1. Introduction
Antarctica provides a unique natural laboratory to investigate the evolutionary processes behind environmental adaptation. Antarctic microorganisms have developed extraordinary survival strategies, including the ability to resist cold temperatures, oxidative stress, and UV radiation, to scavenge iron present in limited concentrations, and to detoxify hazardous compounds, such as heavy metals and pollutants [
1]. They have also shown the ability to produce different pigments and biopolymers in response to environmental stress factors [
1]. Cellulose forms the basic structural foundation of the primary cell wall of green plants, algae, and fungi [
2] and it is the main constituent of natural fibers such as cotton [
3]. It is a polysaccharide consisting of a linear chain of several β (1→4) linked D-glucose units and represents the most abundant organic polymer on Earth [
3]. Cellulose is also present in bacteria, the so-called nanocellulose [
4,
5]. The biosynthesis of Bacterial cellulose (BC) has been observed many years ago by ancient Chinese growing the Kombucha tea mushroom, a syntrophic colony of acetic acid bacteria and yeast, which metabolizes sugar to produce a slightly acidic tea-colored drink and forms a thick cellulosic mat at its surface [
6]. BC was then reported by Brown in 1886, who identified the growth of a non-branched pellicle with a structure chemically equivalent to that of plant cellulose [
7].
In recent year, BC has been evaluated as a promising polymer for biotechnological application. For example, can be used in the food industry, serving as a novel biological material and edible packaging [
8]. In the medical field, BC finds use as a wound dressing material, artificial skin, vascular grafts, scaffolds for tissue engineering, artificial blood vessels, wound pads, and dental implants. Furthermore, BC has industrial applications, such as acting as sponges to collect leaking oil and as materials for absorbing toxins [
9]. In both plants and bacteria, cellulose is synthesized by cellulose synthase enzymes (CesAs). This complex varies considerably by kingdom; however, it shares a conserved catalytic subunit termed BcsA (bacterial cellulose synthase subunit A) in prokaryotes and CesA in eukaryotes [
10]. Plants' CesA derived from the bacterial cellulose synthase upon the endosymbiosis event that led to the formation of chloroplasts [
11]. Very few bacterial species can synthesize BC, and they include
Komagataeibacter xylinus (previously known as
Gluconacetobacter xylinus), some
Agrobacterium spp.,
Azotobacter,
Rhizobium spp.,
Sarcina,
Alcaligenes, and
Pseudomonas genera [
4].
K. xylinus is the most studied and the most efficient BC producer [
12]. It is an aerobic gram-negative bacterium with fermentation activity in the pH range of 3-7 and a growth temperature range of 25-30°C, using saccharides as a carbon source [
13].
K. xylinus can use various sugars and produces relatively high yields of cellulose in liquid medium [
14,
15].
In the present study, we report BC biosynthesis from a marine strain isolated from a bacterial consortium associated with the Antarctic ciliate
Euplotes focardii [
16,
17] and named
Pseudomonas sp. ef1.
E. focardii is a free-swimming ciliate, endemic to the oligotrophic coastal sediments of Terra Nova Bay, and is classified as an obligatory psychrophilic stenothermal organism [
18,
19,
20,
21].
Pseudomonas sp. ef1 was previously demonstrated to be able to transform heavy metals such as copper, nickel, and silver into nanoparticles showing antibiotic activity [
22,
23,
24], and to produce unique pyoverdins [
25]. In this work, we tested
Pseudomonas sp. ef1 for BC production. We found that this strain produces BC using synthesis media conditions that are unique. BC was characterized through chemical analyses and compared with that obtained from the most efficient BC producer
K. xylinus.
Pseudomonas BC was synthesized with different shapes and structural characteristics as visualized under SEM analysis. Additionally, a putative
Pseudomonas sp. ef1 cellulose synthase was identified and characterized.
2. Results
2.1. BC Production
We evaluated the capacity of
Pseudomonas sp. ef1 to synthesize BC through fermentation in standard HS medium containing 1.5% glucose, which is commonly utilized for
K. xylinus, the most well-known BC producer. BC production by
Pseudomonas sp. ef1 was assessed in static and shaking conditions. Under static conditions, the BC produced by
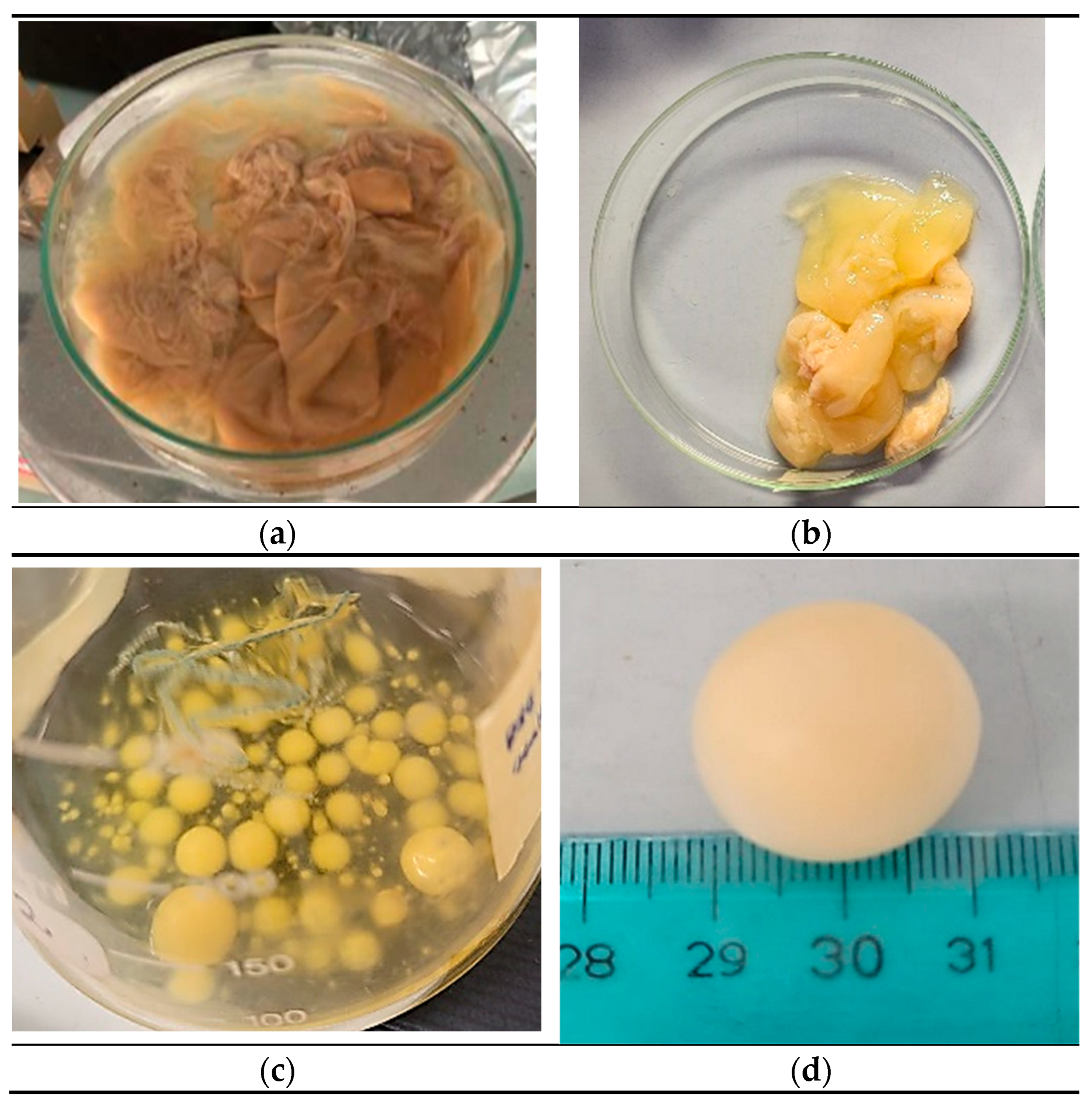
Pseudomonas sp. ef1 exhibited a sheet-like form and was more dispersed in water (
Figure 1A), differing from the BC produced by
K. xylinus, which had a more gelatinous appearance (
Figure 1B).
Pseudomonas sp. ef1 can generate BC at a pH 6.5 and temperatures ranging from 22 to 24°C. In contrast, the optimal conditions for BC production in
K. xylinus are a pH 6 and temperatures between 28 and 30°C. No BC production was detected under shaking conditions.
Since
Pseudomonas sp. ef1 is a marine bacterium, we also examined BC production in artificial seawater supplemented with 1.5% glucose at 22-24°C, both in static and shaking conditions. We started from cultures grown in yeast extract (1%) or nutrient broth liquid medium (as described under Material and Methods). After 5-6 days of incubation under shaking at 100 rpm, we observed the formation of spherical flocculates (
Figure 1C and 1D), with sizes ranging from 3 to 20 mm in diameter. Under these media conditions, BC production under static incubation was not observed. The water content was estimated to be 79%.
2.2. Fourier-Transform Infrared (FTIR) Spectroscopic Characterization of BC
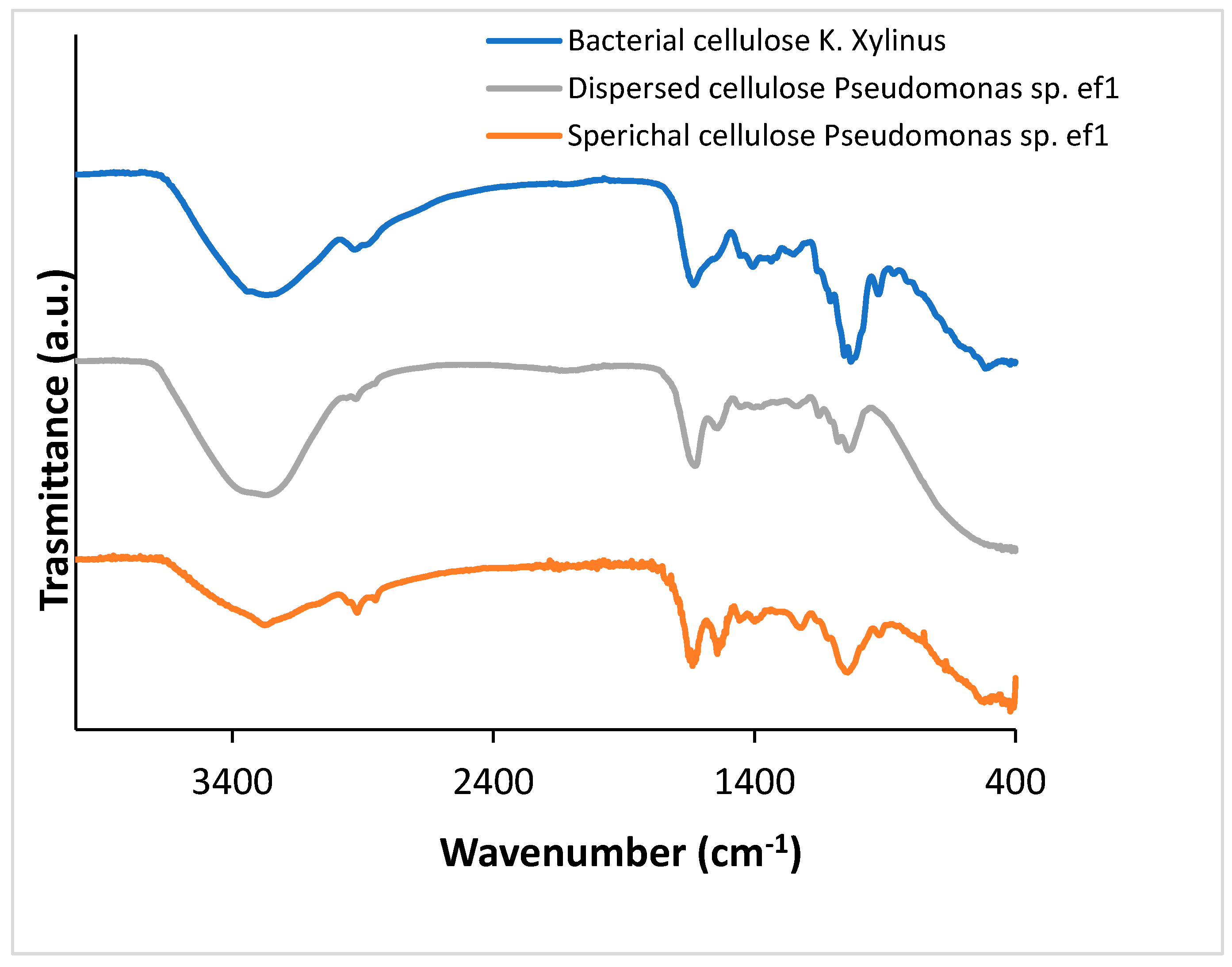
To confirm that the obtained products correspond to BC, Fourier-Transform Infrared Spectroscopy (FTIR) analysis was performed. FTIR spectra were acquired from solid samples after drying both the spherical BC (
Figure 2, gray line) and the dispersed sheet-like BC (
Figure 2, orange line) and compared to cellulose produced by
K.
xylinus (
Figure 2, blue line). Both cellulose types, spherical and dispersed, exhibited IR spectra comparable to those of K.
xylinus and standard plant-derived cellulose, as reported by [
26] Abderrahim (2015). The broad adsorption band at 3330 cm cm⁻¹ is attributed to the -OH stretching, the vibrations in the range 2850-2940 cm⁻¹ are relative to the C-H stretching in the bacterial cellulose structure. The peak at around 1600 cm⁻¹ can be attributed to the residual water, present in the cellulose network. Several bands have been detected at 1460, 1390, 1320 and 930 cm⁻¹ attributed to C−H stretching of CH
2 and CH
3 groups, that could indicate the possibility of a methylated cellulose. Furthermore, a strong band is visible at around 1050 cm⁻¹ that corresponds to C-O-C and C-O-H vibrations [
27,
28,
29].
2.3. Scanning Electron Microscope (SEM) Analysis of Bacterial Cellulose
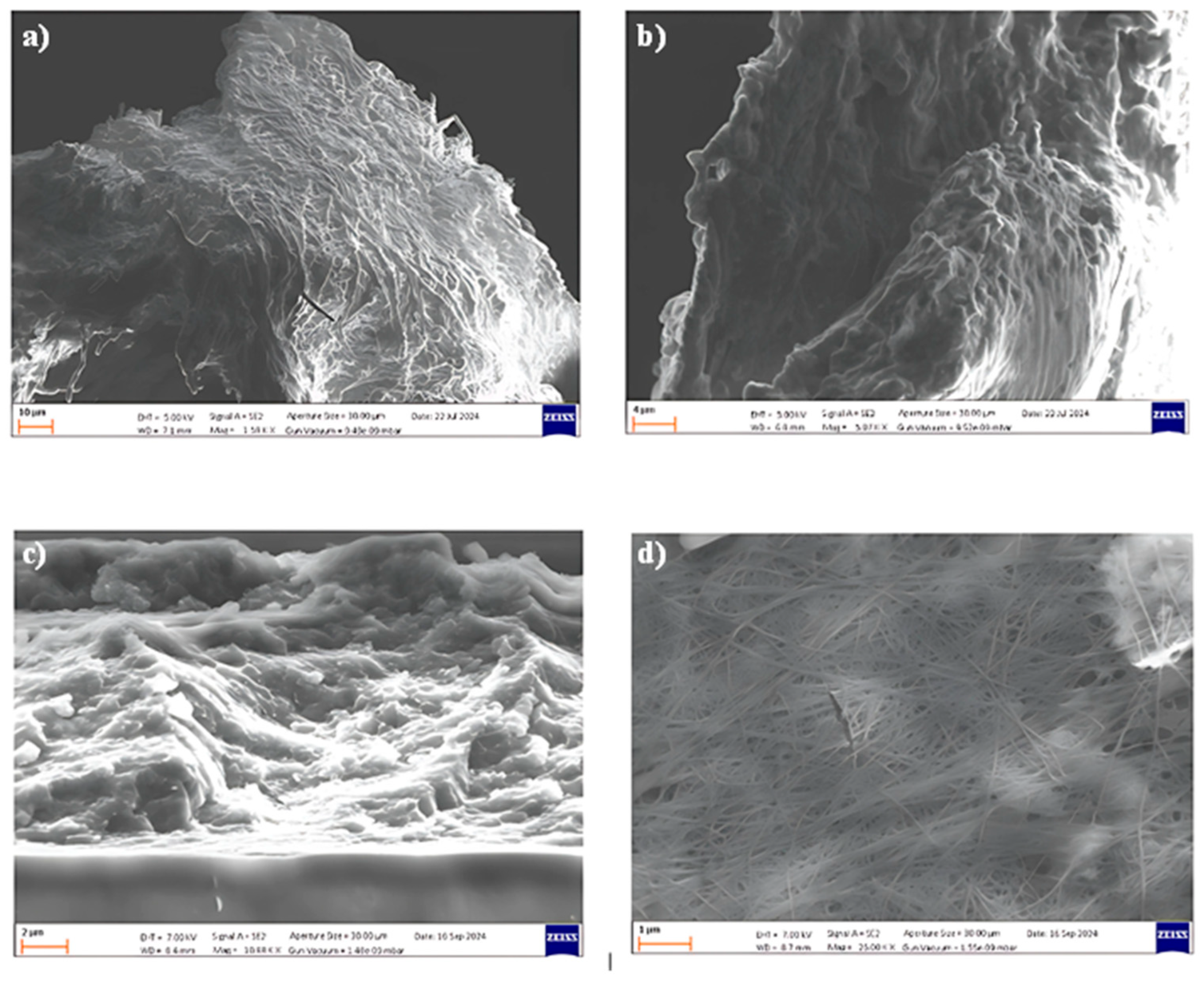
The morphologies of the BC produced by
Pseudomonas sp. ef1 and
K. xylinus samples were evaluated by FE-SEM measurements (
Figure 3).
SEM images reveal that the spherical BC produced by
Pseudomonas sp. ef1 (
Figure 3 A and B) consists of filamentous structures with diameters below 1 µm, similar those observed in BC synthesized by
K. xylinus (Figure3 D). However, the fibrous network of
Pseudomonas sp. ef1 appears less uniform and less distinctly organized compared to the well-defined and homogeneous architecture of
K. xylinus-derived BC.
In contrast, the sheet-like BC morphology observed in
Pseudomonas sp. ef1 (
Figure 3 C) presents a disordered and loosely connected structure. This irregular arrangement may contribute to its enhanced dispersibility in aqueous environments, suggesting potential advantages for applications requiring high water solubility or colloidal stability.
2.4. Identification of the Cellulose Synthase and Structural Prediction
To identify the enzyme(s) involved in
Pseudomonas sp. ef1 BC synthesis, we conducted a TBLASTN search in the corresponding genome using the BC synthase operon protein sequences from
K. xylinus as the query. The accession numbers of the query sequences are listed in
Table S1. Only the
K. xylinus cellulose synthase catalytic subunit (acc. # AHI24410.1), putative cellulose synthase 2 (acc. # AHI24410.1), and cellulose synthase 2 (AHI26282.1) showed a confident match with a single sequence in the
Pseudomonas sp. ef1 genome in TBLASTN result that correspond to the catalytic subunit A. The other sequences from the
K. xylinus operon did not show any confident matches. These findings are summarized in
Table S1. Therefore, we focussed our attention on the characterization of this sequence.
Blast search analysis revealed that the identified protein is composed by two different domains: the Scw11 superfamily that includes the Exo-beta-1,3-glucanase family, and the BcsA superfamily, that includes cellulose synthase catalytic subunit A (
Figure S1). Therefore, we named the protein as putative cellulose synthase catalytic subunit A (hereafter called pBCSA) from
Pseudomonas sp. ef1.
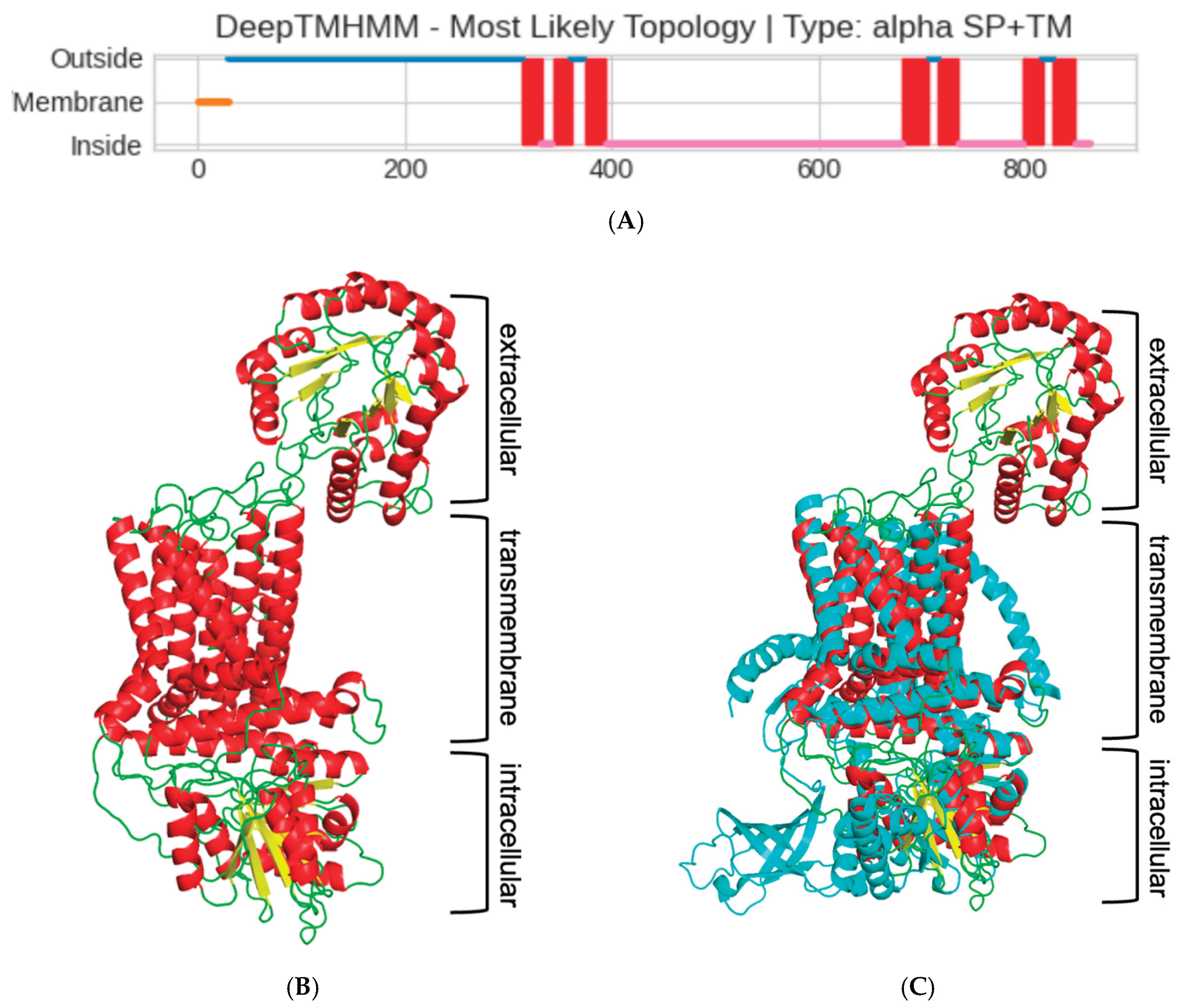
Since the cellulose synthase is known to be a transmembrane (TM) protein, the next step in characterizing the
Pseudomonas sp. ef1 pBCSA involved predicting the topology of the TM regions. This prediction was carried out using DeepTMHMM, a deep learning model designed to calculate the likelihood of each residue being part of the extracellular, transmembrane, or intracellular region [
30]. According to the prediction, a long extracellular domain spanning the first 300 residues (blue line, Fig 4A), three TM-helixes spanning residues from 300 to 400 (red squares in Fig 4A), a long intracellular domain (pink line in Fig 4A), and additional four TM-helixes at the C-terminal domain were identified.
The prediction of the long extracellular domain in the N-terminus was unexpected. To confirm the presence of this domian, we proceeded with the prediction of the 3D structure using RoseTTaFold deep learning tool. Given the constraints on residue length for the protein 3D modelling capabilities of this tool, the
Pseudomonas sp. ef1 pBCSA amino acid sequence was segmented into three distinct overlapping sections based on transmembrane region predictions. These segments are defined as residues 1-350, 300-490, and 420-860, corresponding respectively to the extracellular domain, the initial three transmembrane helices, and the intracellular domain linked to the final four transmembrane helices. The final 3D model is reported in
Figure 4B: the modelling confirmed the presence of the additional extracellular domain, not usually present in most of the BCS subunit A. The presence of this additional domain is even more evident in the superposition of the model with cellulose synthase A subunit of
Cereibacter sphaeroides [
31], that has been used as model template (5EJZ(MMDB) in iCn3D), reported in
Figure 4C.
Some strains of K. xylinus possess operons which encodes a single long BcsAB fusion protein. By contrast, the extracellular domain of the putative Pseudomonas sp. ef1 pBCSA corresponds to the Exo-beta-1,3-glucanase family. These enzymes are known to play a key role in the degradation of beta-1,3-glucans, which are polysaccharides found in the cell walls of fungi, some bacteria, and plants. However, these proteins have also a role in biofilm formation and modification (see discussion).
3. Discussion
Cellulose is the key component of plant cell walls and the most abundant biopolymer on Earth [
32]. While most cellulose is being produced by plant cellulose synthase complexes, this enzyme clearly has bacterial origin: there is no doubt that its genes have been acquired by plants from cyanobacterial ancestors of their chloroplasts [
33]. We isolated a marine Antarctic
Pseudomonas strain able to produce BC from glucose, in energy safe conditions. The produced BC possesses different morphology, according to the protocol used. In HS medium, the produced BC appears as sheet like product, whereas in medium containing either yeast extract or nutrient broth and sea water under shaking conditions, the product appears a spherical flocculates.
K. xylinus cultures grown in liquid media are remarkably efficient at producing a surface pellicle composed entirely of pure cellulose fibers [
4]. The cellulose biosynthesis process in this bacterium is controlled by a four-gene
bcsABCD operon. Among the corresponding proteins, BcsA and BcsB are essential for in vitro cellulose-synthesizing (BCS) activity. However, all four proteins—BcsA, BcsB, BcsC, and BcsD—are necessary to achieve optimal cellulose production in vivo. This suggests that BcsC and BcsD play critical roles in exporting glucan chains and organizing them into fibers at the cell surface. Certain strains of
K. xylinus also possess a second
bcs operon, which encodes a single, elongated BcsAB fusion protein, along with two additional genes,
bcsX and
bcsY, whose functions remain uncharacterized [
34].
Genomic data revealed unexpected diversity of cellulose synthase operons even in closely related bacteria, indicating substantial differences in cellulose secretion mechanisms [34. In Pseudomonas sp. ef1 we identified a putative cellulose synthase subunit A that possesses an extracellular domain represented by a member of the Exo-beta-1,3-glucanase family, differently from some K. xylinus possess strains that possess a single long BcsAB fusion protein. Exo-beta-1,3-glucanase family are enzymes that play a key role in the degradation of beta-1,3-glucans, which are polysaccharides found in the cell walls of fungi, some bacteria, and plants. However, these proteins have additional biological roles, including antifungal defense mechanisms by degrading fungal cell walls, or in biofilm formation and modulation. The unusual structure of the putative BCS A subunit may account for the sheet like and water-soluble cellulose organization that is obtained by incubating Pseudomonas sp. ef1in HS medium at pH 6.5. This BCS A subunit structure is shared also by other cellulose producing Pseudomonas strains. However, these strains also possess a standard operon organization. By contrast, Pseudomonas sp. ef1 appears to have lost the standard operon organization, likely following its adaptation to the Antarctic host environment. The water-soluble form of the Pseudomonas ef1 BC makes it particularly well-suited for coating applications, especially in food packaging, as it eliminates the need for additional processing steps—such as homogenization—before spreading it onto other materials.
Bacterial synthesis of cellulose is seen as a convenient and effective way to produce stable recyclable fibers for use in wound dressing and in a variety of emerging nanotechnologies. Furthermore, BC has industrial applications, such as acting as sponges to collect leaking oil and as materials for absorbing toxins [
35]. Exploring new methods for cellulose synthesis, beyond traditional vegetable sources, will aid in the development of innovative and renewable materials.
4. Materials and Methods
4.1. Strains Culturing and Genome Sequencing
DNA used for sequencing was extracted from a culture of Pseudomonas sp. ef1 grown overnight in 10 mL of LB (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl) at 23°C under shaking. Cells were harvested by centrifugation and DNA was extracted by using the ‘DNeasy PowerSoil Pro Kit’ (Qiagen), following the protocol provided by the manufacturer. Genomic DNA of Pseudomonas sp. ef1 was sequenced with nanopore technology and using a native barcoding approach (since genomic DNA of Pseudomonas sp. ef1 was sequenced with other non-related DNA samples). Sequencing was carried out using a MinION Mk1B and a R9.4 flow cell (Oxford Nanopore Technology, ONT). Basecalling was performed with MinKNOW (v24.02.16) using the super accurate model. Sequencing reads were assembled using EPI2ME (v5.1.14) and the workflow ‘wf-bacterial-genomes’ (v1.2.0) provided by ONT.
4.2. BC Production and Purification
BC production was conducted in Hestrin-Schramm (HS) medium (20 g/L glucose, 5 g/L yeast extract, 5 g/L peptone, 2.7 g/L disodium hydrogen phosphate, 1.15 g/L citric acid) at pH 6.5, under both static and shaking conditions for 3-5 day, at temperatures of 22-24°C. Alternatively, Pseudomonas sp. ef1 BC was produced in artificial seawater (22 ‰) supplemented with 1.5% (w/v) glucose at 22-24°C, both in static and shaking conditions, starting from cultures grown in yeast extract (1% w/v) or nutrient broth liquid medium. K. xylinus BC production was conducted in HS medium at pH 6 and temperatures of 28-30°C. The inoculum consisted of Pseudomonas sp. ef1 cells obtained from LB agar plates. The resulting biofilm was collected using a filter, washed multiple times with sterile deionized water, and incubated at 80°C for 4 hours while soaked in sterile deionized water to completely clean BC from bacteria.
4.3. BC Characterization
Functional groups identification of the bacterial cellulose material was characterized by FT-IR analysis using a Perkin-Elmer System 2000 spectrometer (Waltham, MA, USA equipped with Pike GladiATR technology. Prior to the analysis, the samples were dried in an oven at 40 °C.
Surface morphology and microstructural features of the dried bacterial cellulose samples, coated with a thin layer of chromium, were examined using a Sigma FE-SEM (Zeiss, Germany), operated at 5–15 kV.
4.4. Identification of the Cellulose Synthase Enzymes, Transmembrane (TM) Regions Prediction and Homology Modeling
Pseudomonas sp. ef1 putative cellulose synthase was identified by local TBLASTN search in the corresponding genome using the BC synthase operon protein sequences from
K. xylinus as the query. The accession numbers of the query sequences are listed in
Table S1. The prediction of transmembrane (TM) proteins was conducted using DeepTMHMM, a tool that utilizes deep neural networks [
36]. To obtain the structural model of cellulose synthase, the corresponding sequence was divided into three regions corresponding to three structural domains: the extracellular (residues 1 to 350), transmembrane (residues 300 to 490), and the intracellular domain (residues 420 to 860). A model for each domain was obtained using RoseTTaFold, the deep learning tool of Rosetta [
37]. The resulting models were manually overlapped using PyMol 3.1 (PyMOL | pymol.org).
5. Patents
The manuscript is related to patents: n 102020000031769, 102022000018663, n 102021000017333.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Figure S1: Blast search result using the putative cellulose synthase A from Pseudomonas sp. Ef1.as query.; Table S1: tBlastN results of the Pseudomonas sp. Ef1 genome analysis to serach for BC synthesis genes using as quary the BC synthesis operon sequence from Komagataeibacter xylinus E25 strain
Author Contributions
For research articles with several authors, a short paragraph specifying their individual contributions must be provided. The following statements should be used “Conceptualization, S.P, M.Z. and R.T.; methodology, M.C.B, M.D.S, M.Z, A.V. and F.C..; validation, S.P, M.Z., A.V. and R.T.; formal analysis, M.C.B, M.D.S, M.Z, and F.C.; data curation, S.P, M.Z. and R.T.; writing—original draft preparation, S.P..; writing—review and editing, all authors.; funding acquisition, S.P. and R.G.. All authors have read and agreed to the published version of the manuscript.”
Funding
Please add: This research was funded by S.P. and R.G., by Fondi di Ricerca di Ateneo, (FAR).
Data Availability Statement
Pseudomonas sp ef1 is available at GCA_007293365.1
Acknowledgments
This paper and related research have been conducted during and with the support of the Italian national inter-university PhD course in Sustainable Development and Climate change (link:
www.phd-sdc.it)
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Ramasamy, K.P.; Mahawar, L.; Rajasabapathy, R.; Rajeshwari, K.; Miceli, C.; Pucciarelli, S. Comprehensive insights on environmental adaptation strategies in Antarctic bacteria and biotechnological applications of cold adapted molecules. Front Microbiol. 2023, 14, 1197797. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Brigham, C. Biopolymers: biodegradable alternatives to traditional plastics. In Green Chemistry: an Inclusive Approach, Török, B., Dransfield, T., Eds.; Elsevier Inc., Amsterdam, 2018; 753–770. [CrossRef]
- Felgueiras, C.; Azoia, N.G.; Gonçalves, C.; Gama, M. Dourado, F. Trends on the Cellulose-Based Textiles: Raw Materials and Technologies. Front. Bioeng. Biotechnol 2021, 9, 608826. [Google Scholar] [CrossRef]
- Lahiri, D.; Nag, M.; Dutta, B.; Dey, A.; Sarkar, T.; Pati, S.; Edinur, H.A.; Abdul Kari, Z.; Mohd Noor, N.H.; Ray, R.R. Bacterial Cellulose: Production, Characterization, and Application as Antimicrobial Agent. Int J Mol Sci. 2021, 22, 12984. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Krasteva, P.V.; Bernal-Bayard, J.; Travier, L.; et al. Insights into the structure and assembly of a bacterial cellulose secretion system. Nat. Commun. 2017, 8, 2065. [Google Scholar] [CrossRef] [PubMed]
- Marsh, A.J.; et al. Sequence-based analysis of the bacterial and fungal compositions of multiple Kombucha (tea fungus) samples. Food microbiology. 2014, 38, 171–178. [Google Scholar] [CrossRef]
- Poddar, M.K.; Dikshit, P.K. Recent development in bacterial cellulose production and synthesis of cellulose based conductive polymer nanocomposites. Nano Select. 2021, 2, 1605–1628. [Google Scholar] [CrossRef]
- Sayah, I.; Gervasi, C.; Achour, S.; Gervasi, T. Fermentation Techniques and Biotechnological Applications of Modified Bacterial Cellulose: An Up-to-Date Overview. Fermentation 2024, 10, 100. [Google Scholar] [CrossRef]
- Wang, J.; Tavakoli, J.; Tang, Y. Bacterial Cellulose Production, Properties and Applications with Different Culture Methods—A Review. Carbohydr. Polym. 2019, 219, 63–76. [Google Scholar] [CrossRef]
- McNamara, J.T.; Morgan, J.L.; Zimmer, J.A. Molecular description of cellulose biosynthesis. Annu Rev Biochem. 2015, 84, 895–921. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Little, A.; Schwerdt, J.G.; Shirley, N.J.; Khor, S.F.; Neumann, K.; O'Donovan, L.A.; Lahnstein, J.; Collins, H.M.; Henderson, M.; Fincher, G.B.; Burton, R.A. Revised Phylogeny of the Cellulose Synthase Gene Superfamily: Insights into Cell Wall Evolution. Plant Physiol. 2018, 177, 1124–1141. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- El-Saied, H.; Basta, A.H.; Gobran, R.H. Research Progress in Friendly Environmental Technology for the Production of Cellulose Products (Bacterial Cellulose and Its Application). Polym Plast Technol Eng. 2004, 43, 797–820. [Google Scholar] [CrossRef]
- Castro, C; Zuluaga, R.; Putaux, J.-L.; Caro, G.; Mondragon, I.; Gañán, P. Structural characterization of bacterial cellulose produced by Gluconacetobacter swingsii sp. from Colombian agroindustrial wastes. Carbohydr. Polym 2011, 84, 96–102. [Google Scholar] [CrossRef]
- Ross, P.; Mayer, R.; Benziman, M. Cellulose biosynthesis and function in bacteria. Microbiol Rev. 1991, 55, 35–58. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sani, A.; Dahman, Y. Improvements in the production of bacterial synthesized biocellulose nanofibres using different culture methods. J. Chem. Technol. Biotechnol. 2010, 85, 151–164. [Google Scholar] [CrossRef]
- Pucciarelli, S.; Devaraj, R.R.; Mancini, A.; Ballarini, P.; Castelli, M.; Schrallhammer, M.; Petroni, G.; Miceli, C. Microbial consortium associated with the Antarctic marine ciliate Euplotes focardii: An investigation from genomic sequences. Microb. Ecol. 2015, 70, 484–497. [Google Scholar] [CrossRef]
- Ramasamy, K.P.; Telatin, A.; Mozzicafreddo, M.; Miceli, C.; Pucciarelli, S. Draft genome sequence of a new pseudomonas sp. Strain, ef1, Associated with the psychrophilic Antarctic ciliate Euplotes focardii. Microbiol. Resour. Announ. 2019, 8, e00867-19. [Google Scholar] [CrossRef]
- Yang, G.; De Santi, C.; de Pascale, D.; Pucciarelli, S.; Miceli, C. Characterization of the First Eukaryotic Cold-Adapted Patatin-Like Phospholipase from the Psychrophilic Euplotes focardii: Identification of Putative Determinants of Thermal-Adaptation by Comparison with the Homologous Protein from the Mesophilic Euplotes crassus. Biochimie 2013, 95, 1795–1806. [Google Scholar]
- Mozzicafreddo, M.; Pucciarelli, S.; Swart, E.C.; Piersanti, A.; Emmerich, C.; Migliorelli, G.; Ballarini, P.; Miceli, C. The macronuclear genome of the Antarctic psychrophilic marine ciliate Euplotes focardii reveals new insights on molecular cold adaptation. Sci Rep. 2021, 11, 18782. [Google Scholar] [CrossRef]
- Pischedda, A.; et al. Antarctic marine ciliates under stress: superoxide dismutases from the psychrophilic Euplotes focardii are cold-active yet heat tolerant enzymes. Sci Rep 2018, 8, 14721. [Google Scholar] [CrossRef]
- Pucciarelli, S.; La Terza, A.; Ballarini, P.; Barchetta, S.; Yu, T.; Marziale, F.; Passini, V.; Methé,, B.; Detrich HW,, I.I.I.; Miceli, C. Molecular cold-adaptation of protein function and gene regulation: The case for comparative genomic analyses in marine ciliated protozoa. Mar. Genom. 2009, 2, 57–66.
- John, M.S.; Nagoth, J.A.; Zannotti, M.; Giovannetti, R.; Mancini, A.; Ramasamy, K.P.; Miceli, C.; Pucciarelli, S. Biogenic Synthesis of Copper Nanoparticles Using Bacterial Strains Isolated from an Antarctic Consortium Associated to a Psychrophilic Marine Ciliate: Characterization and Potential Application as Antimicrobial Agents. Mar Drugs. 2021, 19, 263. [Google Scholar] [CrossRef]
- John, M.S.; Nagoth, J.A.; Ramasamy, K.P.; Mancini, A.; Giuli, G.; Miceli, C.; Pucciarelli, S. Synthesis of Bioactive Silver Nanoparticles Using New Bacterial Strains from an Antarctic Consortium. Marine Drugs 2022, 20, 558. [Google Scholar] [CrossRef]
- Nagoth, J.A.; John, M.S.; Ramasamy, K.P.; Mancini, A.; Zannotti, M.; Piras, S.; Giovannetti, R.; Rathnam, L.; Miceli, C.; Biondini, M.C.; Pucciarelli, S. Synthesis of Bioactive Nickel Nanoparticles Using Bacterial Strains from an Antarctic Consortium. Mar. Drugs 2024, 22, 89. [Google Scholar] [CrossRef] [PubMed]
- Zannotti, M.; Di Sessa, M.; Biondini, M. C., Vassallo, A., Ferraro, S., Angeloni, S., Ricciutelli, M., Pucciarelli, S., Giovannetti, R., Towards an easy production of novel pyoverdines by an antarctic Pseudomonas strain: a spectroscopic and HPLC-MS/MS characterization study. Dyes Pigm. 2026, 244, 113096. [CrossRef]
- Abderrahim, B. , Abderrahman, E. , Mohamed, A. , Fatima, T. , Abdesselam, T. , & Krim, O. Kinetic Thermal Degradation of Cellulose, Polybutylene Succinate and a Green Composite: Comparative Study. World Journal of Environmental Engineering 2015, 3, 95–110.
- Oliveira, R.L.; Vieira, J.G.; Barud, H.S.; Assunção, R.M.N.; Filho, G.R.; Ribeiro, S.J.L.; Messadeqq, Y. Synthesis and Characterization of Methylcellulose Produced from Bacterial Cellulose under Heterogeneous Condition, J. Braz. Chem. Soc. 2015, 26. [Google Scholar] [CrossRef]
- Yim, S.M.; Song, J.E.; Kim, H.R. Production and characterization of bacterial cellulose fabrics by nitrogen sources of tea and carbon sources of sugar, Process Biochem. 2017, 59, Part A, 26-36, ISSN 1359-5113. [CrossRef]
- Fatima, A.; Ortiz-Albo, P.; Neves, L.A.; Nascimento, F.X.; Crespo, J.G. Biosynthesis and characterization of bacterial cellulose membranes presenting relevant characteristics for air/gas filtration, J. Membr. Sci. 2023, 674, 121509. [Google Scholar] [CrossRef]
- Jeppe Hallgren Konstantinos, D. Tsirigos, Mads D. Pedersen, José Juan Almagro Armenteros, Paolo Marcatili, Henrik Nielsen, Anders Krogh and Ole Winther (2022).
- Morgan, J.; McNamara, J. & Zimmer, J. Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat Struct Mol Biol 2014, 21, 489–496. [Google Scholar] [CrossRef]
- Baghaei, B.; Skrifvars, M. All-Cellulose Composites: A Review of Recent Studies on Structure, Properties and Applications. Molecules 2020, 25, 2836. [Google Scholar] [CrossRef]
- Nobles, D.R.; Brown, R.M. Jr. The pivotal role of cyanobacteria in the evolution of cellulose synthases and cellulose synthase-like proteins. Cellulose. 2004, 11, 437–448. [Google Scholar] [CrossRef]
- Römling, U.; Galperin, M.Y. Bacterial cellulose biosynthesis: diversity of operons, subunits, products and functions. Environ. Microbiol. 2015, 17, 4107–4121. [Google Scholar] [CrossRef]
- Wang, J.; Tavakoli, J.; Tang, Y. Bacterial Cellulose Production, Properties and Applications with Different Culture Methods—A Review. Carbohydr. Polym. 2019, 219, 63–76. [Google Scholar] [CrossRef]
- Hallgren, J.; Tsirigos, K.D.; Pedersen, M.D.; Armenteros, J.J.A.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks bioRxiv 2022. [CrossRef]
- Baek, M.; Di Maio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; Millán, C.; Park, H.; Adams, C.; Glassman, C.R.; De Giovanni, A.; Pereira, J.H.; Rodrigues, A.V.; van Dijk, A.A.; Ebrecht, A.C.; Opperman, D.J.; Sagmeister, T.; Buhlheller, C.; PavkovKeller, T.; Rathinaswamy, M.K.; Dalwadi, U.; Yip, C.K.; Burke, J.B.; Garcia, K.C.; Grishin, N.V.; Adams, P.D.; Read, R.J.; Baker, D. Accurate prediction of protein structures and interactions using a 3-track network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).