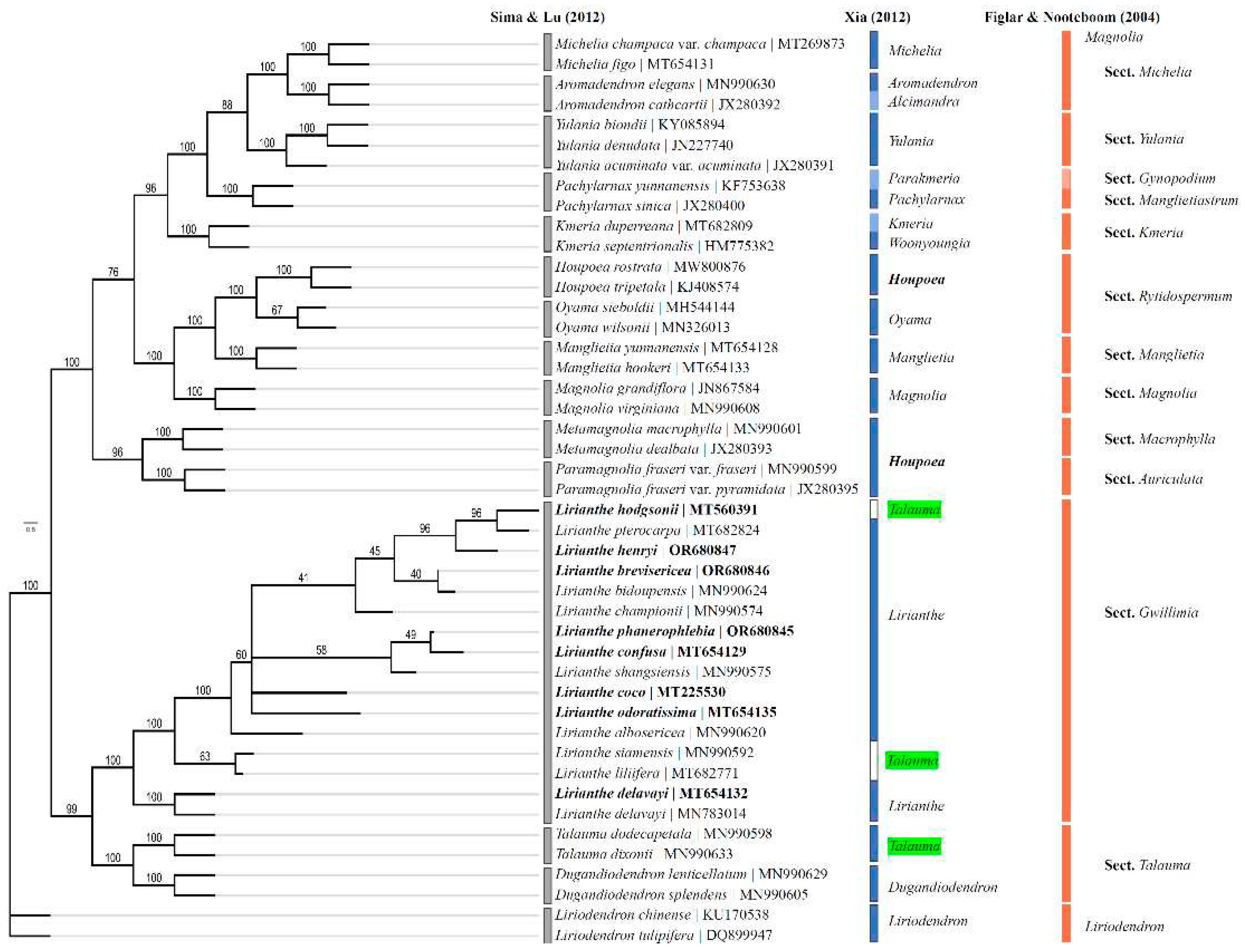
2.6. Phylogenetic analysis
2.7. Plant Morphology
3. Discussion
4. Materials and Methods
4.1. Sample, DNA extraction and sequencing
4.2. Chloroplast genome assembly and annotation
4.3. The analysis of structures and characteristics of CPGs
4.4. Comparitive analysis of chloroplast genomes
4.5. Phylogenetic analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Azuma, H.; ,Garcia-Franco, J.G.; Rico-Gray, V.; Thien, L.B. Molecular phylogeny of the Magnoliaceae: the biogeography of tropical and temperate disjunctions. Am. J. Bot. 2001, 88, 2275-85. [CrossRef]
- Wang, Y.; Liu, B.; Nie, Z.; Chen, H.; Chen, F.; Figlar, R.B.; Wen.J. Major clades and a revised classification of Magnolia and Magnoliaceae based on whole plastid genome sequences via genome skimming. J. Syst. Evol. 2020, 58, 673-695. [CrossRef]
- Dandy, J. E. The genera of Magnolieae. Bulletin of Miscellaneous Information (Royal Botanic Gardens, Kew). 1927, 7, 257-264. [CrossRef]
- Law, Y.W. A preliminary study on the taxonomy of the family Magnoliaceae." J. Syst. Evol. 1984, 22, 89-109.
- Nooteboom, H.P. Notes on Magnoliaceae with a revision of Pachylarnax and Elmerrillia and the Malesian species of Manglietia and Michelia. Blumea: Biodiversity, Evolution and Biogeography of Plants. 1985, 31, 65-121.
- Xia, N.H. A new classification system of family Magnoliaceae. Paper presented at the Proceedings of the Second International Symposium on the Family Magnoliaceae. Wuhan, 2012.
- Sima, Y.K and Lu. S.G. A new system for the family Magnoliaceae. Paper presented at the Proceedings of the Second International Symposium on the Family Magnoliaceae. Wuhan, 2012.
- Figlar, R. B. and Nooteboom, H.P. Notes on Magnoliaceae IV. Blumea: Biodiversity, Evolution and Biogeography of Plants. 2004, 49 , 87-100. [CrossRef]
- Li, J. A cladistic analysis of Magnoliaceae. Acta Botanica Yunnanica (Renamed as Plant Diversity). 1997, 19, 342-56.
- Sima, Y.; Yu, H.; Ma, H.; Hao, J.; Chen, S.; Li, S.; Fu, Y. New combinations in Magnoliaceae (in Chinese). Journal of West China Forestry Science, 2020, 49, 29-40.
- Yang, C.; Deng, J. Zhou, H.; Li, H.; Zheng, Z. The new records of Magnoliaceae in Guizhou province (in Chinese). Guizhou Forestry Science and Technology. 2016, 44, 21-23.
- Yang, C.; Deng, J. Zhou, H. Study on the resources and ornamental characteristics of the native Magnoliaceaes in Guizhou province (in Chinese). Guizhou Forestry Science and Technology, 2017, 45, 19-23.
- Yuan, C.; Yang,Y.; Dai, X.; Li, H.; Yang, C.; Wu, Y.; Wang J. Magnoliaceae plants and its distribution pattern in Guizhou province (in Chinese). Journal of West China Forestry Science, 2017, 46, 68-75.
- Wang, Y.; Wang, D. The complete chloroplast genome and phylogenetic analysis of Manglietia ventii (Magnoliaceae). Mitochondrial DNA Part B-Resour. 2022, 7, 196-198. [CrossRef]
- Figlar, R.B.; and Nooteboom, H.P. Magnolia classification. https://www.magnoliasociety.org/Classification (accessed 2023.11.20).
- Xia, N.; Liu, Y.; Nooteboom, H.P. Magnoliaceae. In Flora of China. Vol. 7, edited by Wu Z.Y. and Raven P.H., 48-91. Beijing: Science Press &: St. Louis: Missouri Botanical Garden Press, 2008. [CrossRef]
- Shui, Y.; Sima, Y.; Wen, J.; Chen, W. Vouchered flora of southeast Yunnan (Vol. 1). Kunming: Yunnan Technology and Sciences Housing, 2009.
- Li, Y.; Li, D. Conservation value and exploitation foreground of the Magnoliaceae plants in Yunnan. Jouranl of Beijing Forestry University, 1999, 21, 29-35.
- Sima, Y.; Ma, H.; Xu, T.; Yang, J.; Zhang, D. The chemical composition of volatile oils from leaves of three species of Lirianthe Spach in Magnoliaceae and its systematic signifcance (in Chinese). Journal of West China Forestry Science, 2018, 47, 7-14,+29.
- Azuma, H., García-Franco, J.G., Rico-Gray, V., Thien, L.B. Molecular phylogeny of the Magnoliaceae: the biogeography of tropical and temperate disjunctions. Am. J. Bot. 2001, 88, 2275-2285. [CrossRef]
- Kim, S.; Suh, Y. Phylogeny of Magnoliaceae based on ten chloroplast dna regions. J. Plant Biol. 2013, 56, 290-305. [CrossRef]
- Wollaeger, H.M. Genetic variability in Magnolia Acuminata (L.) populations in the Eastern United States. Bachelor thesis, Wittenberg University, 2011.
- Chen, S.; Wu, T.; Ma, H.; Fu, Y.; Zhu, Y.; Hao, J.; Jiang, H.; Sima, Y. The complete chloroplast genome sequence of Lirianthe Hodgsonii, a tree species of Magnoliaceae as least concern. Mitochondrial DNA Part B-Resour. 2020, 5, 3064-3066. [CrossRef]
- Redwan, R. M.; Saidin, A.; and Kumar, S.V. Complete chloroplast genome sequence of Md-2 pineapple and its comparative analysis among nine other plants from the Subclass commelinidae. BMC Plant Biol. 2015, 15, 196. [CrossRef]
- Li, X.; Yang, Y.; Henry, R.J.; Rossetto, M.; Wang, Y.; Chen, S. Plant DNA barcoding: from gene to genome. Biol. Rev. Camb. Philos. Soc. 2015, 90, 157-166. [CrossRef]
- Li, H.T.; Luo, Y.; Gan, L.; Ma, P.F.; Gao, L.M.; Yang, J.B.; Cai, J.; Gitzendanner, M.A.; Fritsch, P.W.; Zhang, T.; Jin, J.J.; Zeng, C.X.; Wang, H.; Yu, W.B.; Zhang, R.; van der Bank, M.; Olmstead, R.G.; Hollingsworth, P.M.' Chase, M.W.; Soltis, D.E.; Soltis, P.S.; Yi, T.; Li, D.Z. Plastid phylogenomic insights into relationships of all flowering plant families. BMC Biol. 2021, 19, 232. [CrossRef]
- Nguyen, V.B.; Linh Giang, V.N.; Waminal, N.E.; Park, H.S.; Kim, N.H.; Jang, W.; Lee, J.; Yang, T.J. Comprehensive comparative analysis of chloroplast genomes from seven Panax species and development of an authentication system based on species-unique single nucleotide polymorphism markers. J. Ginseng Res. 2020, 44, 135-44. [CrossRef]
- Kim, K.; Lee, S.C.; Lee, J.; Lee, H.O.; Joh, H.J.; Kim, N.H.; Park, H.S.; Yang, T.J. Comprehensive survey of genetic diversity in chloroplast genomes and 45s nrDNAs within Panax ginseng species. PLoS One, 2015, 10, e0117159. [CrossRef]
- Lang, C.; Weber, N.; Moeller, M.; Schramm, L.; Schelm, S.; Kohlbacher, O.; Fischer, M. Genetic authentication: differentiation of hazelnut cultivars using polymorphic sites of the chloroplast genome. Food Control. 2021, 130, 108344. [CrossRef]
- Sima, Y.; Wu, T.; Fu, Y.; Hao, J.; Chen, S. Complete chloroplast genome sequence of Lirianthe coco (Loureiro) N. H. Xia & C. Y. Wu (Magnoliaceae), a popular ornamental species. Mitochondrial DNA B Resour. 2020. 5, 2410-2412. [CrossRef]
- Cai, Z.; Penaflor, C.; Kuehl, J.V.; Leebens-Mack, J.; Carlson, J.E.; dePamphilis, C.W.; Boore, J.L.; Jansen, R.K. Complete plastid genome sequences of Drimys, Liriodendron, and Piper: implications for the phylogenetic relationships of magnoliids. BMC Evol. Biol. 2006, 6, 77. [CrossRef]
- Xu, Q.; Li, Z; Wu, N.; Yang, J.; Yuan, L.; Zhao, T.; Sima, Y.; & Xu, T. Comparitive analysis of the chloroplast genomes of three Houpoea plants. Genes. 2023, 14, 1262. [CrossRef]
- Yang, T.; Aishan, S.; Zhu, J.; Qin, Y.; Liu, J.; Liu, H.; Tie, J.; Wang, J.; Qin, R. Chloroplast genomes and phylogenetic analysis of three Carthamus (Asteraceae) species. Int. J. Mol. Sci. 2023, 24, 15634. [CrossRef]
- Guo, Y.Y.; Yang, J.X.; Li, H.K.; Zhao, H.S. Chloroplast genomes of two species of Cypripedium: expanded genome size and proliferation of at-biased repeat sequences. Front Plant Sci. 2021, 12, 609729. [CrossRef]
- Lin, X.; Lee, S.Y.; Ni, J.; Zhang, X.; Hu, X; Zou, P.; Wang, W.; Liu, G. Comparative analyses of chloroplast genome provide effective molecular markers for species and cultivar identification in Bougainvillea. Int. J. Mol. Sci. 2023, 24, 15138. [CrossRef]
- Zhang, H.; Li, C.; Miao, H.;Xiong, S. Insights from the complete chloroplast genome into the evolution of Sesamum indicum L. PLoS One. 2013, 11, e80508. [CrossRef]
- China Plant BOL Group, Li, D.Z.; Gao, L.M.; Li, H.T.; Wang, H.; Ge, X.J.; Liu, J.Q.; Chen, Z.D.; Zhou, S.L.; Chen, S.L.; Yang, J.B.; Fu, C.X.; Zeng, C.X.; Yan, H F.; Zhu, Y.J.; Sun, Y.S.; Chen, S.Y.; Zhao, L.; Wang, K.; Yang, T.; Duan, G.W. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. USA. 2011, 108, 19641-19646. [CrossRef]
- Deng, Y.F. (2679) Proposal to Conserve the Name Talauma Fistulosa (Lirianthe Fistulosa, Magnolia Fistulosa) (Magnoliaceae) with a conserved type. Taxon, 2019, 68, 405-406. [CrossRef]
- Daniell, H.; Lin, C.S.; Yu, M.; & Chang, W.J. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [CrossRef]
- Wolf, P.G.; Roper, J.M.; & Duffy, A.M. The evolution of chloroplast genome structure in ferns. Genome, 2010, 53, 731-738. [CrossRef]
- Liang, A.; Luo, W.; Li, Z.; Sima, Y.; Xu, T. The complete chloroplast genome sequence of Magnolia delavayi (Magnoliaceae), a rare ornamental and medical tree endemic to China. Mitochondrial DNA B Resour. 2020, 5, 883-884. [CrossRef]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; dePamphilis, C.W.; Yi, T.S.; Li, D.Z. Getorganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [CrossRef]
- Shi, L.; Chen, H.; Jiang, M.; Wang, L.; Wu, X.; Huang, L.; Liu, C. Cpgavas2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019, 47, W65-W73. [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; Thierer, T.; Ashton, B.; Meintjes, P.; Drummond, A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 2012, 28, 1647-1649. [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W. X.; Wang, G.T. Phylosuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour.2020, 20, 348-355. [CrossRef]
- Tamura, K; Stecher, G.; Kumar, S. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022-3027. [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; & Miyata, T. MAFFT a novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 2002, 30, 3059-3066. [CrossRef]
- Rozas, J.; Rozas, R. DnaSP, DNA sequence polymorphism: an interactive program for estimating population genetics parameters from dna sequence data. Comput Appl. Biosci. 1995, 11, 621-625. [CrossRef]
- Amiryousefi, A.; Hyvönen, J.; & Poczai, P. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018, 34, 3030-3031. [CrossRef]
- Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014, 30, 1312-1313. [CrossRef]
| Species | L. brevisericea | L. coco | L. confusa | L. delavayi | L. henryi | L. hodgsonii | L. odoratissima | L. phanerophlebia |
|---|---|---|---|---|---|---|---|---|
| Name Abbr. | Lbre | Lcoc | Lcon | Ldel | Lhen | Lhod | Lodo | Lpha |
| Accession No. | OR680846 | MT225530 | MT654129 | MT654132 | OR680847 | MT560391 | MT654135 | OR680845 |
| Total Length (bp) | 159,811 | 159,828 | 159,833 | 159,714 | 159,760 | 159,693 | 159,819 | 159,548 |
| LSC (bp) | 87,933 | 87,958 | 87,965 | 87,877 | 87,891 | 87,846 | 87,961 | 87,671 |
| SSC (bp) | 18,758 | 18,760 | 18,752 | 18,761 | 18,757 | 18,745 | 18,772 | 18,761 |
| IR (bp) | 26,560 | 26,555 | 26,558 | 26,538 | 26,556 | 26,551 | 26,543 | 26,558 |
| Total Genes | 130 | |||||||
| CDS | 85 | |||||||
| tRNA | 37 | |||||||
| rRNA | 8 | |||||||
| Total GC% | 39.28 | 39.28 | 39.27 | 39.29 | 39.28 | 39.28 | 39.27 | 39.29 |
| LSC (GC%) | 37.98 | 37.97 | 37.97 | 37.97 | 37.97 | 37.97 | 37.97 | 37.98 |
| SSC (GC%) | 34.36 | 34.37 | 34.39 | 34.46 | 34.38 | 34.37 | 34.32 | 34.41 |
| IR (GC%) | 43.17 | 43.18 | 43.16 | 43.18 | 43.18 | 43.18 | 43.18 | 43.17 |
| A (%) | 47908 (29.98) | 47915 (29.98) | 47913 (29.98) | 47862 (29.97) | 47868 (29.96) | 47,851 (29.96) | 47908 (29.98) | 47799 (29.96) |
| C (%) | 31980 (20.01) | 31983 (20.01) | 31979 (20.01) | 31972 (20.02) | 31980 (20.02) | 31,958 (20.01) | 31977 (20.01) | 31940 (20.02) |
| G (%) | 30790 (19.27) | 30795 (19.27) | 30790 (19.26) | 30779 (19.27) | 30777 (19.26) | 30,774 (19.27) | 30782 (19.26) | 30741 (19.27) |
| T (%) | 49133 (30.74) | 49135 (30.74) | 49151 (30.75) | 49101 (30.74) | 49135 (30.76) | 49,110 (30.75) | 49152 (30.75) | 49068 (30.75) |
| Species | Lbre | Lcoc | Lcon | Ldel | Lhen | Lhod | Lodo | Lpha | |
| SSR | A | 17 | 13 | 13 | 17 | 15 | 16 | 16 | 17 |
| C | 2 | 2 | 3 | 3 | 3 | 3 | 2 | 3 | |
| G | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | |
| T | 21 | 23 | 22 | 25 | 24 | 24 | 21 | 24 | |
| TA | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | |
| TC | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |
| Total No. | 44 | 42 | 43 | 49 | 46 | 48 | 42 | 48 | |
| Tandem Repeats | 16 | 18 | 17 | 17 | 16 | 16 | 17 | 17 | |
| Dispersed Repeats | Complement | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Forward | 16 | 16 | 15 | 18 | 15 | 16 | 17 | 17 | |
| Palindromic | 27 | 25 | 26 | 25 | 26 | 26 | 25 | 25 | |
| Reverse | 7 | 9 | 9 | 7 | 8 | 8 | 8 | 8 | |
| Total No. | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | |
| Species | Leaves | Flowers | Fruits |
|---|---|---|---|
| L. brevisericea | Abaxially yellowish-gray sericeous; leaf blade midveins adaxially impressed, lateral veins 12–19 pairs; stipular scars reaching apex of petioles. | Erect; tepals 9, white; gynoecia densely yellowish-gray sericeous. | Mature carpels dehiscent along dorsal sutures |
| L. coco | Glabrous, leaf blade midveins adaxially impressed, lateral veins 8–16 pairs;; stipular scars reaching apex of petioles. | Pendulous; tepals 9, white; gynoecia glabrous. | Mature carpels dehiscent along dorsal sutures. |
| L. confusa | Abaxially yellowish-white curved trichomes; leaf blade midveins adaxially impressed, lateral veins 10–15 pairs;; stipular scars reaching apex of petioles. | Pendulous; tepals 9, white; gynoecia very densely yellowish-white villose. | Mature carpels dehiscent along dorsal sutures |
| L. delavayi | Abaxially densely interwoven tomentose and white powdery but later only with residual trichomes on veins; leaf blade midveins adaxially impressed, lateral veins 11–16 pairs; stipular scars reaching apex of petioles. | Erect; tepals 9 to 12, white, yellowish-white, pink or red; gynoecia fine yellow villose. | Mature carpels dehiscent along dorsal sutures. |
| L. henryi | Abaxially sparsely appressed pubescent; leaf blade midveins adaxially prominent, lateral veins 14–20 pairs; stipular scars reaching apex of petioles. | Pendulous; tepals 9, white; gynoecia glabrous. | Mature carpels dehiscent along dorsal sutures |
| L. hodgsonii | Glabrous; leaf blade midveins adaxially prominent, lateral veins 10–20 pairs; stipular scars reaching apex of petioles. | Erect; tepals 9, white; gynoecia glabrous. | Mature carpels circumscissile |
| L. odoratissima | Abaxially yellowish-white or grayish-brown curved trichomes; leaf blade midvein adaxially impressed, lateral veins 9–14 pairs; stipular scars reaching apex of petioles. | Pendulous; tepals 9 to 10, white; gynoecia densely grayish-brown pubescent. | Mature carpels dehiscent along dorsal sutures |
| L. phanerophlebia | Glabrous; leaf blade midveins adaxially impressed, lateral veins 11–17 pairs; stipular scars 1/3–1/2 as long as petioles. | Pendulous; tepals 8 to 9, white; gynoecia glabrous or glaucous. | Mature carpels dehiscent along dorsal sutures |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





