1. Introduction
1.1. Research Objective
The nose is a paramount aesthetic feature of the human face, with nasal bone fractures ranking as the most prevalent bone injury in adults and the third most common fracture across the entire body [
1]. Approximately 40% of facial trauma incidents involve fractures of the nasal bones, accounting for approximately 50,000 cases annually in the United States alone [
1,
2]. Despite the perception that nasal bone fractures are minor injuries, prompt and precise diagnosis is crucial [
3]. Delayed treatment may result in functional and cosmetic complications, exacerbating traumatic edema, preexisting nasal deformities, and occult septal injuries [
4,
5]. Consequently, promptly and swiftly achieving an accurate diagnosis determines the need for surgical intervention, which plays a pivotal role in ensuring favorable patient outcomes [
3,
4].
Our study introduces a predictive model based on deep learning techniques, aiming to provide healthcare practitioners with a diagnostic tool that enhances the efficiency and effectiveness of managing patients with nasal trauma visiting the emergency department. Moreover, our model sought to expedite the determination of surgical indications and streamline the decision-making process within a compressed timeframe.
1.2. Research Scope and Methods
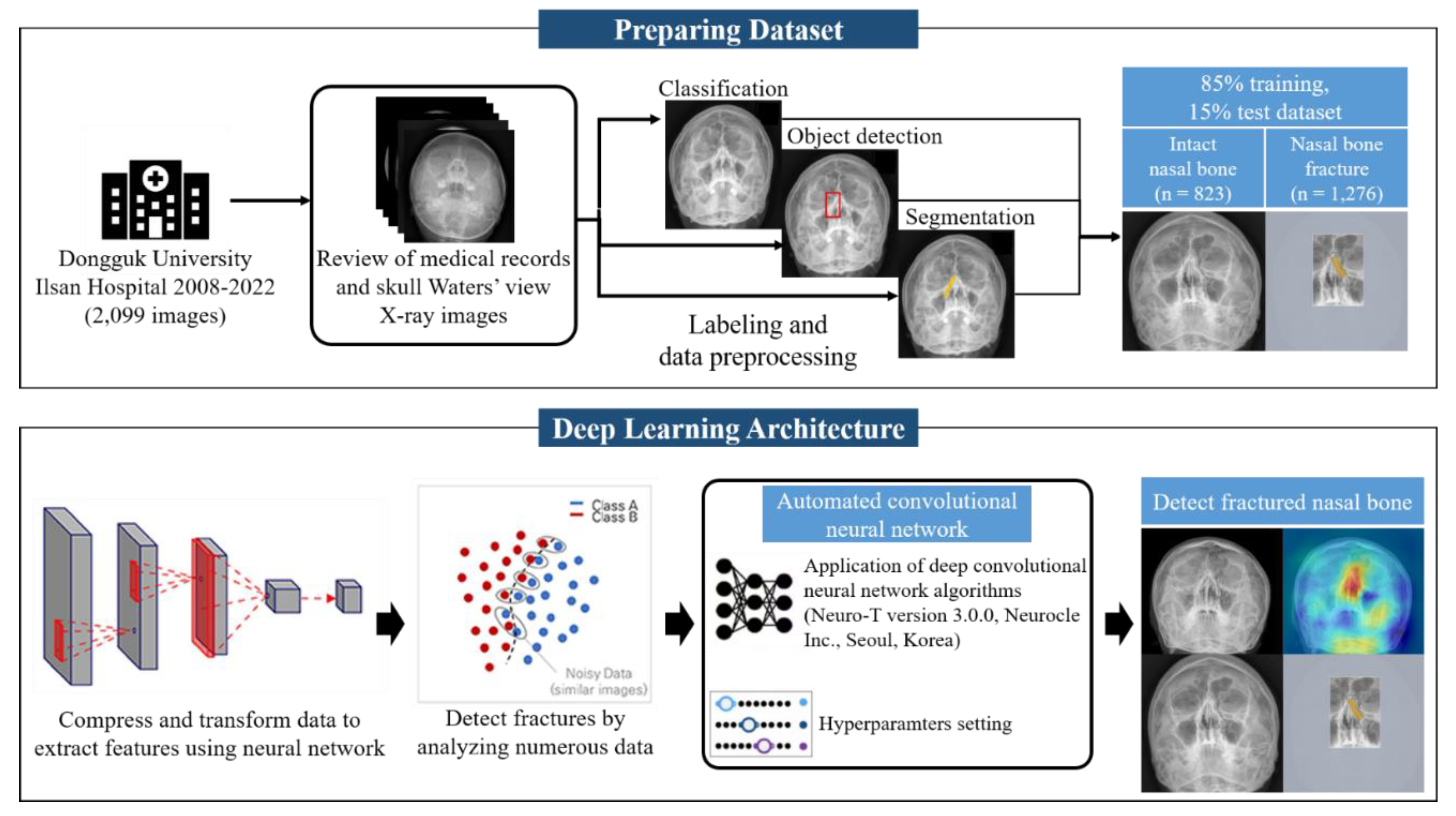
We performed a retrospective review of all patients with facial trauma who underwent occipitomental radiography (Waters’ view) at their initial visit to the emergency room between March 2008 and July 2022.
1.3. Research Scope
A dataset consisting of 2,579 skull Waters view radiographs was collected with the approval of the individual institutional review board (IRB no. DUIH 2023-05-017). A total of 2,099 radiographs were used in this study. Patients treated surgically for nasal bone fractures at the Department of Plastic and Reconstructive Surgery of Dongguk University Ilsan Hospital were included. We excluded 480 radiographs due to issues including an inclination of the vertical axis over 5°and obvious artifacts interfering with the visualization of the nasal bone, such as metal wires of masks or dental braces, were excluded. The total number of images comprised 823 normal and 1,276 fracture radiographs. The training, validation, and test cohorts consisted of 2099, 730, and 315 patients, respectively (
Figure 1).
1.4. Patients and Methods
We identified the surgical indication factors, including displacement angle, size of the bony gap, thickness of soft tissue swelling, septal hematoma, and existence of subcutaneous emphysema. Two experienced clinicians determined the presence or absence of nasal bone fractures. The patients who underwent closed reduction were included in the fracture group. An AI algorithm based on deep learning was engineered, trained, and validated to detect fractures in radiographs.
1.5. Hyperparameter
In the context of the hyperparameter settings for our experiment, a batch size of 20 and 70 epochs, a network architecture consisting of 50 layers, utilization of the Adam optimizer, and an initial learning rate of 0.001 were employed.
2. Theoretical Background
2.1. Diagnosis of Nasal Bone Fracture
Conventional diagnostic approaches for nasal bone fractures typically rely on physical examinations and the clinical expertise of healthcare professionals [
3]. Although clinical symptoms may raise suspicion of nasal fractures, they often require confirmation through facial bone computed tomography (CT) to ascertain both the accurate diagnosis and extent of nasal deformity [3-5]. Nevertheless, these conventional methods suffer from potential drawbacks such as time-intensive procedures, subjectivity, and susceptibility to inter-observer variability [
3]. These limitations can lead to delayed diagnosis and suboptimal treatment outcomes. In particular, in Korea, emergency medicine practitioners, who are responsible for initiating primary radiologic diagnostic assessments for patients with facial trauma, serve a diverse patient population, thus encountering challenges in diagnosing nasal bone fractures through radiography or selecting the appropriate facial bone CT in a timely manner [1-3].
2.2. Artificial Intelligence (AI) in Fracture Diagnosis
Recent advances in artificial intelligence (AI) have yielded promising results in the analysis and prognostication of medical imaging data [6-9]. In this study, we presented an innovative AI-based predictive algorithm tailored to assess the surgical necessity for nasal bone fractures by leveraging Waters’ view radiographs. The Waters view constitutes a standard radiographic projection meticulously designed to visualize the nasal bones and contiguous anatomical structures [
2,
3]. Existing research has primarily focused on lateral-view radiographs and deep learning, or convolutional neural network (CNN) methodologies applied to three-dimensional reconstructed CT images [10-13]. While lateral view radiographs are notably beneficial for the diagnosis and pre- and postoperative assessment of nasal tip fractures, their efficacy in diagnosing unilateral nasal bone fractures, fractures involving deviations, and similar cases is notably limited when relying solely on radiographs [
12].
3. Results
3.1. Patients
Baseline characteristics of the study cohort are summarized in <
Table 1>. The training, validation, and test cohorts consisted of 2099, 730, and 315 patients, respectively (mean age, 38–41 years; male, 55.9–63.3%; fractures, 54.9–60.8%). There were significantly more males in the training and validation cohorts (62.7–63.3%) than in the test sets (55.9%) (P < 0.001). There were no significant differences among the cohorts in mean age (P = 0.112) or proportion of fractures (P = 0.081).
3.2. Diagnostic Performance of Deep Learning Model.
The diagnostic performance measures of the model with the classification method showed 67.09% accuracy, 67.90% precision, 65.80% recall, and an F1 score of 66.80%. The diagnostic performance measures of the model with the object-detection method showed 67.41% accuracy, 67.30% precision, 66.7% recall, and an F1 score of 67.00%. The deep learning model with segmentation labeling demonstrated excellent diagnostic performance, with 97.68% accuracy, 82.2% precision, 88.9% sensitivity, and an 85.4% F1 score (
Table 2).
Figure 3 visualizes the significantly superior performance of the segmentation labeling method compared to classification and object detection. Furthermore, the diagnostic performance measures of the deep learning model with segmentation labeling and quick learning showed 97.34% accuracy, 79.8% precision, 89.4% recall, and 84.3% F1 score. The dataset with the region of interest (ROI) application for the nasal bone region and segmentation of the fractured area amplified the accuracy of the fracture diagnosis. The result was significantly improved and more accurate than that of the classification or object detection labeling method.
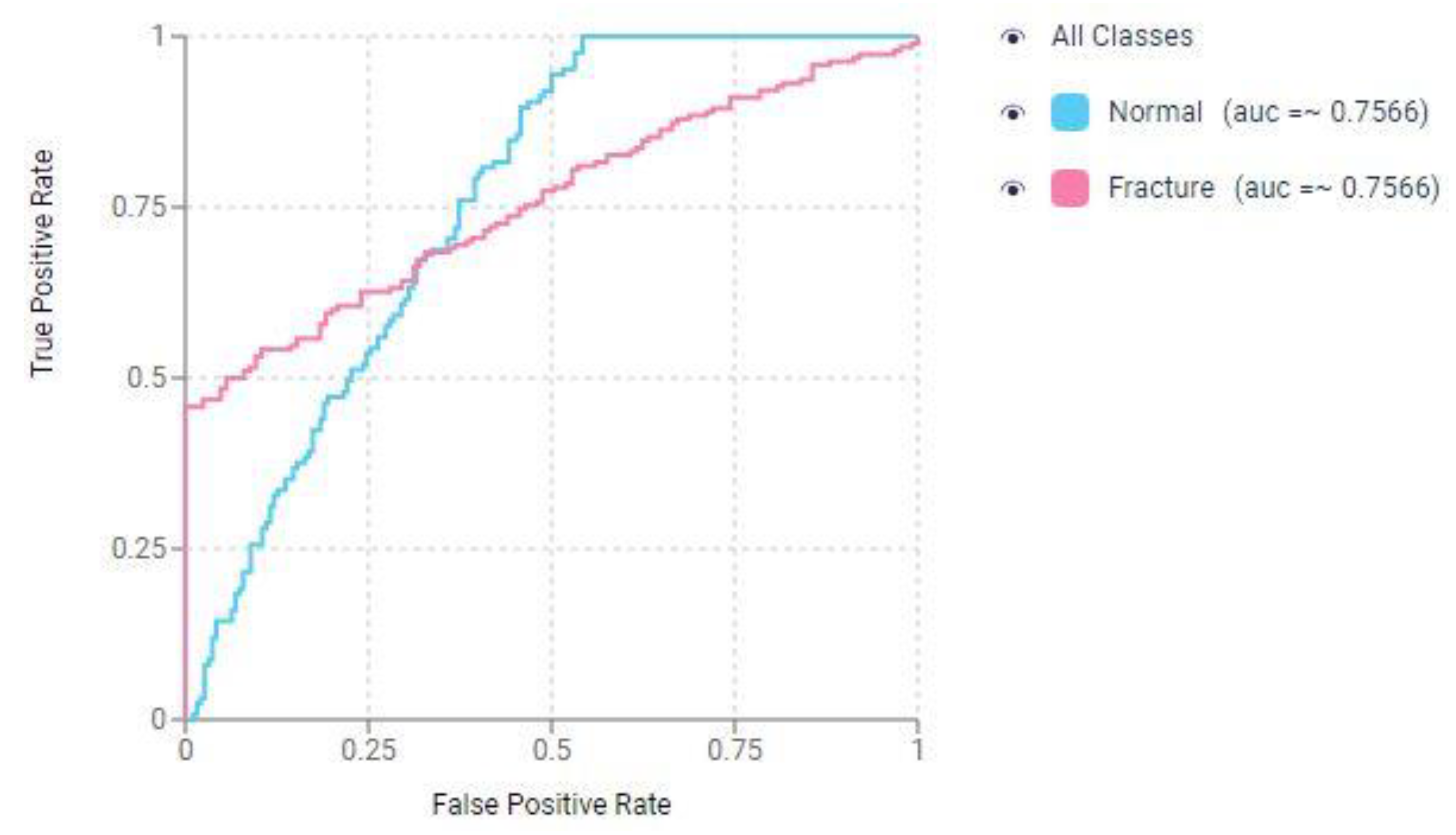
Figure 4 displays the Receiver Operating Characteristic (ROC) curve for the CNN in the classification-based prediction model for nasal bone fracture detection.
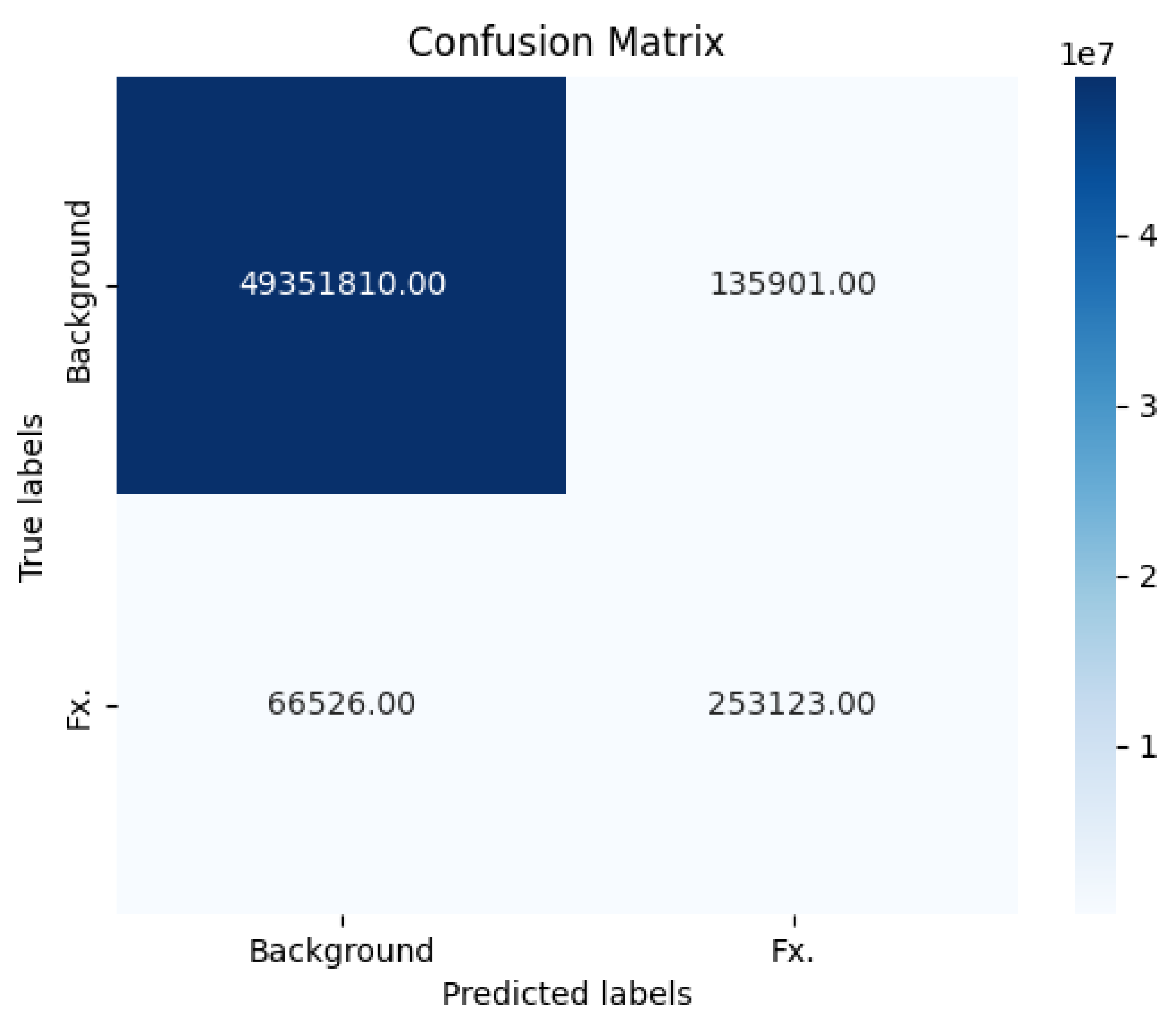
The key performance metrics of our fracture detection model with segmentation labeling are presented in the form of a confusion matrix (
Figure 5). These metrics were calculated based on the following values: 253,123 true positive cases, 66,526 false negative cases, 49,351,810 true negative cases, and 135,901 false positive cases. Our model exhibited a notable level of sensitivity, approximately 0.889, signifying its ability to accurately identify positive cases, or fractures, among the total instances. Additionally, the model demonstrated a high degree of specificity, approximately 0.9973, indicating its proficiency in correctly classifying negative cases, or non-fractures. The Negative Predictive Value (NPV) achieved a rate of 99.87%.
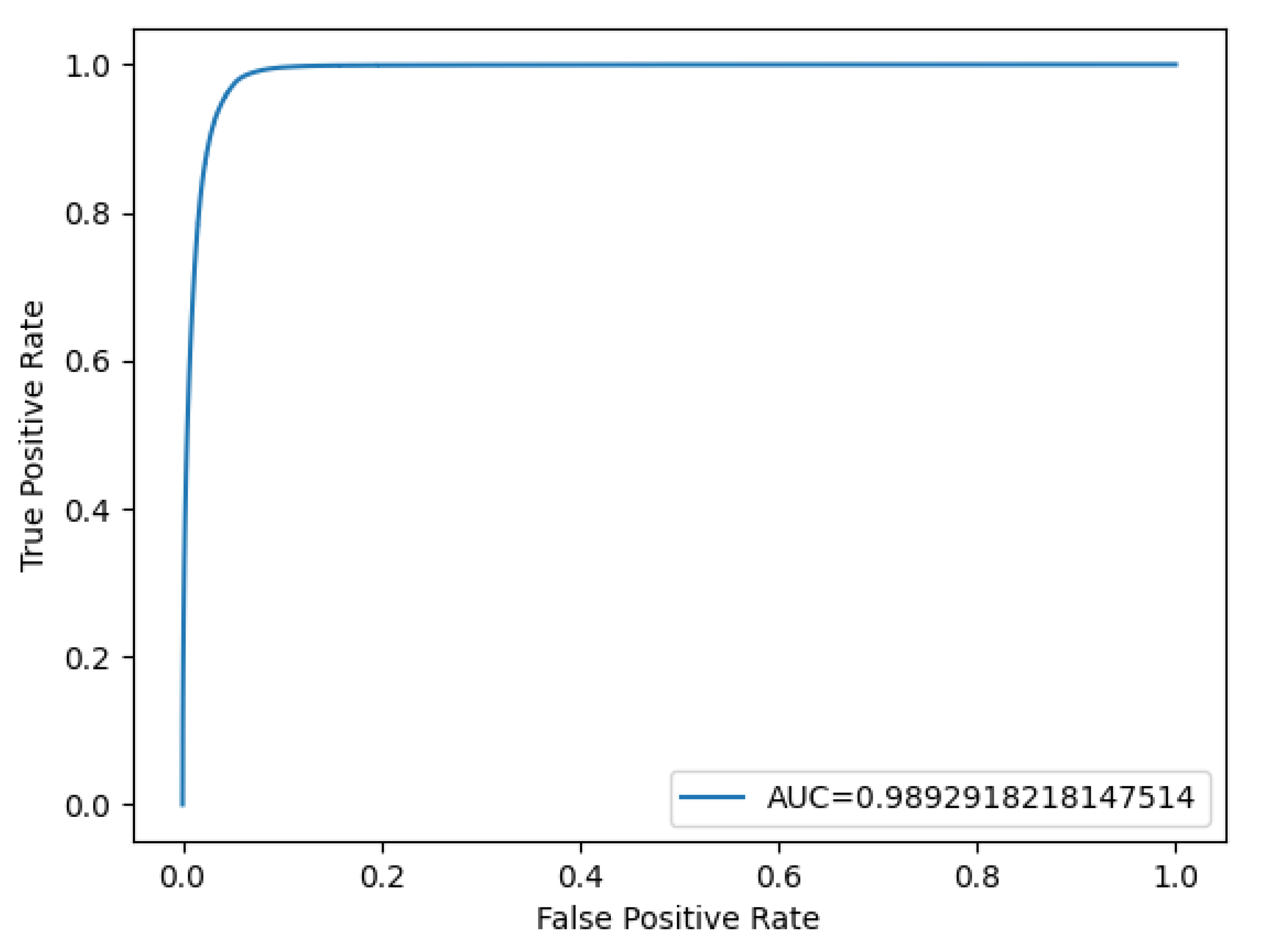
Based on a sensitivity of 88.9% and a specificity of 99.73%, a ROC curve and an Area Under the Curve (AUC) score were generated using the probability values from both the ground truth images and the model’s results (
Figure 6). These results emphasize the model’s robustness and reliability in accurately detecting fractures while minimizing false positives, making it a promising tool for medical applications.
3.3. True Positive and False Positive of Deep Learning Models
The accuracy and F1 score of the deep learning model with segmentation labeling were significantly higher than those of the classification model (85.4% and 77.9%, respectively). Example radiographs of nasal bone fractures correctly diagnosed using gradient-weighted class activation maps (CAM) in the classification model are shown in
Table 3. The nasal bone area is highlighted in the heat map of the X-rays with correct detection. In addition, examples of normal nasal radiographs incorrectly diagnosed as fractures using the deep learning model with classification are shown in
Table 4. The probability for the predicted class (%) is also provided for each sample.
Example images of skull waters’ view radiographs overlaid with the segmentation labeling with correct detection are exhibited in (
Figure 7). The deep-learning model correctly identified the nasal bone fractures.
Figure 8 displays representative cases of incorrectly predicted classes in the test dataset using the no-code platform with the segmentation labeling method. The orange color represents the area labeled by the authors using a segmentation method, while the hatched part corresponds to the pixel area predicted as fractures by the deep-learning model. Based on these results, we proposed a prediction algorithm to determine whether conservative treatment or surgical intervention should be performed.
4. Discussion
The nose is the foremost aesthetic focal point of the human face, and nasal bone fractures constitute the most prevalent bone injuries in adults, ranking as the third most common fracture across the entire body [
1,
2]. Facial trauma incidents involving nasal bone fractures are estimated to comprise approximately 40% of all cases, translating into approximately 50,000 occurrences annually in the United States alone [
1]. Although nasal bone fractures are often regarded as minor injuries, expeditious and accurate diagnosis of these fractures is of paramount significance [3-5]. This is primarily because delayed treatment has been linked to functional and cosmetic complications, the exacerbation of traumatic edema, preexisting nasal deformities, and occult septal injuries [
4]. Consequently, promptly and swiftly achieving an accurate diagnosis determines the need for surgical intervention, which plays a pivotal role in ensuring favorable patient outcomes [
5].
Traditional diagnostic approaches for nasal bone fractures typically rely on physical examinations and the clinical expertise of healthcare professionals [
2]. Although clinical symptoms may raise suspicion of nasal fractures, they often require confirmation through facial bone computed tomography (CT) to ascertain both the accurate diagnosis and extent of nasal deformity [
3]. Nevertheless, these conventional methods suffer from potential drawbacks such as time-intensive procedures, subjectivity, and susceptibility to inter-observer variability [3-5]. These limitations can lead to delayed diagnosis and suboptimal treatment outcomes. In particular, in Korea, emergency medicine practitioners, who are responsible for initiating primary radiologic diagnostic assessments for patients with facial trauma, serve a diverse patient population, thus encountering challenges in diagnosing nasal bone fractures through radiography or selecting the appropriate facial bone CT in a timely manner.
Recent advances in the field of AI have yielded promising results in the analysis and prognostication of medical imaging data [14-21]. In this study, we present an innovative AI-based predictive algorithm tailored to assess the surgical necessity for nasal bone fractures by leveraging Waters’ view radiographs. The Waters view constitutes a standard radiographic projection meticulously designed to visualize the nasal bones and contiguous anatomical structures [
2,
3,
22]. To date, existing research has primarily focused on lateral-view radiographs, and deep learning or CNN methodologies applied to three-dimensional reconstructed CT images [10-13]. While lateral view radiographs are notably beneficial for the diagnosis and pre- and postoperative assessment of nasal tip fractures, their efficacy in diagnosing unilateral nasal bone fractures, fractures involving deviations, and similar cases is notably limited when relying solely on radiographs [
2]. Our study introduces a predictive model based on deep learning techniques, aiming to provide healthcare practitioners with a diagnostic tool that enhances the efficiency and effectiveness of managing patients with nasal trauma visiting the emergency department. Moreover, our model sought to expedite the determination of surgical indications and streamline the decision-making process within a compressed timeframe.
With recent advancements in artificial intelligence systems, active research has been conducted in the craniomaxillofacial area [23-29]. Many studies have reported on orthognathic surgery [
23], cosmetic instrumentation using 3-D images of the face [6-8], and dental image analysis, including implant dentistry [
24]. AI research on facial bone fractures has been largely based on 3-D reconstructed CT) images [10-13]. Even when radiography is used, only a few studies have used the nasal lateral view for nasal bone fractures [
12], and there have been more studies on the zygoma complex [
6].
In this study, we proposed a novel AI-based prediction algorithm for the surgical indication of nasal bone fractures using Waters view radiographs. Our algorithm uses a convolutional neural network (CNN) to analyze radiographs and predict whether surgical intervention is necessary in patients with nasal bone fractures. We trained and validated our algorithm using a dataset of 2,099 patients with nasal bone fractures, who underwent surgical or conservative treatment. The results showed that our AI algorithm achieved high accuracy and reliability, with a sensitivity of 88.9%, a specificity of 99.73%, and an overall accuracy of 97.68%. These results are comparable to those of studies on the diagnosis of nasal bone fractures using machine learning techniques with other radiological modalities [10-12].
Moreover, by utilizing Waters’ view and AI to identify nasal bone fractures, we gained several additional advantages beyond the previously mentioned time efficiency. This mitigates unnecessary labor costs and benefits national finances in terms of health insurance. In South Korea, where the health insurance system continues to face deficit problems, a 3-D facial bone CT scan costs approximately
$335.5, but the national insurance covers
$239.6; therefore, the more unnecessary the CT scan, the more it costs the country. However, Waters’ view radiographs cost approximately
$13.2, and the government covers
$9.4, which is significantly less than the cost of a CT scan, thereby substantially reducing the national financial burden per person [
1,
2,
30].
Furthermore, there are evident health benefits for patients. A typical skull radiograph imparts an effective radiation dose of 0.1 mSv, while a CT scan imparts an effective radiation dose of 10 mSv, signifying a considerable disparity. Radiographs dramatically diminish radiation exposure in patients [
30].
Although our study demonstrated the potential of AI in the diagnosis and prediction of nasal bone fractures, some limitations need to be addressed. First, our dataset was relatively small, which may have limited the generalizability of our findings. Further studies, using larger datasets from multiple institutions, are required to confirm our results. Second, our algorithm was trained and validated using only one type of radiograph from the Waters’ view. Additional studies are required to determine the generalizability of our algorithm to other types of radiographs. Third, the lack of external validation was the most significant limitation. It cannot be excluded that the diagnostic accuracy would be low if data from other radiograph machines at other institutions were used to confirm the diagnosis. Finally, although bilateral nasal bones can be identified, fractures of the nasal tip are difficult to detect. To overcome this limitation, further multicenter studies, secure external validation, and engineering techniques should be developed to filter and distinguish imaging noise more accurately.
5. Conclusions
In conclusion, our study provides a new basis for the efficient diagnosis of nasal bone fractures by interpreting Waters’ view images using AI. This diagnostic prediction model speeds up diagnoses in the emergency department and helps less-experienced doctors order, read, and diagnose imaging tests. It can even help to quickly determine the indications for surgery. Our proposed algorithm has the potential to revolutionize the diagnosis and management of nasal bone fractures, leading to improved patient outcomes and clinical efficiency. Further studies are required to validate these findings.
Author Contributions
Conceptualization, S.R.E. and D.Y.L.; methodology, S.R.E., S.A.L. and D.Y.L.; software, S.R.E., S.A.L. and D.Y.L.; validation, S.R.E., S.A.L. and D.Y.L.; formal analysis, S.R.E., S.A.L. and D.Y.L.; investigation, S.R.E., S.A.L. and D.Y.L.; resources, S.R.E., S.A.L. and D.Y.L.; data curation, S.R.E. and D.Y.L.; writing—original draft preparation, S.R.E. and D.Y.L.; writing—review and editing, S.R.E., S.A.L. and D.Y.L.; visualization, S.R.E. and D.Y.L.; supervision, S.R.E.; project administration, S.R.E. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki and approved by the institutional review board of Dongguk University Ilsan Hospital, which waived the requirement for informed consent (IRB No.: 2023-05-017, Date of approval: 24 May 2023).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study and written informed consent has been obtained from the patients to publish this paper.
Data Availability Statement
The data from this study will be made available by the corresponding authors upon request. Due to privacy and ethical restrictions, the data is not publicly accessible.
Acknowledgments
We extend our gratitude to all colleagues who assisted with participant enrollment in the department of plastic and reconstructive surgery and clinical teams across participating institutions. Their dedicated efforts in patient enrollment and crucial data acquisition were invaluable to this diagnostics study.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| AI |
Artificial intelligence |
| DCNN |
Deep convolutional neural networks |
| CNN |
Convolutional neural network |
| CT |
Computed tomography |
| ROI |
Region of interest |
| ROC |
Receiver operating characteristic |
| NPV |
Negative predictive value |
| AUC |
Area under the curve |
| CAM |
Class activation mapping |
References
- Dong, S.X.; Shah, N.; Gupta, A. Epidemiology of nasal bone fractures. Facial Plast. Surg. Aesthet. Med. 2022, 24, 27–33. [Google Scholar] [CrossRef]
- Chukwulebe, S.; Hogrefe, C. The diagnosis and management of facial bone fractures. Emerg. Med. Clin. North Am. 2019, 37, 137–151. [Google Scholar] [CrossRef]
- Manson, P.N.; Markowitz, B.; Mirvis, S.; Dunham, M.; Yaremchuk, M. Toward CT-based facial fracture treatment. Plast. Reconstr. Surg. 1990, 85, 202–12; [Google Scholar] [CrossRef]
- Erfanian, R.; Farahbakhsh, F.; Firouzifar, M.; Sohrabpour, S.; Irani, S.; Heidari, F. Factors related to successful closed nasal bone reduction: a longitudinal cohort study. Br. J. Oral Maxillofac. Surg. 2022, 60, 974–977. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.; Chen, C.W.; Wu, Y.C.; Chen, K.K.; Lee, S.S. Refinement treatment of nasal bone fracture: A 6-year study of 329 patients. Asian J. Surg. 2015, 38, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.; Leonard, K.W.; Jassal, J.S.; Levin, A.M.; Ramachandra, V.; Jones, L.R. Artificial intelligence in facial plastic surgery: a review of current applications, future applications, and ethical considerations. Facial Plast. Surg. 2023, 39, 454–459. [Google Scholar] [CrossRef] [PubMed]
- Mir, M.A. Artificial intelligence revolutionizing plastic surgery scientific publications. Cureus 2023, 15. [Google Scholar] [CrossRef]
- Wheeler, D.R. Art, Artificial intelligence, and aesthetics in plastic surgery. Plast. Reconstr. Surg. 2021, 148, 529e–530e. [Google Scholar] [CrossRef]
- Torosdagli, N.; Anwar, S.; Verma, P.; Liberton, D.K.; Lee, J.S.; Han, W.W.; Bagci, U. Relational reasoning network for anatomical landmarking. J. Med. Imaging (Bellingham) 2023, 10. [Google Scholar] [CrossRef]
- Seol, Y.J.; Kim, Y.J.; Kim, Y.S.; Cheon, Y.W.; Kim, K.G. A study on 3D deep learning-based automatic diagnosis of nasal fractures. Sensors (Basel) 2022, 22. [Google Scholar] [CrossRef]
- Yang, C.; Yang, L.; Gao, G.D.; Zong, H.Q.; Gao, D. Assessment of artificial intelligence-aided reading in the detection of nasal bone fractures. Technol. Health Care 2023, 31, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Nam, Y.; Choi, Y.; Kang, J.; Seo, M.; Heo, S.J.; Lee, M.K. Diagnosis of nasal bone fractures on plain radiographs via convolutional neural networks. Sci. Rep. 2022, 12. [Google Scholar] [CrossRef] [PubMed]
- Prescher, A.; Meyers, A.; Gerf von Keyserlingk, D. Neural net applied to anthropological material: a methodical study on the human nasal skeleton. Ann. Anat. 2005, 187, 261–269. [Google Scholar] [CrossRef]
- Kuo, R.Y.L.; Harrison, C.; Curran, T.A.; Jones, B.; Freethy, A.; Cussons, D.; Stewart, M.; Collins, G.S.; Furniss, D. Artificial intelligence in fracture detection: A systematic review and meta-analysis. Radiology 2022, 304, 50–62. [Google Scholar] [CrossRef]
- Guermazi, A.; Tannoury, C.; Kompel, A.J.; Murakami, A.M.; Ducarouge, A.; Gillibert, A.; Li, X.; Tournier, A.; Lahoud, Y.; Jarraya, M.; Lacave, E.; Rahimi, H.; Pourchot, A.; Parisien, R.L.; Merritt, A.C.; Comeau, D.; Regnard, N.E.; Hayashi, D. Improving radiographic fracture recognition performance and efficiency using artificial intelligence. Radiology 2022, 302, 627–636. [Google Scholar] [CrossRef]
- Duron, L.; Ducarouge, A.; Gillibert, A.; Lainé, J.; Allouche, C.; Cherel, N.; Zhang, Z.; Nitche, N.; Lacave, E.; Pourchot, A.; Felter, A.; Lassalle, L.; Regnard, N.E.; Feydy, A. Assessment of an AI aid in detection of adult appendicular skeletal fractures by emergency physicians and radiologists: A multicenter cross-sectional diagnostic study. Radiology 2021, 300, 120–129. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, Y.; Shen, Y.W.; Zhang, K.R.; Jiang, Z.K.; Ma, L.T.; Ding, C.; Wang, B.Y.; Meng, Y.; Liu, H. Diagnostic accuracy and potential covariates of artificial intelligence for diagnosing orthopedic fractures: a systematic literature review and meta-analysis. Eur. Radiol. 2022, 32, 7196–7216. [Google Scholar] [CrossRef]
- Anderson, P.G.; Baum, G.L.; Keathley, N.; Sicular, S.; Venkatesh, S.; Sharma, A.; Daluiski, A.; Potter, H.; Hotchkiss, R.; Lindsey, R.V.; Jones, R.M. Deep learning assistance closes the accuracy gap in fracture detection across clinician types. Clin. Orthop. Relat. Res. 2023, 481, 580–588. [Google Scholar] [CrossRef]
- Li, Y.C.; Chen, H.H.; Horng-Shing Lu, H.; Hondar Wu, H.T.; Chang, M.C.; Chou, P.H. Can a deep-learning model for the automated detection of vertebral fractures approach the performance level of human subspecialists? Clin. Orthop. Relat. Res. 2021, 479, 1598–1612. [Google Scholar] [CrossRef]
- Cohen, M.; Puntonet, J.; Sanchez, J.; Kierszbaum, E.; Crema, M.; Soyer, P.; Dion, E. Artificial intelligence vs. radiologist: accuracy of wrist fracture detection on radiographs. Eur. Radiol. 2023, 33, 3974–3983. [Google Scholar] [CrossRef] [PubMed]
- Zech, J.R.; Santomartino, S.M.; Yi, P.H. Artificial intelligence (AI) for fracture diagnosis: an overview of current products and considerations for clinical adoption, from the AJR special series on AI applications. A.J.R. Am. J. Roentgenol. 2022, 219, 869–878. [Google Scholar] [CrossRef]
- Tuan, H.N.A.; Hai, N.D.X.; Thinh, N.T. Shape prediction of nasal bones by digital 2D-photogrammetry of the nose based on convolution and back-propagation neural network. Comput. Math. Methods Med. 2022, 2022, 5938493. [Google Scholar] [CrossRef]
- Seo, J.; Yang, I.H.; Choi, J.Y.; Lee, J.H.; Baek, S.H. Three-dimensional facial soft tissue changes after orthognathic surgery in cleft patients using artificial intelligence-assisted landmark autodigitization. J. Craniofac. Surg. 2021, 32, 2695–2700. [Google Scholar] [CrossRef]
- Jung, S.K.; Kim, T.W. New approach for the diagnosis of extractions with neural network machine learning. Am. J. Orthod. Dentofacial Orthop. 2016, 149, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Han, J.; Xie, B.; Xue, J.; Zhou, H.; Jiang, Y.; Hu, L.; Chen, C.; Zhang, K.; Zhu, F.; Lu, L. The two-stage ensemble learning model based on aggregated facial features in screening for fetal genetic diseases. Int. J. Environ. Res. Public Health 2023, 20, 2377. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Zhuang, X.H.; Wang, M.J.; Song, X.F. Assessment of low-dose paranasal sinus CT imaging using a new deep learning image reconstruction technique in children compared to adaptive statistical iterative reconstruction V (ASiR-V). B.M.C. Med. Imaging 2022, 22, 106. [Google Scholar] [CrossRef] [PubMed]
- Lamassoure, L.; Giunta, J.; Rosi, G.; Poudrel, A.S.; Meningaud, J.P.; Bosc, R.; Haïat, G. Anatomical subject validation of an instrumented hammer using machine learning for the classification of osteotomy fracture in rhinoplasty. Med. Eng. Phys. 2021, 95, 111–116. [Google Scholar] [CrossRef]
- Nakagawa, J.; Fujima, N.; Hirata, K.; Tang, M.; Tsuneta, S.; Suzuki, J.; Harada, T.; Ikebe, Y.; Homma, A.; Kano, S.; Minowa, K.; Kudo, K. Utility of the deep learning technique for the diagnosis of orbital invasion on CT in patients with a nasal or sinonasal tumor. Cancer Imaging 2022, 22, 52. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, C.; Zhang, X.; Deng, W. Synthetic CT generation based on T2 weighted MRI of nasopharyngeal carcinoma (NPC) using a Deep Convolutional Neural Network (DCNN). Front. Oncol. 2019, 9, 1333. [Google Scholar] [CrossRef]
- Smith, E.B.; Patel, L.D.; Dreizin, D. Postoperative computed tomography for facial fractures. Neuroimaging Clin. N. Am. 2022, 32, 231–254. [Google Scholar] [CrossRef]
Figure 1.
A flowchart of image selection process.
Figure 1.
A flowchart of image selection process.
Figure 2.
Schematic Illustration of Convolutional Neural Network (CNN) Applications. The training process centered around a single CNN architecture for the purpose of comparing accuracy performance in the detection of nasal bone fractures, according to the labeling methods applied to the dataset.
Figure 2.
Schematic Illustration of Convolutional Neural Network (CNN) Applications. The training process centered around a single CNN architecture for the purpose of comparing accuracy performance in the detection of nasal bone fractures, according to the labeling methods applied to the dataset.
Figure 3.
Graph comparing the fracture detection performance of AI models by labeling method.
Figure 3.
Graph comparing the fracture detection performance of AI models by labeling method.
Figure 4.
Receiver operating characteristic (ROC) curve of the convolutional neural network (CNN) for detecting nasal bone fractures in the prediction model with classification.
Figure 4.
Receiver operating characteristic (ROC) curve of the convolutional neural network (CNN) for detecting nasal bone fractures in the prediction model with classification.
Figure 5.
The key performance metrics of our fracture detection model with segmentation labeling are presented in the form of a confusion matrix. These metrics were calculated based on the following values: 253,123 true positive cases, 66,526 false negative cases, 49,351,810 true negative cases, and 135,901 false positive cases. Our model exhibited a notable level of sensitivity, approximately 88.9%. The specificity was calculated using true negatives and false positives with a value of 99.73%.
Figure 5.
The key performance metrics of our fracture detection model with segmentation labeling are presented in the form of a confusion matrix. These metrics were calculated based on the following values: 253,123 true positive cases, 66,526 false negative cases, 49,351,810 true negative cases, and 135,901 false positive cases. Our model exhibited a notable level of sensitivity, approximately 88.9%. The specificity was calculated using true negatives and false positives with a value of 99.73%.
Figure 6.
Receiver operating characteristic (ROC) curve of the convolutional neural network (CNN) for detecting nasal bone fractures in the prediction model with segmentation labeling.
Figure 6.
Receiver operating characteristic (ROC) curve of the convolutional neural network (CNN) for detecting nasal bone fractures in the prediction model with segmentation labeling.
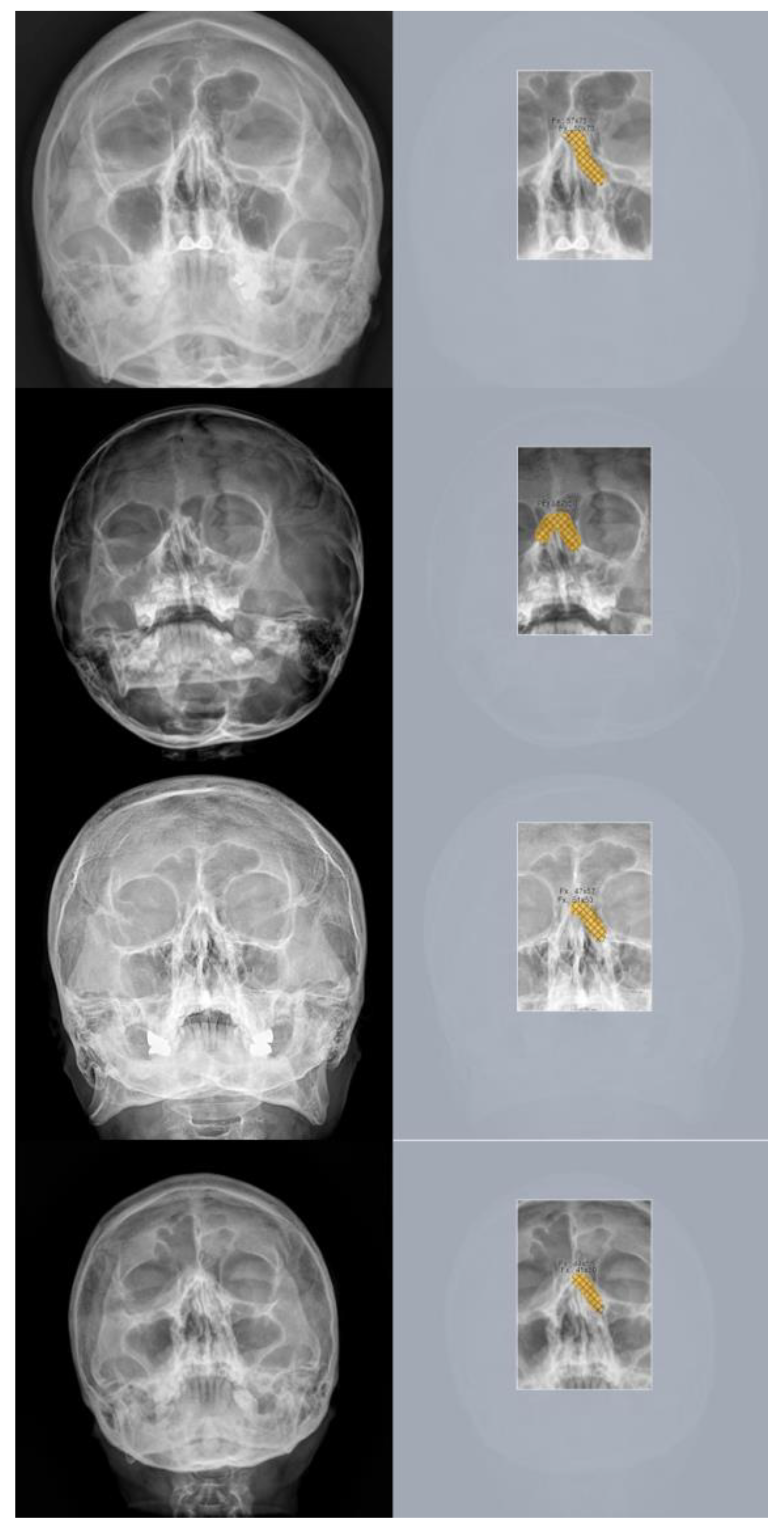
Figure 7.
Example images of skull waters’ view radiographs overlaid with segmentation labeling with correct detection. The deep-learning model correctly identified the nasal bone fractures. (Left) Radiographic images of the Skull Waters’ view with the removal of right/left markings from the corners of the X-ray. (Right) The orange color represents the area labeled by the authors using a segmentation method, while the hatched part corresponds to the pixel area predicted as fractures by the deep-learning model. The images (A,a), (B,b) and (C,c) show typical unilateral nasal bone fractures with fracture lines; images (D,d) show comminuted fractures with soft tissue swelling.
Figure 7.
Example images of skull waters’ view radiographs overlaid with segmentation labeling with correct detection. The deep-learning model correctly identified the nasal bone fractures. (Left) Radiographic images of the Skull Waters’ view with the removal of right/left markings from the corners of the X-ray. (Right) The orange color represents the area labeled by the authors using a segmentation method, while the hatched part corresponds to the pixel area predicted as fractures by the deep-learning model. The images (A,a), (B,b) and (C,c) show typical unilateral nasal bone fractures with fracture lines; images (D,d) show comminuted fractures with soft tissue swelling.
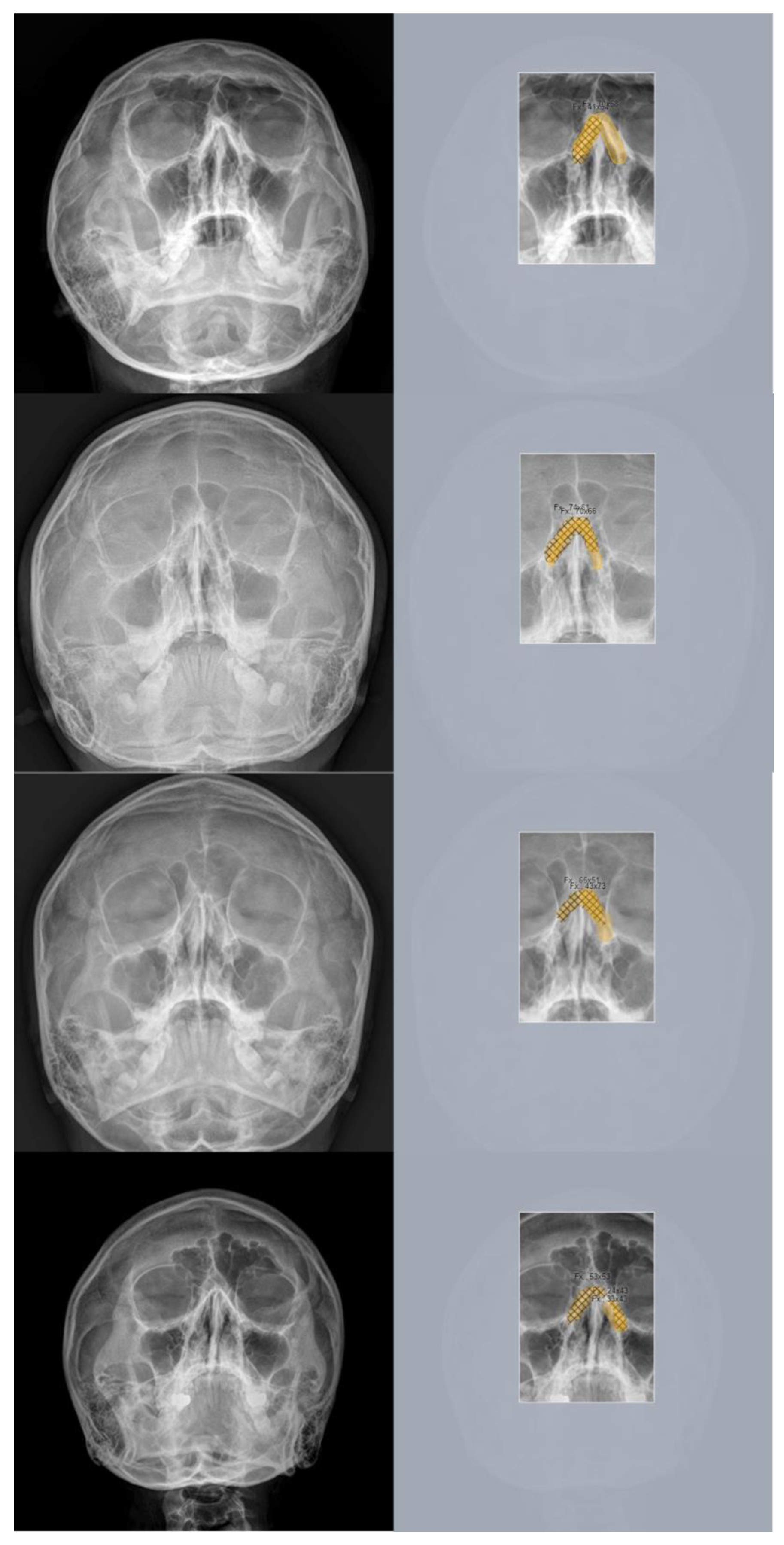
Figure 8.
Representative cases of incorrectly predicted classes in the test dataset using the no-code platform with a segmentation labeling method. (Left) Radiographic images of the Skull Waters’ view with the removal of right/left markings from the corners of the X-ray. (Right) The orange color represents the area labeled by the authors using a segmentation method, while the hatched part corresponds to the pixel area predicted as fractures by the deep-learning model.
Figure 8.
Representative cases of incorrectly predicted classes in the test dataset using the no-code platform with a segmentation labeling method. (Left) Radiographic images of the Skull Waters’ view with the removal of right/left markings from the corners of the X-ray. (Right) The orange color represents the area labeled by the authors using a segmentation method, while the hatched part corresponds to the pixel area predicted as fractures by the deep-learning model.
Table 1.
Baseline characteristics of patients.
Table 1.
Baseline characteristics of patients.
Table 2.
Summary of the fracture detection AI model’s performance according to the labeling methods.
Table 2.
Summary of the fracture detection AI model’s performance according to the labeling methods.
| Labeling methods |
Accuracy |
Precision |
Sensitivity |
F1 Score |
| Classification |
67.09 |
67.90 |
65.80 |
66.80 |
| Object detection |
67.41 |
67.30 |
66.70 |
67.00 |
| Segmentation |
97.68 |
82.2 |
88.9 |
85.4 |
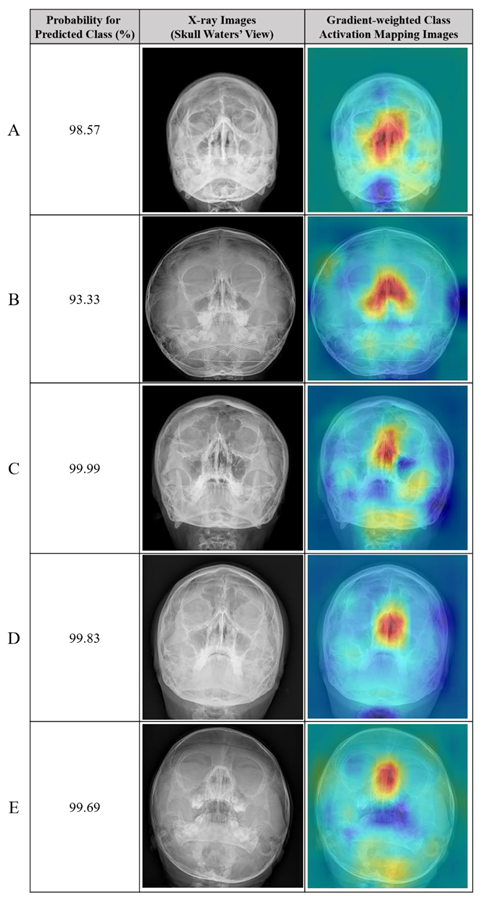
Table 3.
Representative cases of correctly predicted classes (true positive) in the test dataset using the no-code platform with a classification labeling method. (Left) Radiographic images of the Skull Waters’ view with the removal of right/left markings from the corners of the X-ray. (Right) Gradient-weighted class activation mapping (CAM) images. The probability for the predicted class (%) is also provided for each sample. (A,B,C,D) fractures of the unilateral nasal side wall; (E) fracture of the bilateral nasal side walls.
Table 3.
Representative cases of correctly predicted classes (true positive) in the test dataset using the no-code platform with a classification labeling method. (Left) Radiographic images of the Skull Waters’ view with the removal of right/left markings from the corners of the X-ray. (Right) Gradient-weighted class activation mapping (CAM) images. The probability for the predicted class (%) is also provided for each sample. (A,B,C,D) fractures of the unilateral nasal side wall; (E) fracture of the bilateral nasal side walls.
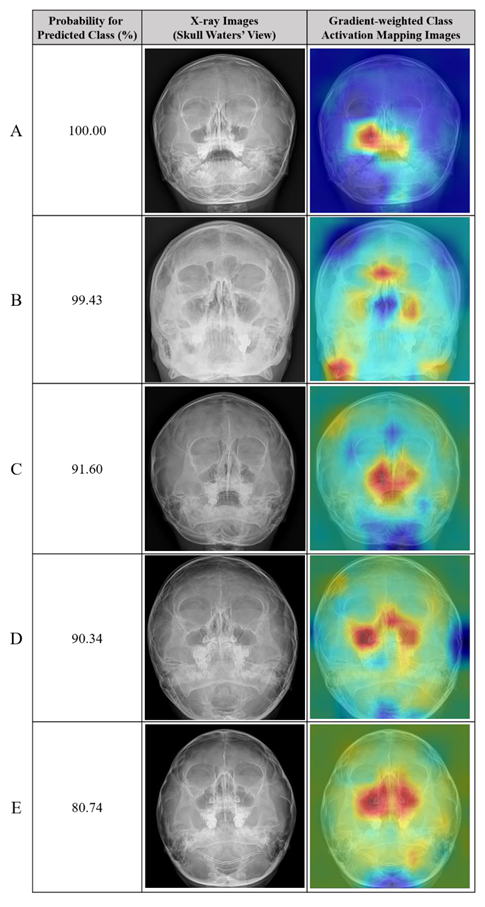
Table 4.
Representative cases of incorrectly predicted classes (false positive) in the test dataset using the no-code platform with a classification labeling method. (Left) Radiographic images of the Skull Waters’ view with the removal of right/left markings from the corners of the X-ray. (Right) Gradient-weighted class activation mapping (CAM) images. The probability for the predicted class (%) is also provided for each sample.
Table 4.
Representative cases of incorrectly predicted classes (false positive) in the test dataset using the no-code platform with a classification labeling method. (Left) Radiographic images of the Skull Waters’ view with the removal of right/left markings from the corners of the X-ray. (Right) Gradient-weighted class activation mapping (CAM) images. The probability for the predicted class (%) is also provided for each sample.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).