1. Introduction
In recent years, artificial snowmaking has become an indispensable component of ski resort operations across Europe, including Poland. As natural snowfall becomes increasingly unpredictable, the reliance on technical snow, which is produced by combining water and compressed air, has grown substantially [
1]. To meet the increasing demand for technical snow production, snowmaking systems utilize various water resources, such as rivers, streams, different types of reservoirs and occasionally treated wastewater. These water sources are often abundant in microorganisms and can serve as microbial vectors into the snowmaking infrastructure and ultimately onto ski slopes [
2,
3]. Previous studies have shown that water used for technical snow production harbors diverse microbial communities, including potentially pathogenic and antibiotic-resistant bacteria [
4,
5]. The microbial quality of these source waters, especially when reclaimed water or surface runoff is involved, poses ecological and public health concerns, particularly as snowmelt may return microorganisms to downstream aquatic ecosystems and/or soil [
6]. Nevertheless, the impacts of the snowmaking process itself, particularly changes in bacterial community structure, molecular mechanisms underlying microbial survival, selection, and adaptation as well as potential microbial dissemination routes, remain poorly understood.
Snow, regardless of origin, represents a climatically sensitive and transient ecosystem that links the atmosphere with the underlying soil. It has a strong impact on the hydrological cycle and interacts with various ecosystems. These characteristics apply to both natural and technical (i.e. ‘artificial’) snow. However, because technical snow has much higher density than natural snow [
7], it melts more slowly than the natural snow. Thereby, it exerts a greater and more prolonged influence on the hydrological cycle and surrounding aquatic environments near ski slopes where it is produced and stored. It has already been demonstrated that snowpack harbors distinct microbial communities, often shaped by environmental origin and physicochemical conditions [
8,
9,
10]. Regardless of the snow type, it creates harsh conditions with limited liquid water and extreme temperatures, creating a habitat with a specific “core” microbiome [
11]. Snow creates a specialized environment for bacterial communities, with technical snow forming an even more unique ecological niche. The conditions that prevail in snow drive molecular-level adaptations such as the expression of cold-shock proteins, antifreeze proteins, and membrane fluidity regulators, enabling microbial persistence in snow environments. All the above can promote the development of a specialized microbiota, the composition of which may differ significantly not only from that of natural snow but also from the microbial communities present in the water used to feed snow cannons.
Although the physicochemical properties of technical snow have been relatively well studied [
10,
12,
13,
14], there is a paucity of research exploring how snow production systems affect microbial community structures and functions. In particular, few studies have evaluated microbial responses at the molecular level or across the full production cycle, from source water to aged snow or meltwater. Snow, especially of technical origin, may act both as a barrier and a vector for microbial dissemination, with potential implications for downstream environmental microbiology and human exposure.
Recent advances in high-throughput sequencing, particularly 16S rRNA gene amplicon profiling, have enabled detailed taxonomic and functional characterization of microbial communities in engineered and natural environments [
15]. Functional prediction tools like FAPROTAX further allow inference of metabolic traits, such as chemoheterotrophy, fermentation, and aromatic compound degradation, based on taxonomic profiles [
16]. These molecular insights are critical for understanding how snowmaking systems shape microbial community structure and function, and how these changes may impact downstream ecosystems [
17].
To date, microbial research in cold environments has primarily focused on natural snow, glaciers or polar systems [
8,
11,
18,
19], while artificially produced snow remains underexplored. Moreover, few studies have integrated high-throughput sequencing (e.g. 16S rRNA gene amplicon profiling) with culture-based microbiological techniques to assess and compare both viable and non-culturable microbial fractions in snow production systems.
This study aims to characterize bacterial communities across the full technical snow production system in a Polish ski resort, using a dual approach: culture-based bacterial isolation and high-throughput 16S rRNA gene amplicon sequencing. Sampling sites included feed water sources, storage reservoirs, snowmaking infrastructure (e.g. snow cannon filters), and different stages of technical snow life (freshly produced, aged, and meltwater), as well as downstream sediments. Functional profiling was conducted using FAPROTAX-based inference to predict key microbial processes associated with snow microbiomes. To our knowledge, this is the first comprehensive microbiological and molecular assessment of the full cycle of artificial snow production and transformation. The findings provide a comprehensive molecular perspective on microbial selection, redistribution and functional transformation in technical snow, with potential implications for microbial transport and ecological processes in downstream aquatic systems.
2. Results and Discussion
To assess how microbial communities change across stages of technical snow production, we integrated culture-based enumeration of bacteria with high-throughput 16SRNA gene sequencing to obtain information about total bacterial communities across all sample types. These analyses were supplemented with functional prediction via FAPROTAX. This dual approach enabled us to capture both viable and non-culturable bacterial fractions and infer metabolic traits relevant to cold adaptation, stress tolerance, and ecological function.
2.1. Abundance of Culturable Bacteria and OTU Richness Across the Snow Production Pipeline
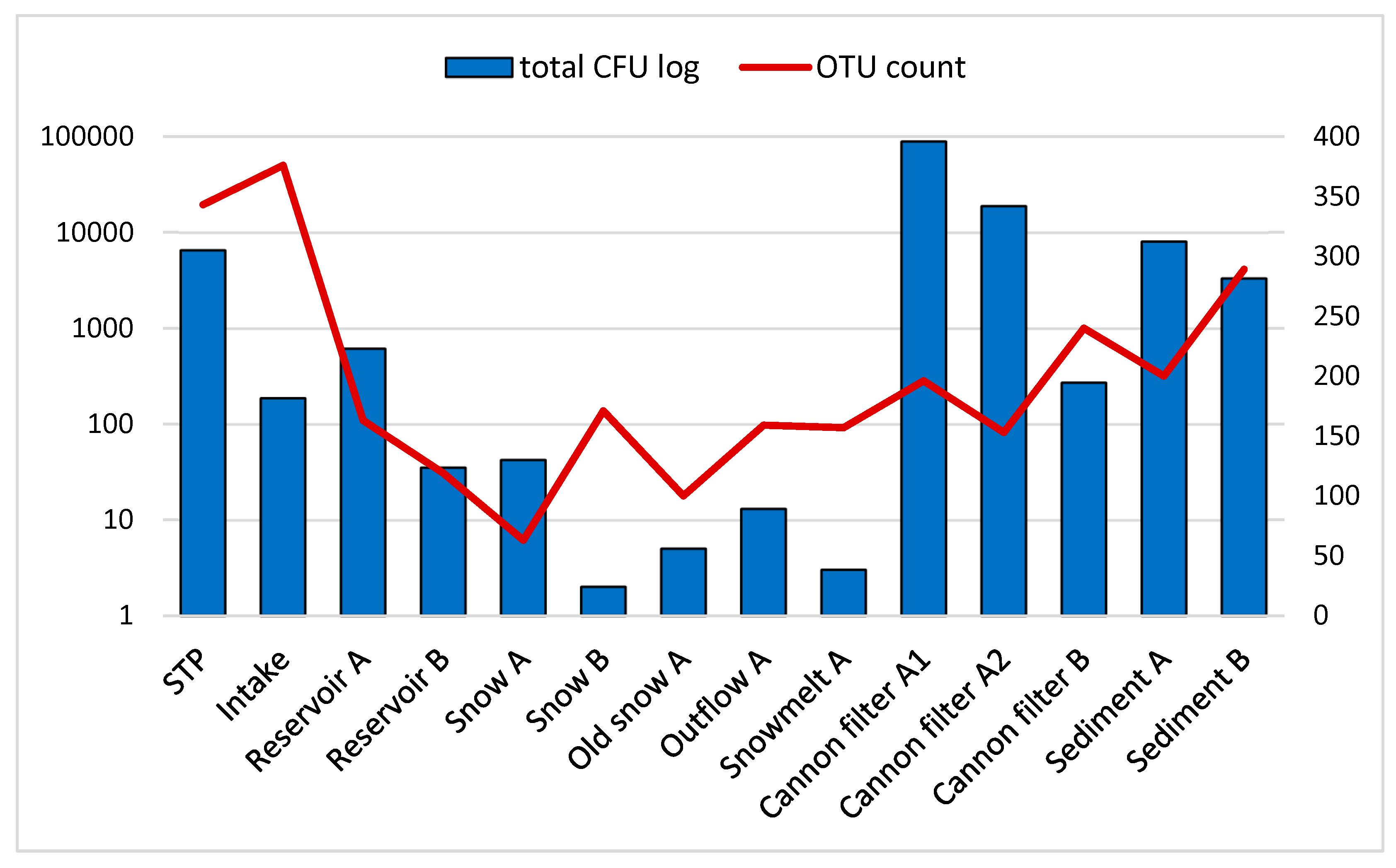
Figure 1 shows total CFU (sum of
E. coli, enterococci, coagulase-positive staphylococci, and mesophilic bacteria) as blue bars (log scale, left axis), while OTU counts based on 16S rRNA gene amplicon sequencing are shown as a red line (right axis). Detailed CFU data for individual bacterial groups are provided in
Supplementary Table S1. The data (
Figure 1,
Supplementary Table S1) illustrate the contrast between culture-based and sequencing-based detection, particularly in snow and meltwater samples, where OTUs remain detectable despite low or absent culturable bacteria. This discrepancy reflects the limitations of culture-based methods, which typically capture less than 1% of environmental bacterial diversity [
20,
21]. It also supports the value of high-throughput molecular methods such as 16S rRNA sequencing in uncovering the microbial diversity, including viable but non-culturable (VBNC) and dead cells retaining DNA signatures in low-biomass, oligotrophic or stressed environments, like snow. Snow cannon filters and sediment samples exhibit the highest microbial loads in both approaches, highlighting their potential as microbial reservoirs or biofilm-rich sites within the system. Presumably, snow cannons may trap organic debris and provide surfaces for microbial attachment, as well as provide hideout from direct UV light exposure, which is favorable for microbial persistence. Similar observations have been reported in engineered aquatic environments, where biofilm accumulation and particle trapping contribute to increased microbial biomass and diversity (Kumar et al. 2018). In contrast, the near absence of culturable bacteria in fresh snow and snowmelt suggests harsh environmental elimination, possibly due to freezing, UV exposure, or nutrient scarcity, which inhibits growth of non-adapted taxa but does not preclude their molecular detection which captures DNA from viable but non-culturable cells and dead cells as well [
4,
11,
19]. This may explain why OTUs are still observed in sites where CFUs are nearly absent.
2.2. Microbial Diversity and Richness
Bacterial diversity varied clearly across the stages of technical snow production (
Table 1). The highest richness and diversity were observed at the WWTP and Intake sites, reflecting the complexity and heterogeneity typical of anthropogenically impacted freshwater systems. This observation is consistent with earlier findings that such environments act as microbial hubs, integrating a wide range of bacterial taxa from diverse sources [
4,
22].
In contrast, reservoirs showed reduced diversity and evenness, likely as a result of environmental filtering and passive treatment processes such as UV exposure or settling [
4]. Both fresh snow samples exhibited low diversity and high dominance, suggesting strong selective pressure during snow formation. Factors such as high-pressure spraying, rapid freezing, and low nutrient content likely favor psychrotolerant or stress-adapted taxa while excluding others [
11,
23,
24]. Interestingly, the cannon filter B and reservoir sediment samples retained relatively high diversity and richness, consistent with their role as microbial accumulation zones or biofilm reservoirs within the system [
25,
26]. Their protected surfaces and stable conditions may promote colonization and retention of a broader microbial community.
To evaluate compositional similarities between samples, we calculated Bray-Curtis dissimilarities based on the 50 most abundant genera (
Figure 2). The calculated dissimilarity values are very high, suggesting clear variation in bacterial community composition across the technical snowmaking system. However, the highest similarities were observed between WWTP and Intake (dissimilarity = 0.26), two snow cannons A1 and A2 (0.47) and between the two reservoirs (0.44). Cannon A1 also showed moderate similarity to reservoir sediment B (0.39). These patterns suggest that spatial proximity, similar environmental conditions, or shared functions (e.g. anthropogenically impacted water, water retention, biofilm formation and freezing temperatures) can support overlapping microbial assemblages. In contrast, snow related samples (Fresh Snow A, Fresh Snow B and Aged Snow A) showed high or nearly complete dissimilarity with source waters (e.g. Fresh Snow A vs. WWTP = 0.97; Fresh Snow A vs. Intake = 0.95; Fresh snow B vs. WWTP = 0.86) underscoring the strong selective pressure during the snow formation processes. These patterns align with previous studies showing that technical and environmental filtering, including UV light disinfection, pressurized spraying and water freezing, significantly alter microbial communities in engineered water systems [
27,
28,
29,
30]. Overall, these results support the model of progressive microbial selection and restructuring from input waters through snowmaking and deposition. Also the fact that snow and melt samples cluster away from input water sources, support the idea that cold adaptation, desiccation tolerance, and low nutrient availability drive microbial divergence in snow environments [
31,
32]. Meanwhile, sediment samples retained greater microbial similarity to each other and to cannon filters, likely due to their role as microbial sieves, enriched in particle-associated and biofilm-forming bacteria [
33].
2.3. Bacterial Community Composition
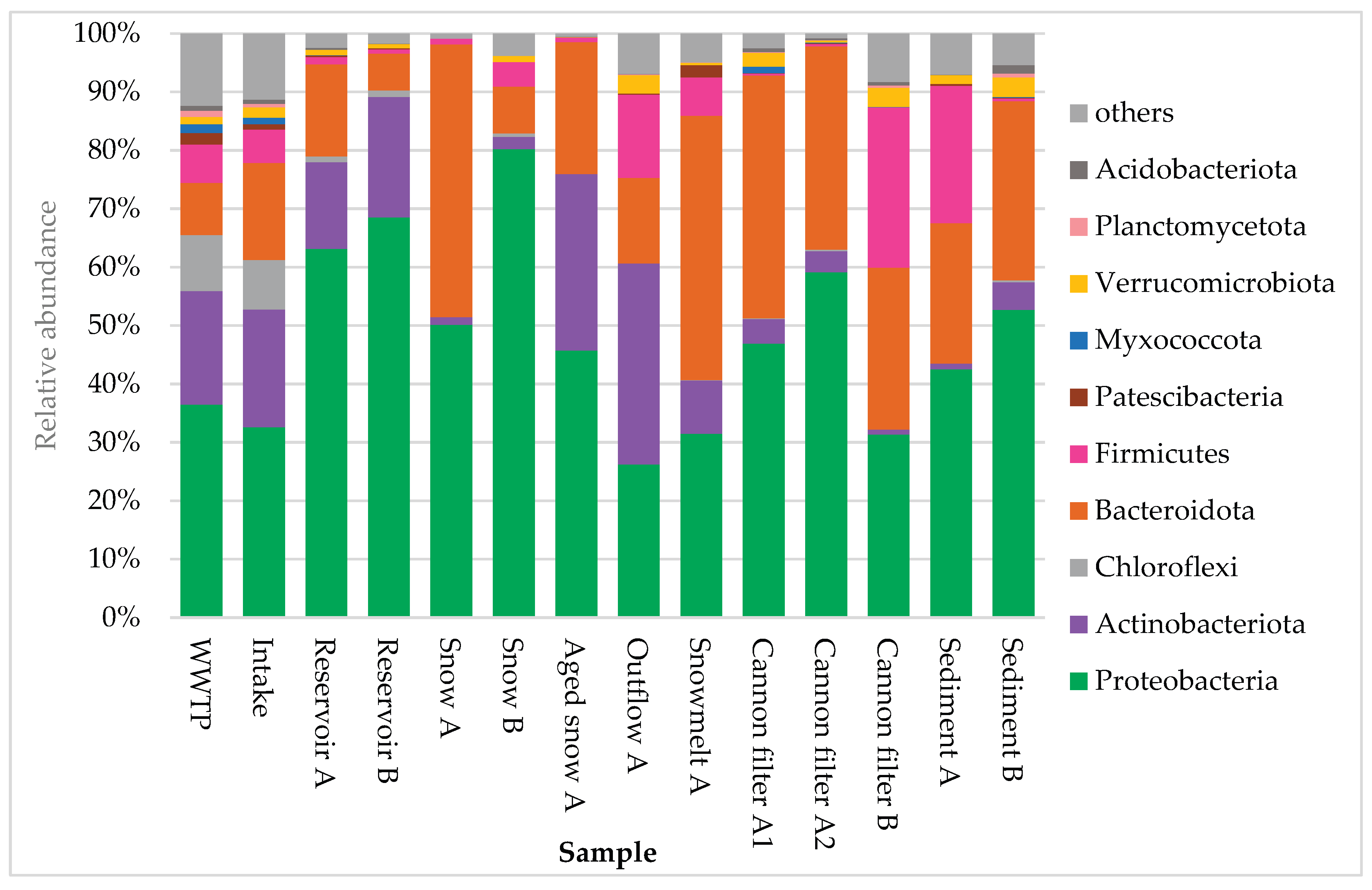
The composition of bacterial communities across the technical snow production system revealed pronounced shifts in dominant taxa, as visualized in
Figure 3 and
Figure 4. At the phylum level (Fig. 3), the most abundant groups across all samples were Proteobacteria
, Bacteroidota
, Actinobacteriota
, and Firmicutes, although their proportions varied substantially by site. Proteobacteria clearly dominated in reservoir samples, where their relative abundance exceeded 50%. They also consistently prevailed in cannon filters, freshly produced technical snow and sediment samples. Snowmelt water, however, was dominated by Bacteroidota (45.3%), with Proteobacteria as the second most abundant phylum (31.5%), indicating a shift in bacterial community composition during thawing.
Aged snow displayed a more balanced community composition, with co-dominance of Proteobacteria (45.8%), Actinobacteriota (30.2%), and Bacteroidota (22.6%). These patterns suggest selective survival or accumulation effects within the snowpack over time. In WWTP and Intake water samples, Proteobacteria and Actinobacteriota were most abundant, consistent with their prevalence in urban and freshwater environments. Meanwhile, cannon filters showed high proportions of Bacteroidota and Proteobacteria, while Firmicutes became co-dominant in the Cannon filter B (27.5%), possibly indicating niche adaptation or biofilm formation in response to operational conditions. Finally, sediment samples (A and B) showed mixed phylum-level composition, with notable variation between the two reservoirs.
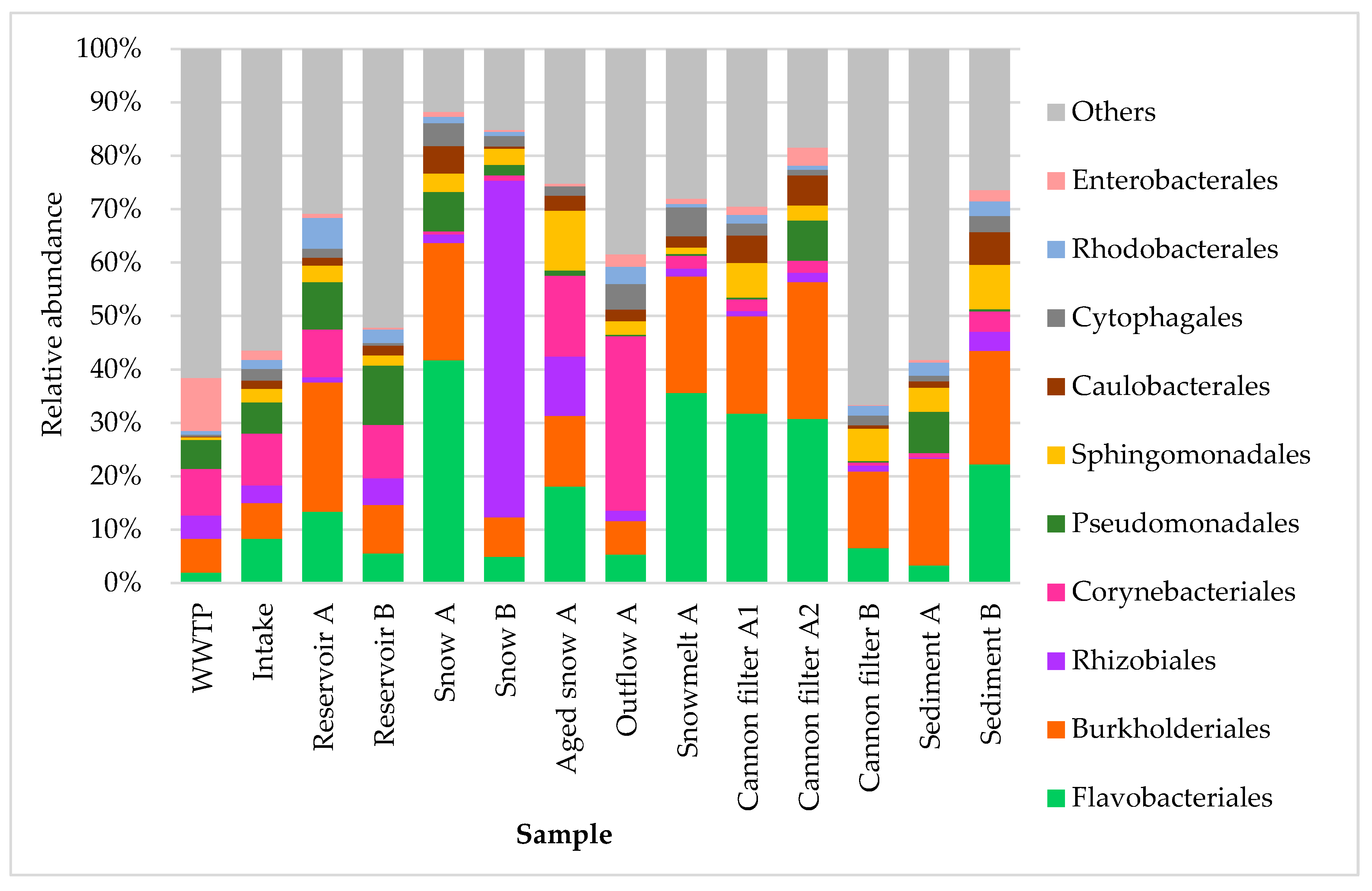
At the order level (Fig. 4), bacterial community composition was even more variable across sample types and individual samples. Flavobacteriales dominated the snow continuum, being most abundant in Fresh snow (41.8%), Aged snow (18.1%) and Snowmelt (35.6%) samples, suggesting their adaptability to cold and/or oligotrophic aquatic environments. They also dominated in snow cannon filters A, reinforcing their role as key snow-associated taxa. Burkholderiales, a diverse order commonly associated with both freshwater and soil environments, was also highly abundant across multiple sites, including reservoirs, cannon filters, snow and sediments. Surprisingly, Rhizobiales, indicative of soil or plant origin, were highly prevalent in Snow B (63.1%), suggesting airborne, soil or plant-related microbial inputs [
26,
32] into the snowpack. Corynebacteriales were the most abundant in the Outflow A sample (32.6%) and the second most abundant in Aged snow A (15.2%), suggesting either their persistence or enrichment over time [
10,
34]. In contrast, the WWTP sample exhibited the highest abundance of Enterobacterales, consistent with their association with anthropogenic sources. Interestingly, Cannon filters A1 and A2 retained detectable levels of Enterobacterales (1.5% and 3.4%, respectively), and they were also present in Outflow A (2.3%) and Reservoir B sediment (2.1%), suggesting partial retention or downstream transport and deposition of wastewater-associated taxa within the system.
These taxonomic shifts likely reflect a combination of environmental filtering (e.g., freezing and thawing, desiccation) treatment effects (e.g. UV light disinfection, aeration in reservoirs) coupled with surface-associated microbial selection in snow cannon filters or in bottom sediments of reservoirs [
25,
26,
35]. All these factors shape the microbial communities involved in technical snow production and transformation, and during snowmelt they may further influence the composition of microbial communities in receiving waters and underlying soil [
24,
36].
2.4. Ecologically Distinct Taxa and Indicators Across Snow Production Stages
One of the aims of this study was to profile bacterial community shifts throughout the five-month cycle of technical snow production and transformation. Several taxa emerged as ecologically or functionally distinct, serving as indicators of particular environments or selective pressures along the process. While the overall taxonomic composition varied across the water–to-snow-to-sediment pathway, the presence of certain genera provided insight into their possible source, selective pressure put thereon and bacterial adaptations to changing conditions.
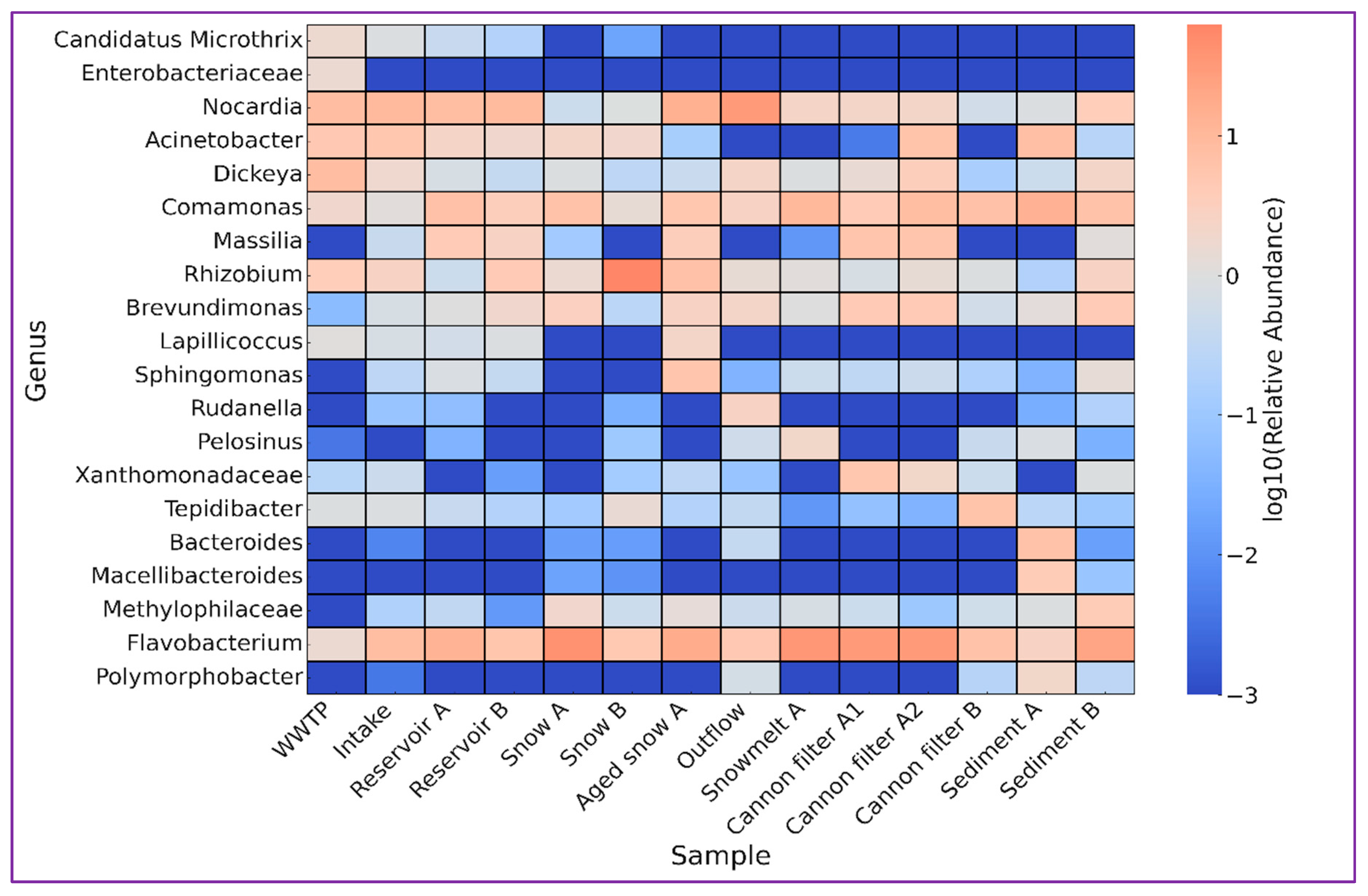
Table 2 and
Figure 5 summarize the key genera selected based on their abundance patterns, ecological significance, and known environmental niches. Analysis of their abundance within the examined anthropogenically impacted environment offered a deeper understanding of microbial dynamics and possible effects on further elements of the receiving compartments. Wastewater-associated bacteria such as Candidatus
Microthrix and unidentified genera of
Enterobacteriaceae family were most abundant by the wastewater treatment plant (WWTP), but nearly absent at downstream sites, indicating their dilution or removal during subsequent stages. The river water intake for technical snowmaking (Intake site) showed a community typical of moderate anthropogenic impact, including
Nocardia (9.36%) or potentially pathogenic freshwater opportunists
Acinetobacter (5.12%) and biofilm-forming
Flavobacterium (7.80%) [
23]. Technical reservoirs, which are subsequent step in technical snow production, appeared to facilitate both attenuation of anthropogenic indicators and enrichment of resilient taxa such as
Comamonas,
Massilia [
37] or
Flavobacterium [
23], suggesting their dual role as both biological filters and selective habitats for cold-adapted bacteria.
Snow cannon filters, another step in technical snow production, selected strongly for surface-associated, biofilm-forming or stress-tolerant taxa. These included members of
Xanthomonadaceae family,
Flavobacterium, Brevundimonas, but also taxa adapted to harsh conditions, like e.g.
Massilia (early colonizers of oligotrophic habitats, isolated from glacial meltwater, [
37,
38]), or anaerobic
Tepidibacter [
39]. Likely, the filters provide surfaces with periodic water flow, nutrient scarcity and physical trapping, which on one hand allow for the purification of contaminated water and on the other - shape the downstream bacterial community. Freshly produced technical snow showed simple microbial communities, dominated by
Flavobacterium (snow A) or
Rhizobium cluster (snow B). It suggests that physical stress of snow production, i.e. high-pressure water spraying, air mixing, freezing) favors specific groups, likely these with fast response to shock and rapid surface attachment ability [
23,
32]. The ageing of snow on ski slopes is reflected by the community composition observed in samples such as Aged snow A and Snowmelt A, where the bacterial community diversified. In these samples,
Flavobacterium and
Comamonas remained abundant, but additional taxa with distinct habitat preferences started to appear. These included
Rhizobium (soil or plant origin),
Brevundimonas (oligotrophic environments), and
Lapillicoccus, typically isolated from harsh, nutrient-poor extreme environments, such as Antarctic soils after long incubation periods [
40]. Cold-adapted genera like
Sphingomonas [
11,
31,
38], and anaerobes commonly found in anoxic environments such as
Pelosinus [
41] were also present, reflecting post-depositional ecological shift and interaction with surrounding environment.
Another aspect to consider relevant in terms of microbial input into the environment as a result of technical snow production, is the fact that changing the ecological balance never remains without consequences for downstream ecosystems. Several taxa identified in the samples most likely to contribute to the microbial composition downstream (i.e. snowmelt water, reservoir outflow and sediments) show moderate to high relevance in terms of pathogenic potential to humans, animals or plants, and may have important ecological impact.
Acinetobacter, prevalent in Sediment A, includes species known as nosocomial pathogens with strong biofilm-forming ability and resistance to disinfection, posing a potential risk if released during reservoir maintenance or flushing [
42,
43]. Environmental taxa, like
Comamonas (highly prevalent in Sediment A and Snowmelt water) or
Brevundimonas (enriched in Outflow A and Sediment B), adapt to nutrient-poor environments, contribute to biofilm formation and include species recently reported as important opportunistic pathogens [
44,
45]. Members of the genus
Dickeya, known phytopathogens, and
Flavobacterium, which includes opportunistic pathogens to humans and fish and species with algicidal activity, may affect soil or aquatic health if introduced into vegetated zones or irrigation systems [
46,
47,
48]. Two taxa within the
Clostridium sensu stricto complex were detected in multiple samples
. Clostridium sensu stricto 1 includes well-known human and animal pathogens, such as
C. perfringens and
C. botulinum, capable of forming resistant spores and surviving harsh conditions, posing a latent risk upon environmental release [
49].
Clostridium sensu stricto 13, while less characterized, is associated with fermentative activity in sediments and anaerobic digesters, and may signal eutrophic or anoxic conditions [
50]. The persistence of these
Clostridium groups in cannon filters and sediments raises concerns about long-term microbial dissemination, particularly during snowmelt or sediment displacement. These findings underline the importance of monitoring microbial communities not only for direct pathogenic threats but also for their environmental consequences following snowmelt or sediment dispersal.
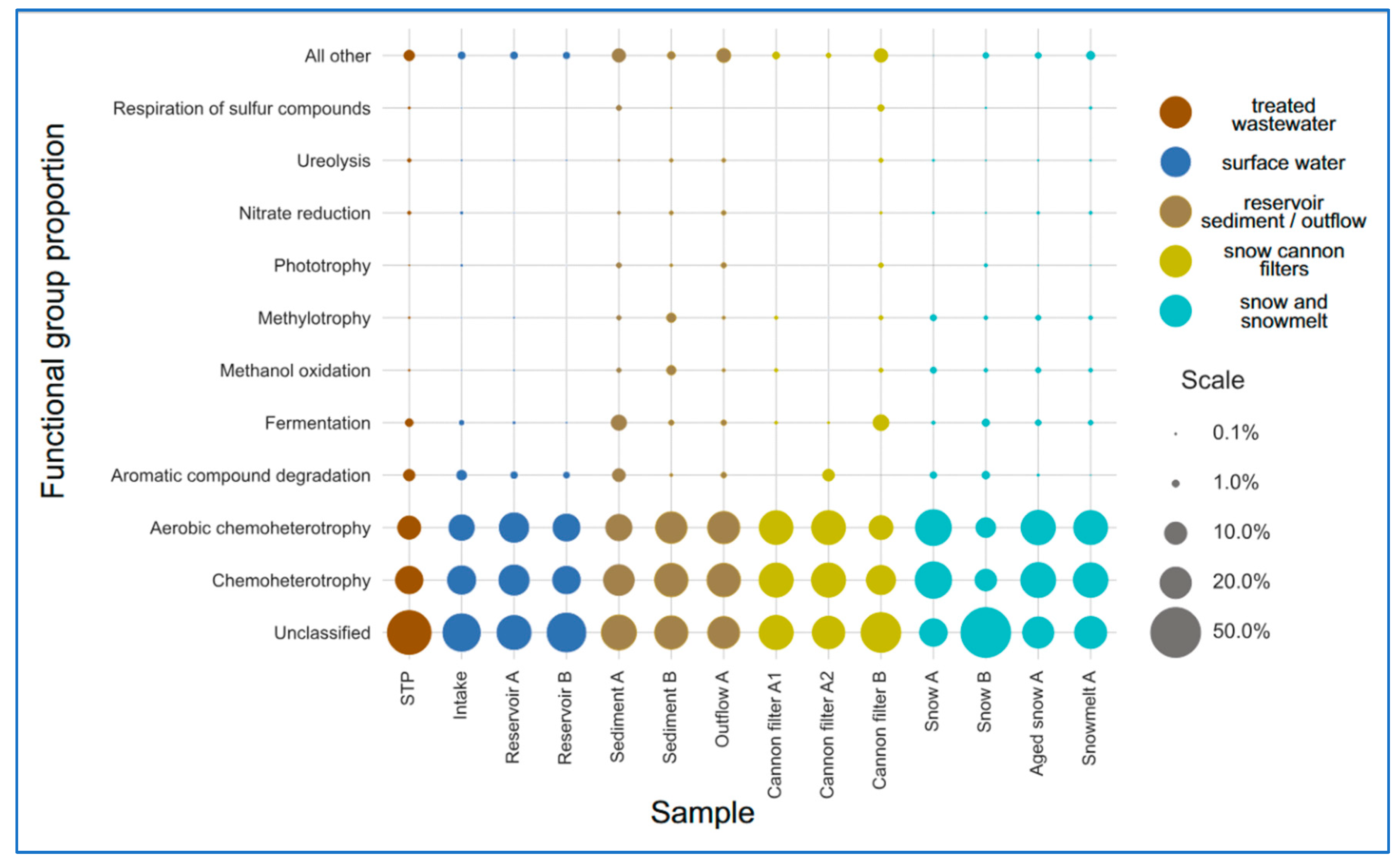
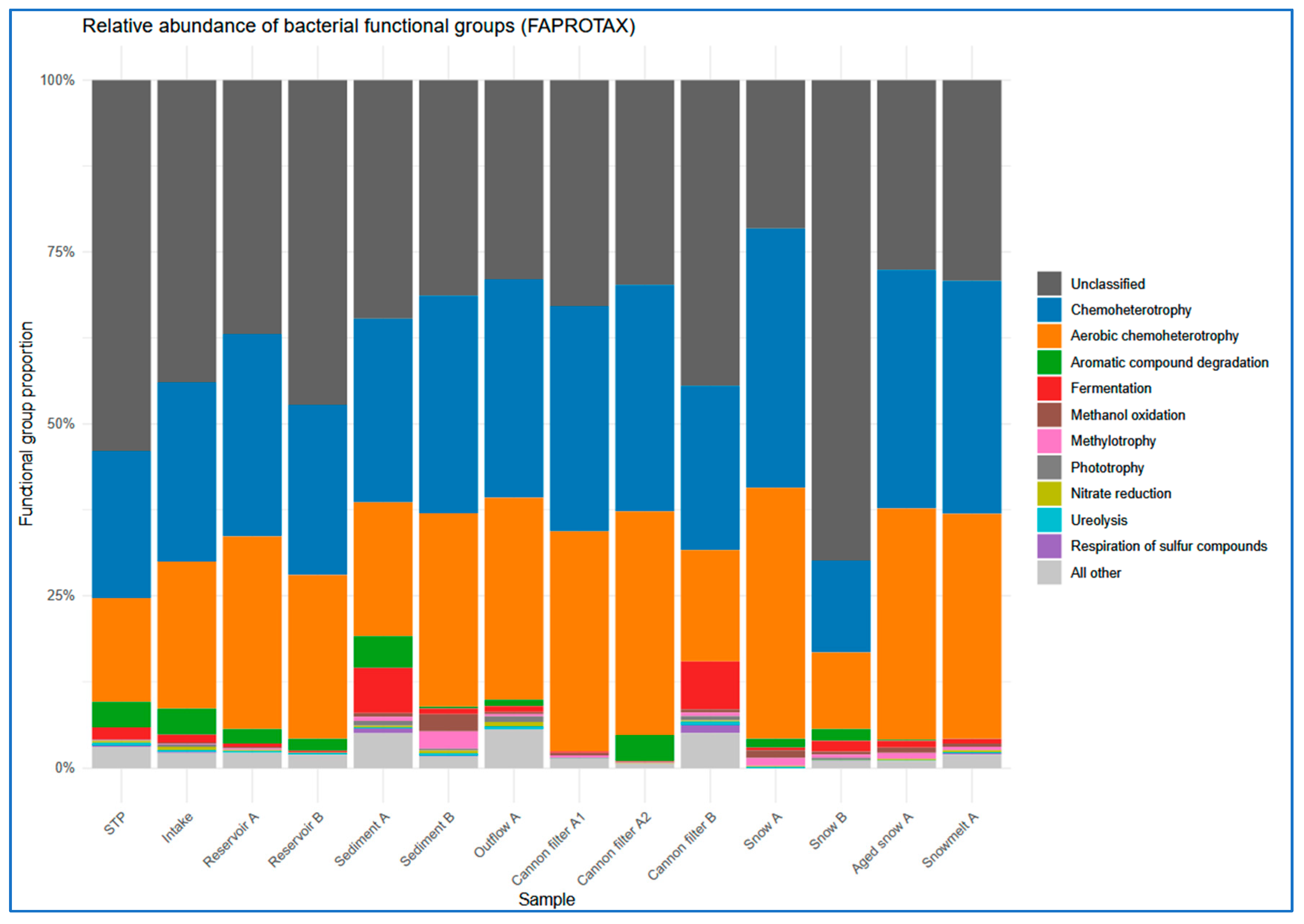
2.5. Functional Prediction of Bacterial Communities Inferred by FAPROTAX
To infer the metabolic potential of bacterial communities, 16S rRNA-based taxonomic profiles were analyzed using the FAPROTAX database. This approach allowed to assign putative ecological functions based on the known metabolism traits.
The most prevalent functional categories across all compartments were chemoheterotrophy and aerobic chemoheterotrophy, consistent with heterotrophic bacterial dominance (
Figure 6,
Figure 7,
Supplementary Table S2). These functions were particularly enriched in snow-associated samples such as Snow A (0.377 and 0.364, respectively) and Aged Snow A (0.346 and 0.335), followed by Snowmelt A (0.339 and 326), indicating the presence of metabolically active, cold-adapted heterotrophs capable of surviving harsh, nutrient-limited conditions. Aromatic compound degradation-associated taxa were detected in most compartments (Fig. 6) but were especially enriched in Sediment A (0.046), snow cannon filter A2 (0.038), and STP (0.037), suggesting possible retention or accumulation of pollutant-degrading taxa within infrastructure components. Fermentation was among the less abundant but environmentally relevant functions. Fermentative potential was the highest in cannon filter B (0.069) and sediment (0.065) (
Supplementary Table S2,
Figure 7) samples, suggesting that these compartments may be characterized by low-oxygen or anoxic conditions, promoting anaerobic metabolic processes [
51]. In contrast, surface water and fresh snow samples showed significantly lower values (below 0.015).
Interestingly, the predicted functional profiles revealed contrasts between the sampling sites. Snow A exhibited low number (only nine) of distinct functional groups, dominated by chemoheterotrophy and aerobic respiration, indicating a low-complexity microbial community shaped by strong selection pressures during snow production. In contrast, Sediment A and Cannon Filter B showed much higher functional richness (31 and 26 functions, respectively;
Supplementary Table S2), reflecting the capacity of these environments to accumulate and sustain metabolically diverse bacterial populations. On the other hand, Snow B had the highest relative abundance of taxa classified as “others” by FAPROTAX (68%), suggesting a prevalence of taxa not well-represented in current databases or possessing poorly annotated metabolic capabilities. This divergence may indicate microbial input from alternative sources (e.g., aerosols, biofilms), differential infrastructure contamination, or variability in snowmaking processes. Taken together, these findings highlight the heterogeneity of technical snow environments and their distinct roles in shaping microbial functional potential.
Overall, the FAPROTAX results revealed clear stage-specific functional profiles, indicating that snowmaking systems not only shape taxonomic community structure but also modulate microbial metabolic potential with implications for downstream biogeochemical cycling.
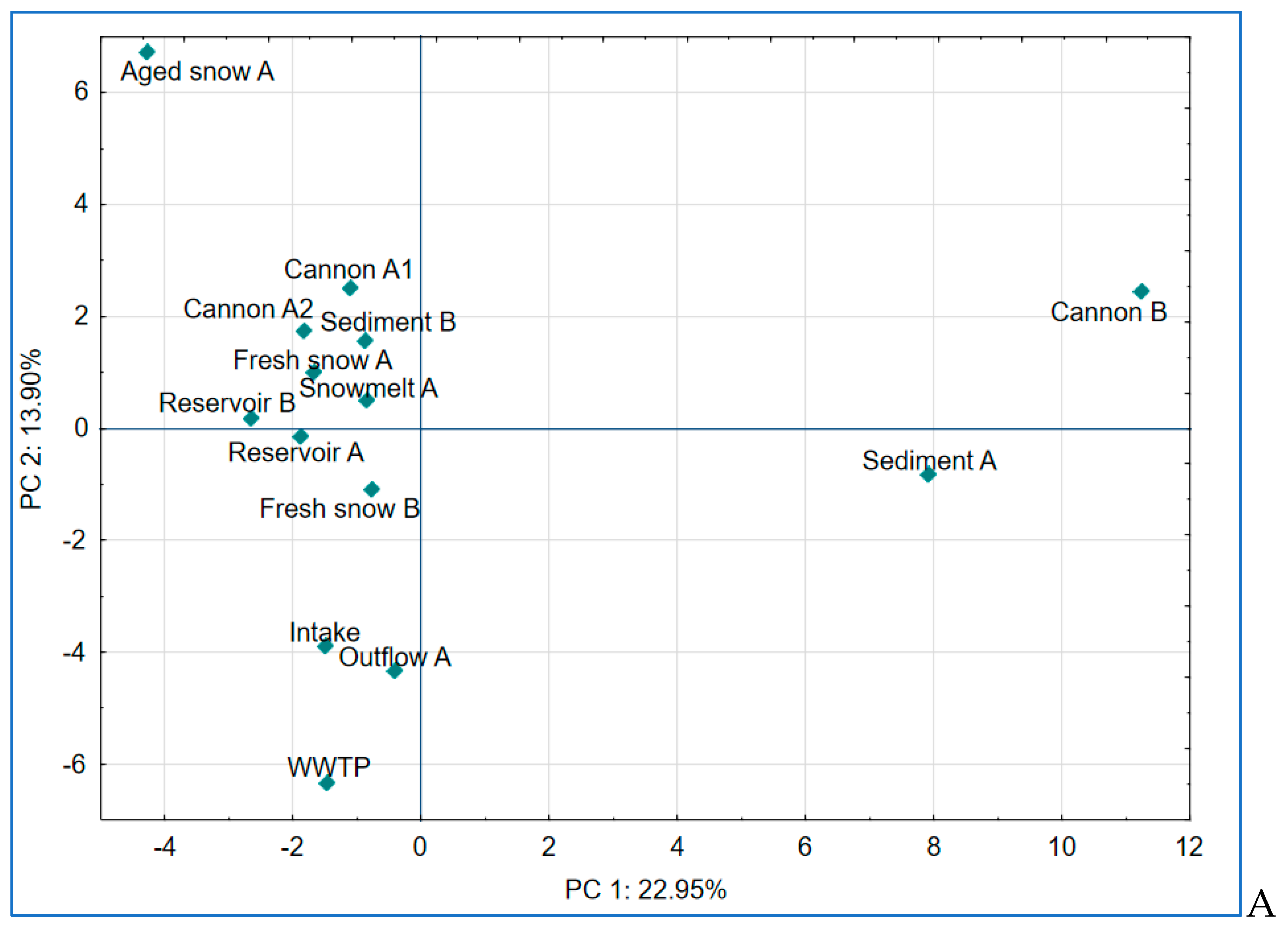
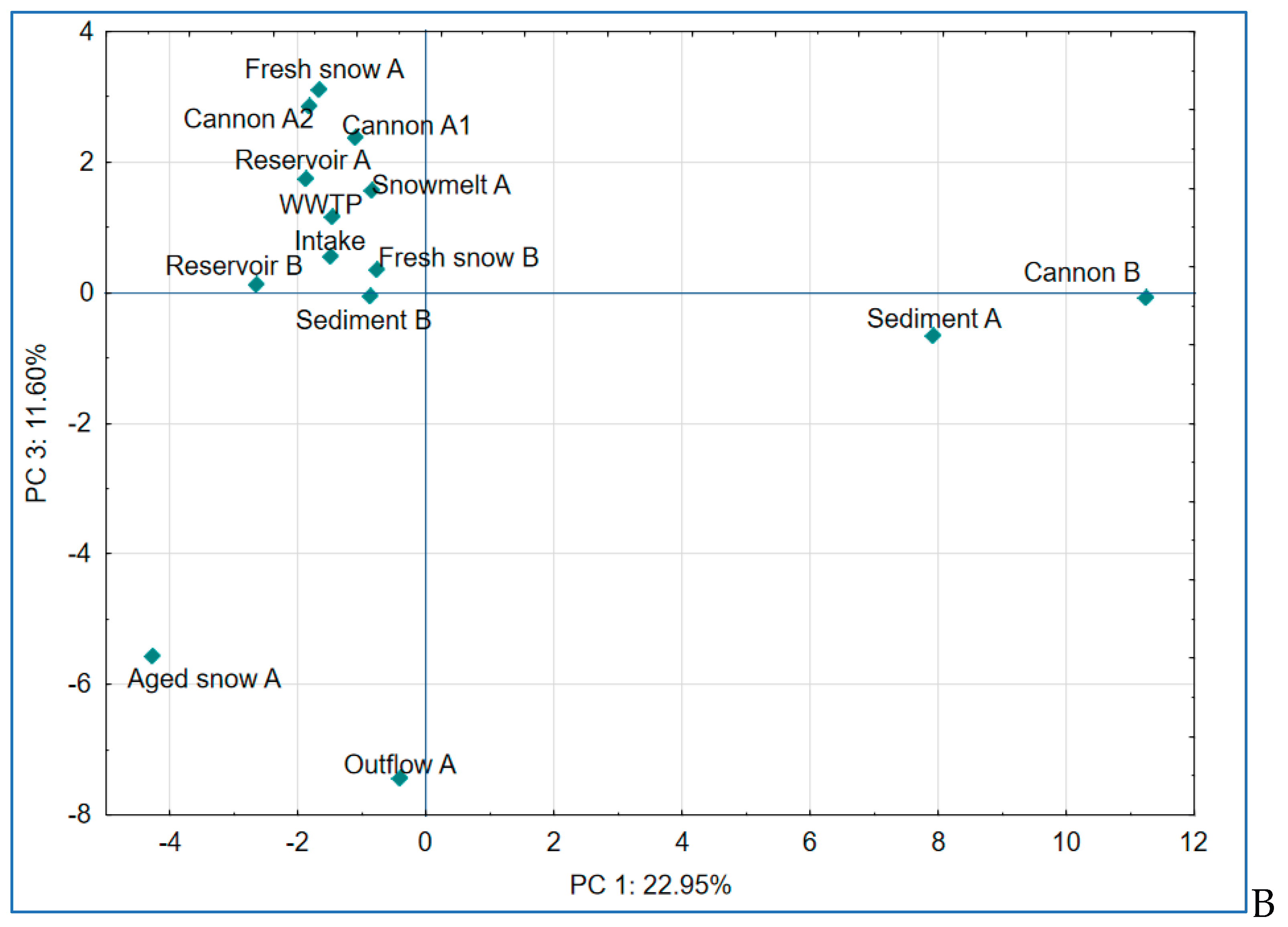
2.6. Principal Component Analysis of Microbial Communities
To further explore the dynamics in bacterial community composition, we conducted principal component analysis (PCA) on the relative abundance of dominant genera. The first three components explained 48.5% of the total variance (
Figure 8 A and B).
PC1 (explaining 22.95% of variance) reflected variation in anaerobic, sludge or sediment-associated communities with highest contributions from
Paludibacter,
Sporomusa and
Ruminiclostridium (members of Clostridia). These are anaerobic bacteria, typically dwelling in sediments or biofilm-associated environments (in our study: snow cannons and sediment) [
52,
53].
PC2 (13.90% of variance) separated samples influenced by biofilm-forming and cold-adapted taxa (i.e. Aged snow with taxa such as
Sphingomonas,
Dyadobacter) [
11,
54] from those dominated by treatment-resistant organisms such as
Candidatus Microthrix and
Caldilineaceae (samples WWTP, Intake, Outflow), suggesting a filtering effect during snow production and water treatment processes.
Finally, PC3 explained 11.60% of variance and reflected transitions possibly linked to site-specific microbial adaptations that occur in time. The effect presented by PC3 is mostly influenced by
Nocardia,
Rudanella and
Paenibacillus, soil-associated, organic matter-degrading taxa [
55,
56]. The genus
Paenibacillus has been also reported to dwell in nutrient-poor conditions, the trait that is shared with
Pedobacter, which also significantly contributed to PC3 and is typically described as psychrotolerant and oligotrophic bacterium [
57,
58].
The patterns described above are consistent with the suggestions we proposed earlier in this section, as well as with other studies that emphasize the selective pressure exerted on bacterial communities by technical processes and environmental conditions (including UV light disinfection, pressurized water spraying and freezing) in engineered systems [
4,
59]. They also underscore the ecological relevance of cold-tolerant and oligotrophic taxa in snow and glacier-like environments [
11,
31].
3. Materials and Methods
3.1. Site Description and Sample Collection
The collection of samples was conducted in one of the Polish ski resorts, in the Polish Carpathians. Due to a non-disclosure agreement with the facility operators, the exact location cannot be provided. The snowmaking infrastructure at this site includes water intake from a mountain river, water storage in technical reservoirs, and a snow cannon network distributed across ski slopes.
A total of fourteen samples were collected before, during and after the 2023/2024 snowmaking season (i.e. from November 2023 to April 2024) to represent the full spectrum of the technical snow production and transformation process. The samples included (
Table 3):
In all cases, the samples were collected in three instantaneous replications that formed the final pooled sample. The samples of water and sediment were collected into sets of autoclaved 1000 ml sterile polyethylene bottles, while snow samples were collected by first scratching the superficial layer, followed by the collection of snow with a snow corer (a 1.0 m-long, 10 cm-wide tube) into double sterile plastic string bags, where it melted. Then snowmelt water was transferred into sets of 1000 ml sterile polypropylene bottles and analyzed. The snow cannon biofilm and sediment was collected by scratching with sterile spatulas and swabs. The collected material was placed in 100 ml sterile Falcon tubes. Sediments were homogenized prior to processing.
3.2. Culture-Based Microbial Analysis of Samples
To assess the abundance of viable and potentially health-relevant bacteria, four microbial groups were enumerated in all samples: Escherichia coli, fecal enterococci (Enterococcus faecalis / E. faecium), coagulase-positive staphylococci and total mesophilic bacteria. The following culture media were used for each group: Tryptone Bile X-glucuronide (TBX) agar (Biomaxima, Lublin, Poland) for E. coli, Slanetz-Bartley agar (Biomaxima, Lublin, Poland) for enterococci, Baird-Parker agar (Biomaxima, Lublin, Poland) for staphylococci and Tryptic Soy Agar (TSA, Biomaxima, Lublin, Poland)) for total mesophilic heterotrophic bacteria.
For water and snowmelt water samples, E. coli and enterococci were quantified using the membrane filtration method, with 100 ml of each sample filtered through a 0.22 µm membrane filter (Sartorius, Germany) and transferred onto the respective media. Plates were incubated at 37 °C for enterococci and 44 °C for E. coli according to standard microbiological protocols. Staphylococci and heterotrophic mesophilic bacteria in liquid samples were assessed by the pour plate method, in which 1 ml of each sample was plated into sterile Petri dishes and overlaid with the appropriate medium. Incubation was carried out at 37 °C for 24–48 hours depending on the target group. Solid samples, including reservoir sediments and biofilms from snow cannon filters, were analyzed by preparing serial tenfold dilutions from 1 g of homogenized material in sterile saline solution. Subsequently, 1 ml of each dilution was plated onto the respective media using the pour plate technique.
Colonies characteristic of each group were counted and the results were expressed as the numbers of colony forming units (CFU) per ml or 100 ml of liquid samples or per gram for solid samples. All microbiological procedures were conducted under aseptic conditions to avoid cross-contamination.
3.3. DNA Extraction and 16S rRNA Sequencing
For molecular analysis of the total bacterial community, genomic DNA was extracted from all samples using standard protocols optimized for environmental matrices. For liquid samples (water and snowmelt), a volume of 500 ml was filtered through 0.22 µm sterile membrane filter (Sartorius, Germany). Filters were immediately transferred to sterile 60mm Petri plates and stored at –20 °C until DNA extraction. For solid samples (sediment and snow cannon biofilm), approximately 0.5 g of material was used for each extraction.
DNA extraction was performed using the Genomic Mini AX Bacteria + extraction kit (A&A Biotechnology, Gdańsk, Poland), following the manufacturer’s protocol. DNA was purified using Anty-Inhibitor Kit (A&A Biotechnology, Gdańsk, Poland) and DNA concentration was measured fluorometrically on a Qbit 4 Fluorometer (ThermoFisher Scientific, Waltham, WA, USA). Amplicon libraries targeting the hypervariable V3–V4 region of the 16S rRNA gene were prepared following the Illumina 16S Metagenomic Sequencing Library Preparation Guide (Part #15044223 Rev. B, Illumina, San Diego, CA, USA). A two-step PCR protocol was used for amplification, employing the following primers: forward primer
: 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′ and reverse primer 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′ [
60]. PCR reactions were performed using Herculase II Fusion DNA Polymerase and indexed with the Nextera XT Index Kit v2 (Agilent Technologies, Santa Clara, CA, USA). The resulting libraries were quality-checked and pooled before sequencing. Sequencing was conducted on the Illumina MiSeq platform (2 × 300 bp paired-end) at Macrogen Inc
. (Seoul, Republic of Korea).
3.4. Bioinformatics and Statistical Methods
The normality of data was assessed using the Shapiro–Wilk test. Since the distributions were approximately normal, parametric tests were applied in subsequent analyses. Differences in bacterial counts as well as functional traits between sample types and among individual samples were evaluated using one-way analysis of variance (ANOVA), followed by post-hoc comparisons using the least significant difference (LSD) method. A significance threshold of p < 0.05 was applied in all statistical tests.
Principal component analysis (PCA) was used to investigate relationships between culturable bacterial counts and the relative abundance of bacterial genera identified through Illumina sequencing. The number of components retained was determined using the Kaiser criterion, with factors exhibiting eigenvalues greater than 1.0 considered for interpretation. All statistical analyses were performed using Statistica software, version 13 (TIBCO Software Inc., Palo Alto, CA, USA).
Sequencing data targeting the V3–V4 region of the 16S rRNA gene were taxonomically classified by aligning reads against the Greengenes database (version 13; 97% similarity threshold, minimum score 40). Sequences were clustered into operational taxonomic units (OTUs) and assigned to taxonomic levels down to genus using the QIIME2 pipeline. To assess differences in bacterial community composition across samples, Bray–Curtis dissimilarity was calculated using relative abundance data of bacterial taxa. The resulting pairwise dissimilarity matrix was used to evaluate beta diversity between sample types. A clustered heatmap was generated with hierarchical clustering applied to rows (samples) based on Bray–Curtis distances. The relative abundance of dominant bacterial phyla and genera was calculated and visualized using R software (version 4.4.2) with the packages: vegan [
61], ggplot2 [
62] and pheatmap [
63].
Functional prediction was performed using the FAPROTAX database (Functional Annotation of Prokaryotic Taxa) as implemented in the Python script version [v.1.2.4]) [
16]. The normalized OTU table obtained after taxonomic assignment was used as a input. Functional annotations were inferred based on taxonomic affiliation and matched to ecological functions. The output was used to generate relative abundance plots to visualize predicted metabolic potentials across the sampling stages.
4. Conclusions
This study provides the first comprehensive molecular-level characterization of microbial communities across the full cycle of technical snow production and transformation from water intake to snowmelt water and post-season reservoir sediments. By integrating high-throughput 16S rRNA gene sequencing with functional prediction using FAPROTAX, we demonstrated that each stage of the snowmaking system – from treated wastewater that contaminates river intake, through reservoirs and snow cannons to snow deposited on the slopes and sediments that accumulate throughout the season – act as a distinct ecological and functional filter, progressively shaping the microbial communities and metabolic potential.
Our results highlight the dual role of technical snowmaking infrastructure. On one hand it can reduce microbial richness and create a selective environment that reduces many anthropogenic bacteria, but on the other – it is a temporary reservoir for biofilm-forming, cold-adapted and stress-tolerant taxa. Functional predictions revealed consistent dominance of aerobic chemoheterotrophic processes, while specialized functions such as methanol oxidation and fermentation were confined to specific compartments like sediments and cannon filters. Importantly, certain genera identified in sediments and snowmelt (e.g. Acinetobacter, Comamonas, Clostridium, Flavobacterium) may possess both environmental relevance and potential pathogenic or resistance-associated traits, highlighting the need for further genomic investigation.
Given that snowmelt, reservoir outflow and sediments may reintroduce these microbes into natural or semi-natural environments, our findings underscore the importance of molecular monitoring of microbial community dynamics and functional potentials not only from a public health perspective but also to understand possible long-term impacts of technical snow production processes on aquatic ecosystems.
Future studies could consider seasonal variation, antibiotic resistance genes, virulence factors, and compare natural versus artificial snow systems to better assess the ecological and public health consequences of using technical snowmaking in ski resorts and other engineered environments.
Supplementary Materials
The following supporting information can be downloaded at: Preprints.org, Table S1: Data values obtained from the analyses; Table S2: FAPROTAX results
Author Contributions
Conceptualization, AL-B., P.B. and K.S.; methodology, A.L.-B, P.B., B.G., K.B. and K.S.; software, B.G., K.B.; validation, A.L.-B., B.G. K.B. and K.S.; formal analysis, A.L-B. and P.B.; investigation, K.B., N.C. and A.R.; resources, A.L.-B., P.B. and B.G..; data curation, A.L-B.; writing—original draft preparation, A.L.-B.; writing—review and editing, P.B., B.G.,K.B., N.C. and A.R; visualization, A.L.-B. and P.B.; supervision, A.L-B.; project administration, A.L-B. and K.S..; funding acquisition, A.L.-B. and K.S. All authors have read and agreed to the published version of the manuscript.”
Funding
This research was funded by the statutory measures of the University of Agriculture in Kraków
Institutional Review Board Statement
Not applicable
Informed Consent Statement
Not applicable.
Data Availability Statement
The data are available on request from the corresponding author.
Acknowledgments
The Authors would like to thank PhD e.g. Piotr Boroń for his help in sample collection.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- François, H.; Samacoïts, R.; Bird, D.N.; Köberl, J.; Prettenthaler, F.; Morin, S. Climate Change Exacerbates Snow-Water-Energy Challenges for European Ski Tourism. Nature Climate Change 2023, 13, 935–942. [Google Scholar] [CrossRef]
- Lenart-Boroń, A.; Bojarczuk, A.; Żelazny, M. Impact of Construction and Functioning of a Newly Built Ski Slope on the Quality of Nearby Stream Water. Applied Sciences 2023, 13. [Google Scholar] [CrossRef]
- de Jong, C. Artificial Production of Snow. Encyclopedia of Earth Sciences Series 2011, Part 3, 61–66. [Google Scholar] [CrossRef]
- Stankiewicz, K.; Boroń, P.; Prajsnar, J.; Żelazny, M.; Heliasz, M.; Hunter, W.; Lenart-Boroń, A. Second Life of Water and Wastewater in the Context of Circular Economy - Do the Membrane Bioreactor Technology and Storage Reservoirs Make the Recycled Water Safe for Further Use? The Science of the total environment 2024, 921, 170995. [Google Scholar] [CrossRef] [PubMed]
- Stankiewicz, K.; Boroń, P.; Prajsnar, J.; Lenart-Boroń, A. Is Our Winter Experience Safe? Micropollutant Risks for Artificial Snowing. Science of The Total Environment 2025, 968, 178876. [Google Scholar] [CrossRef]
- Lagriffoul, A.; Boudenne, J.-L.; Absi, R.; Ballet, J.-J.; Berjeaud, J.-M.; Chevalier, S.; Creppy, E.E.; Gilli, E.; Gadonna, J.-P.; Gadonna-Widehem, P.; et al. Bacterial-Based Additives for the Production of Artificial Snow: What Are the Risks to Human Health? Science of The Total Environment 2010, 408, 1659–1666. [Google Scholar] [CrossRef]
- Rixen, C.; Stoeckli, V.; Ammann, W. Does Artificial Snow Production Affect Soil and Vegetation of Ski Pistes? A Review. Perspectives in Plant Ecology, Evolution and Systematics 2003, 5, 219–230. [Google Scholar] [CrossRef]
- Malard, L.A.; Šabacká, M.; Magiopoulos, I.; Mowlem, M.; Hodson, A.; Tranter, M.; Siegert, M.J.; Pearce, D.A. Spatial Variability of Antarctic Surface Snow Bacterial Communities. Frontiers in Microbiology 2019, Volume 10.
- Wunderlin, T.; Ferrari, B.; Power, M. Global and Local-Scale Variation in Bacterial Community Structure of Snow from the Swiss and Australian Alps. FEMS Microbiology Ecology 2016, 92, fiw132. [Google Scholar] [CrossRef]
- Keuschnig, C.; Vogel, T.M.; Barbaro, E.; Spolaor, A.; Koziol, K.; Björkman, M.P.; Zdanowicz, C.; Gallet, J.-C.; Luks, B.; Layton, R.; et al. Selection Processes of Arctic Seasonal Glacier Snowpack Bacterial Communities. Microbiome 2023, 11, 35. [Google Scholar] [CrossRef]
- Tighe, S.W.; Vellone, D.L.; Tracy, K.M.; Lynch, D.B.; Finstad, K.H.; Mcllelan, M.C.; Dragon, J.A. Microbiome and Microbial Profiling of Arctic Snow Using Whole Genome Sequencing, Psychrophilic Culturing, and Novel Sampling Techniques. Journal of biomolecular techniques : JBT 2025, 36. [Google Scholar] [CrossRef] [PubMed]
- Baloh, P.; Els, N.; David, R.O.; Larose, C.; Whitmore, K.; Sattler, B.; Grothe, H. Assessment of Artificial and Natural Transport Mechanisms of Ice Nucleating Particles in an Alpine Ski Resort in Obergurgl, Austria. Frontiers in microbiology 2019, 10, 2278. [Google Scholar] [CrossRef] [PubMed]
- Casagrande Bacchiocchi, S.; Zerbe, S.; Cavieres, L.A.; Wellstein, C. Impact of Ski Piste Management on Mountain Grassland Ecosystems in the Southern Alps. Science of The Total Environment 2019, 665, 959–967. [Google Scholar] [CrossRef]
- Křenová, Z.; Bílek, O.; Zýval, V. Does Artificial Snow Fertilise the Soil of Mountain Meadows in the Krkonoše National Park? European Journal of Environmental Sciences 2020, 10, 32–41. [Google Scholar] [CrossRef]
- G., L.A.; Marica, Z.; Chiara, D.; Federico, R.; Davide, R.; Manuela, M.; Giorgio, G.; Giacomo, M.; Simone, G.; Luca, B.; et al. QIIME2 Enhances Multi-Amplicon Sequencing Data Analysis: A Standardized and Validated Open-Source Pipeline for Comprehensive 16S RRNA Gene Profiling. Microbiology Spectrum 2025, 0, e01673-25. [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling Function and Taxonomy in the Global Ocean Microbiome. Science (New York, N.Y.) 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Leho, T.; Mads, A.; Sten, A.; Benjamin, C. Perspectives and Benefits of High-Throughput Long-Read Sequencing in Microbial Ecology. Applied and Environmental Microbiology 2021, 87, e00626–21. [Google Scholar] [CrossRef]
- Christner, B.C.; Mosley-Thompson, E.; Thompson, L.G.; Zagorodnov, V.; Sandman, K.; Reeve, J.N. Recovery and Identification of Viable Bacteria Immured in Glacial Ice. Icarus 2000, 144, 479–485. [Google Scholar] [CrossRef]
- Carpenter, E.J.; Lin, S.; Capone, D.G. Bacterial Activity in South Pole Snow. Applied and environmental microbiology 2000, 66, 4514–4517. [Google Scholar] [CrossRef]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic Identification and in Situ Detection of Individual Microbial Cells without Cultivation. Microbiological Reviews 1995, 59, 143–169. [Google Scholar] [CrossRef]
- Martiny, A.C. High Proportions of Bacteria Are Culturable across Major Biomes. The ISME Journal 2019, 13, 2125–2128. [Google Scholar] [CrossRef]
- Meziti, A.; Tsementzi, D.; Ar. Kormas, K.; Karayanni, H.; Konstantinidis, K.T. Anthropogenic Effects on Bacterial Diversity and Function along a River-to-Estuary Gradient in Northwest Greece Revealed by Metagenomics. Environmental Microbiology 2016, 18, 4640–4652. [Google Scholar] [CrossRef]
- Ríos-Castillo, A.G.; Thompson, K.D.; Adams, A.; de Mateo, M.; Rodríguez-Jerez, J.J. Biofilm Formation of \textit{Flavobacterium Psychrophilum} on Various Substrates. Aquaculture Research 2018, 49, 3830–3837. [Google Scholar] [CrossRef]
- Monaco, P.; Divino, F.; Naclerio, G.; Bucci, A. Microbial Community Analysis with a Specific Statistical Approach after a Record Breaking Snowfall in Southern Italy. Annals of Microbiology 2020, 70, 63. [Google Scholar] [CrossRef]
- Sorensen, P.O.; Beller, H.R.; Bill, M.; Bouskill, N.J.; Hubbard, S.S.; Karaoz, U.; Polussa, A.; Steltzer, H.; Wang, S.; Williams, K.H.; et al. The Snowmelt Niche Differentiates Three Microbial Life Strategies That Influence Soil Nitrogen Availability During and After Winter. Frontiers in microbiology 2020, 11, 871. [Google Scholar] [CrossRef]
- Maccario, L.; Carpenter, S.D.; Deming, J.W.; Vogel, T.M.; Larose, C. Sources and Selection of Snow-Specific Microbial Communities in a Greenlandic Sea Ice Snow Cover. Scientific Reports 2019, 9, 2290. [Google Scholar] [CrossRef]
- Zhao, Z.; Sun, W.; Ke, Y.; Zhang, Y.; Wang, X. Microbial Dynamics at Different Stages of Drinking Water Treatment Systems. Environmental Science: Water Research & Technology 2025, 11, 1401–1427. [Google Scholar] [CrossRef]
- Kauser, I.; Ciesielski, M.; Poretsky, R.S. Ultraviolet Disinfection Impacts the Microbial Community Composition and Function of Treated Wastewater Effluent and the Receiving Urban River. PeerJ 2019, 7, e7455. [Google Scholar] [CrossRef]
- Harding, T.; Jungblut, A.D.; Lovejoy, C.; Vincent, W.F. Microbes in High Arctic Snow and Implications for the Cold Biosphere. Applied and environmental microbiology 2011, 77, 3234–3243. [Google Scholar] [CrossRef]
- Volkert, M.; Ananta, E.; Luscher, C.; Knorr, D. Effect of Air Freezing, Spray Freezing, and Pressure Shift Freezing on Membrane Integrity and Viability of Lactobacillus Rhamnosus GG. Journal of Food Engineering 2008, 87, 532–540. [Google Scholar] [CrossRef]
- Zhang, D.-C.; Busse, H.-J.; Liu, H.-C.; Zhou, Y.-G.; Schinner, F.; Margesin, R. Sphingomonas Glacialis Sp. Nov., a Psychrophilic Bacterium Isolated from Alpine Glacier Cryoconite. International Journal of Systematic and Evolutionary Microbiology 2011, 61, 587–591. [Google Scholar] [CrossRef] [PubMed]
- Bowers, R.M.; McLetchie, S.; Knight, R.; Fierer, N. Spatial Variability in Airborne Bacterial Communities across Land-Use Types and Their Relationship to the Bacterial Communities of Potential Source Environments. The ISME Journal 2011, 5, 601–612. [Google Scholar] [CrossRef]
- Moncada, C.; Hassenrück, C.; Gärdes, A.; Conaco, C. Microbial Community Composition of Sediments Influenced by Intensive Mariculture Activity. FEMS Microbiology Ecology 2019, 95, fiz006. [Google Scholar] [CrossRef] [PubMed]
- Kragh, K.N.; Tolker-Nielsen, T.; Lichtenberg, M. The Non-Attached Biofilm Aggregate. Communications Biology 2023, 6, 898. [Google Scholar] [CrossRef]
- Hervé, V.; Morelle, J.; Lambourdière, J.; Lopez, P.J.; Claquin, P. Together throughout the Year: Seasonal Patterns of Bacterial and Eukaryotic Microbial Communities in a Macrotidal Estuary. Environmental Microbiome 2025, 20, 8. [Google Scholar] [CrossRef]
- Villeneuve, K.; Turcotte-Blais, V.; Lazar, C.S. Effect of Snowmelt on Groundwater Bacterial Community Composition and Potential Role of Surface Environments as Microbial Seed Bank in Two Distinct Aquifers from the Region of Quebec, Canada. Microorganisms 2023, 11. [Google Scholar] [CrossRef]
- Guo, B.; Liu, Y.; Gu, Z.; Shen, L.; Liu, K.; Wang, N.; Xing, T.; Liu, H.; Zhou, Y.; Li, J. Massilia Psychrophila Sp. Nov., Isolated from an Ice Core. International journal of systematic and evolutionary microbiology 2016, 66, 4088–4093. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, A.; Antony, R.; Samui, G.; Thamban, M. Microbial Communities and Their Potential for Degradation of Dissolved Organic Carbon in Cryoconite Hole Environments of Himalaya and Antarctica. Microbiological Research 2018, 208, 32–42. [Google Scholar] [CrossRef]
- Slobodkin, A.I.; Tourova, T.P.; Kostrikina, N.A.; Chernyh, N.A.; Bonch-Osmolovskaya, E.A.; Jeanthon, C.; Jones, B.E. Tepidibacter Thalassicus Gen. Nov., Sp. Nov., a Novel Moderately Thermophilic, Anaerobic, Fermentative Bacterium from a Deep-Sea Hydrothermal Vent. International journal of systematic and evolutionary microbiology 2003, 53, 1131–1134. [Google Scholar] [CrossRef]
- Pulschen, A.A.; Bendia, A.G.; Fricker, A.D.; Pellizari, V.H.; Galante, D.; Rodrigues, F. Isolation of Uncultured Bacteria from Antarctica Using Long Incubation Periods and Low Nutritional Media. Frontiers in microbiology 2017, 8, 1346. [Google Scholar] [CrossRef]
- Ray, A.E.; Connon, S.A.; Neal, A.L.; Fujita, Y.; Cummings, D.E.; Ingram, J.C.; Magnuson, T.S. Metal Transformation by a Novel Pelosinus Isolate From a Subsurface Environment. Frontiers in Microbiology 2018, Volume 9-.
- Lenart-Boroń, A.; Boroń, P.; Kulik, K.; Prajsnar, J.; Żelazny, M.; Chmiel, M.J. Anthropogenic Pollution Gradient along a Mountain River Affects Bacterial Community Composition and Genera with Potential Pathogenic Species. Scientific Reports 2022, 12, 1–14. [Google Scholar] [CrossRef]
- Harb, M.; Hong, P.Y. Molecular-Based Detection of Potentially Pathogenic Bacteria in Membrane Bioreactor (MBR) Systems Treating Municipal Wastewater: A Case Study. Environmental Science and Pollution Research 2017, 24, 5370–5380. [Google Scholar] [CrossRef] [PubMed]
- Ryan, M.P.; Sevjahova, L.; Gorman, R.; White, S. The Emergence of the Genus Comamonas as Important Opportunistic Pathogens. Pathogens (Basel, Switzerland) 2022, 11. [Google Scholar] [CrossRef]
- Ryan, M.P.; Pembroke, J.T. Brevundimonas Spp: Emerging Global Opportunistic Pathogens. Virulence 2018, 9, 480–493. [Google Scholar] [CrossRef]
- Hugouvieux-Cotte-Pattat, N.; Condemine, G.; Gueguen, E.; Shevchik, V.E. Dickeya Plant Pathogens. In eLS; 2020; pp. 1–10 ISBN 9780470015902.
- Zaheen, Z.; War, A.F.; Ali, S.; Yatoo, A.M.; Ali, M.N.; Ahmad, S.B.; Rehman, M.U.; Paray, B.A. Chapter 7 - Common Bacterial Infections Affecting Freshwater Fish Fauna and Impact of Pollution and Water Quality Characteristics on Bacterial Pathogenicity. In Bacterial Fish Diseases; Dar, G.H., Bhat, R.A., Qadri, H., Al-Ghamdy, K.M., Hakeem, K.R.B.T.-B.F.D., Eds.; Academic Press, 2022; pp. 133–154 ISBN 978-0-323-85624-9.
- Loch, T.P.; Faisal, M. Emerging Flavobacterial Infections in Fish: A Review. Journal of advanced research 2015, 6, 283–300. [Google Scholar] [CrossRef]
- Lawson, P.A.; Citron, D.M.; Tyrrell, K.L.; Finegold, S.M. Reclassification of Clostridium Difficile as Clostridioides Difficile (Hall and O’Toole 1935) Prévot 1938. Anaerobe 2016, 40, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, D.; McGowan, A.M.; Gibson, J.S.; Lanyon, J.M.; Horsman, S.; Seddon, J.M. Faecal Bacterial Communities Differ amongst Discrete Foraging Populations of Dugongs along the East Australian Coast. FEMS Microbiology Ecology 2024, 100, fiae051. [Google Scholar] [CrossRef] [PubMed]
- Morris, B.E.L.; Henneberger, R.; Huber, H.; Moissl-Eichinger, C. Microbial Syntrophy: Interaction for the Common Good. FEMS Microbiology Reviews 2013, 37, 384–406. [Google Scholar] [CrossRef] [PubMed]
- Wörner, S.; Pester, M. Microbial Succession of Anaerobic Chitin Degradation in Freshwater Sediments. Applied and environmental microbiology 2019, 85. [Google Scholar] [CrossRef]
- Fang, H.; Chen, Y.; Huang, L.; He, G. Analysis of Biofilm Bacterial Communities under Different Shear Stresses Using Size-Fractionated Sediment. Scientific reports 2017, 7, 1299. [Google Scholar] [CrossRef]
- Kumar, S.; Deep Chandra, S.; Mamta, B.; and Goel, R. Plant Growth Promoting Potential of Psychrotolerant Dyadobacter Sp. for Pulses and Finger Millet and Impact of Inoculation on Soil Chemical Properties and Diazotrophic Abundance. Journal of Plant Nutrition 2018, 41, 1035–1046. [Google Scholar] [CrossRef]
- Wanger, A.; Chavez, V.; Huang, R.S.P.; Wahed, A.; Actor, J.K.; Dasgupta, A. Chapter 6 - Overview of Bacteria. In; Wanger, A., Chavez, V., Huang, R.S.P., Wahed, A., Actor, J.K., Dasgupta, A.B.T.-M. and M.D. in P., Eds.; Elsevier, 2017; pp. 75–117 ISBN 978-0-12-805351-5.
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.-C. Current Knowledge and Perspectives of Paenibacillus: A Review. Microbial Cell Factories 2016, 15, 203. [Google Scholar] [CrossRef]
- Dahal, R.H.; Chaudhary, D.K.; Kim, D.-U.; Kim, J. Nine Novel Psychrotolerant Species of the Genus Pedobacter Isolated from Arctic Soil with Potential Antioxidant Activities. International Journal of Systematic and Evolutionary Microbiology 2020, 70, 2537–2553. [Google Scholar] [CrossRef] [PubMed]
- Osman, J.R.; Dubow, M.S. Bacterial Communities on the Surface of Oligotrophic (Nutrient-Poor) Soils. Current Topics in Biotechnology 2019, 9, hal–02067677. [Google Scholar]
- Saini, S.; Tewari, S.; Dwivedi, J.; Sharma, V. Biofilm-Mediated Wastewater Treatment: A Comprehensive Review. Materials Advances 2023, 4, 1415–1443. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic acids research 2013, 41, e1. [Google Scholar] [CrossRef]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; Wagner, H.; Barbour, M.; Bedward, M.; Bolker, B.; et al. Package ‘ Vegan .’ 2025.
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; 2nd ed.; Springer International Publishing: Cham, Switzerland, 2016. [Google Scholar]
- Kolde, R. Package “Pheatmap”: Pretty Heatmaps. Version 1.0.12 2019, 1–8. [Google Scholar]
Figure 1.
Comparison of total culturable bacterial counts (CFU) and OTU richness across various sampling sites involved in technical snow production.
Figure 1.
Comparison of total culturable bacterial counts (CFU) and OTU richness across various sampling sites involved in technical snow production.
Figure 2.
Clustered heatmap of Bray-Curtis dissimilarity values among bacterial across all sampling sites in the technical snow production system. Bray–Curtis dissimilarities were calculated based on relative abundance data of 50 most abundant bacterial taxa. Lower values (purple) indicate higher community similarity, while higher values (yellow) represent greater dissimilarity. Hierarchical clustering was applied to rows (samples) to reveal community structure groupings.
Figure 2.
Clustered heatmap of Bray-Curtis dissimilarity values among bacterial across all sampling sites in the technical snow production system. Bray–Curtis dissimilarities were calculated based on relative abundance data of 50 most abundant bacterial taxa. Lower values (purple) indicate higher community similarity, while higher values (yellow) represent greater dissimilarity. Hierarchical clustering was applied to rows (samples) to reveal community structure groupings.
Figure 3.
Relative abundance of ten dominant bacterial phyla in samples collected from various stages of technical snow production and the related aquatic systems. Each bar represents the taxonomic composition at the phylum level, based on 16S rRNA gene sequencing, expressed as percentage of total reads per sample. The graph presents ten most abundant taxa, while “others” represent the group of less abundant ones.
Figure 3.
Relative abundance of ten dominant bacterial phyla in samples collected from various stages of technical snow production and the related aquatic systems. Each bar represents the taxonomic composition at the phylum level, based on 16S rRNA gene sequencing, expressed as percentage of total reads per sample. The graph presents ten most abundant taxa, while “others” represent the group of less abundant ones.
Figure 4.
Relative abundance of ten dominant bacterial orders in samples collected from various stages of technical snow production and the related aquatic systems. Each bar represents the taxonomic composition at the order level based on 16S rRNA gene sequencing, expressed as percentage of total reads per sample. The graph presents ten most abundant taxa, while “others” represent the group of less abundant ones.
Figure 4.
Relative abundance of ten dominant bacterial orders in samples collected from various stages of technical snow production and the related aquatic systems. Each bar represents the taxonomic composition at the order level based on 16S rRNA gene sequencing, expressed as percentage of total reads per sample. The graph presents ten most abundant taxa, while “others” represent the group of less abundant ones.
Figure 5.
Heatmap showing the log₁₀-transformed relative abundance of selected bacterial taxa across different sampling sites involved in the technical snow production cycle. The analyzed taxa represent ecologically or functionally distinct groups, including these typical of wastewater (e.g. Candidatus Microthrix, Enterobacteriaceae), cold-adapted or oligotrophic bacteria (e.g. Sphingomonas, Lapillicoccus), and genera associated with sediments or biofilms (e.g. Comamonas, Brevundimonas). The scale bar indicates the relative abundance values after logarithmic transformation. Sampling sites include treated wastewater discharge site (WWTP), river water intake for technical snowmaking, water storage reservoirs (A and B), snow cannon filters (A1, A2, B), fresh and aged snow samples, snowmelt, and post-season sediments.
Figure 5.
Heatmap showing the log₁₀-transformed relative abundance of selected bacterial taxa across different sampling sites involved in the technical snow production cycle. The analyzed taxa represent ecologically or functionally distinct groups, including these typical of wastewater (e.g. Candidatus Microthrix, Enterobacteriaceae), cold-adapted or oligotrophic bacteria (e.g. Sphingomonas, Lapillicoccus), and genera associated with sediments or biofilms (e.g. Comamonas, Brevundimonas). The scale bar indicates the relative abundance values after logarithmic transformation. Sampling sites include treated wastewater discharge site (WWTP), river water intake for technical snowmaking, water storage reservoirs (A and B), snow cannon filters (A1, A2, B), fresh and aged snow samples, snowmelt, and post-season sediments.
Figure 6.
Relative abundance of predicted bacterial functional groups across sample categories, based on FAPROTAX analysis. Samples were grouped into functional stages of technical snow production: wastewater treatment plant (STP), intake water, reservoirs, snow (fresh, aged), snowmelt, cannon filters, outflow, and sediments. The most abundant functions include chemoheterotrophy and aerobic chemoheterotrophy. The category “other” includes less specific or unassigned functions.
Figure 6.
Relative abundance of predicted bacterial functional groups across sample categories, based on FAPROTAX analysis. Samples were grouped into functional stages of technical snow production: wastewater treatment plant (STP), intake water, reservoirs, snow (fresh, aged), snowmelt, cannon filters, outflow, and sediments. The most abundant functions include chemoheterotrophy and aerobic chemoheterotrophy. The category “other” includes less specific or unassigned functions.
Figure 7.
Site-specific distribution of ten most prevalent FAPROTAX-predicted functions. Each bar represents a single sampling site, showing the relative contribution of the most dominant metabolic functions.
Figure 7.
Site-specific distribution of ten most prevalent FAPROTAX-predicted functions. Each bar represents a single sampling site, showing the relative contribution of the most dominant metabolic functions.
Figure 8.
Results of principal component analysis (PCA) based on bacterial community composition at the genus or the closest level across all sampled stages of the examined technical snow production system. (A) PCA plot showing separation along principal components PC 1 and PC 2, which explain 22.95% and 13.90% of the total variance, respectively; (B) PCA plot showing separation along principal components PC 1 and PC 3, explaining 22.95% and 11.60% of the total variance, respectively.
Figure 8.
Results of principal component analysis (PCA) based on bacterial community composition at the genus or the closest level across all sampled stages of the examined technical snow production system. (A) PCA plot showing separation along principal components PC 1 and PC 2, which explain 22.95% and 13.90% of the total variance, respectively; (B) PCA plot showing separation along principal components PC 1 and PC 3, explaining 22.95% and 11.60% of the total variance, respectively.
Table 1.
Diversity indices calculated for the bacterial community composition across samples collected throughout the technical snow production system. The indices include: Shannon diversity (H), Simpson diversity (Ds), Simpson’s dominance (λ), Pielou diversity (PIE) and Pielou evenness (J), Margalef richness (Dm) and observed species richness (number of OTUs). A heatmap-style coloring scheme was used, with shades of blue indicating lower values and shades of red indicating higher values for each diversity metric.
Table 1.
Diversity indices calculated for the bacterial community composition across samples collected throughout the technical snow production system. The indices include: Shannon diversity (H), Simpson diversity (Ds), Simpson’s dominance (λ), Pielou diversity (PIE) and Pielou evenness (J), Margalef richness (Dm) and observed species richness (number of OTUs). A heatmap-style coloring scheme was used, with shades of blue indicating lower values and shades of red indicating higher values for each diversity metric.
| |
H |
Ds |
λ |
PIE |
J |
Dm |
Species richness |
| WWTP |
1.753 |
0.961 |
0.039 |
0.961 |
0.709 |
62.682 |
296 |
| Intake |
1.833 |
0.967 |
0.033 |
0.967 |
0.731 |
68.267 |
321 |
| Reservoir A |
1.402 |
0.927 |
0.073 |
0.927 |
0.646 |
31.775 |
148 |
| Reservoir B |
1.211 |
0.851 |
0.149 |
0.851 |
0.592 |
24.424 |
111 |
| Fresh snow A |
1.086 |
0.804 |
0.196 |
0.804 |
0.603 |
13.755 |
63 |
| Fresh snow B |
0.878 |
0.598 |
0.402 |
0.598 |
0.399 |
33.808 |
159 |
| Old snow A |
1.367 |
0.926 |
0.074 |
0.926 |
0.690 |
21.013 |
96 |
| Outflow |
1.403 |
0.886 |
0.114 |
0.886 |
0.649 |
31.197 |
145 |
| Snowmelt |
1.300 |
0.856 |
0.144 |
0.856 |
0.597 |
32.467 |
151 |
| Cannon filter A1 |
1.415 |
0.887 |
0.113 |
0.887 |
0.628 |
38.265 |
179 |
| Cannon filter A2 |
1.288 |
0.883 |
0.117 |
0.883 |
0.599 |
29.954 |
141 |
| Cannon filter B |
1.831 |
0.974 |
0.026 |
0.974 |
0.792 |
43.557 |
205 |
| Sediment A |
1.688 |
0.959 |
0.041 |
0.959 |
0.757 |
36.213 |
169 |
| sediment B |
1.592 |
0.934 |
0.066 |
0.934 |
0.667 |
50.499 |
243 |
Table 2.
Key bacterial taxa characteristic of individual snow production and post-production stages.
Table 2.
Key bacterial taxa characteristic of individual snow production and post-production stages.
| Genus |
Typical Habitat |
Ecological/Functional Remarks |
Sampling Site of Highest Relative Abundance |
Max. Observed Abundance |
| Candidatus Microthrix |
activated sludge systems; wastewater treatment plants |
associated with sludge bulking and foaming issues |
WWTP |
1.67 |
| Enterobacteriaceae |
wastewater, wastewater treatment plants |
typical indicators of wastewater contamination |
WWTP |
1.58 |
| Nocardia |
soil, water, nutrient-poor environments |
survives nutrient-poor conditions, opportunistic pathogens |
Outflow Reservoir A |
31.05 |
| Acinetobacter |
various environments |
includes potential pathogens, biofilm forming |
Sediment A |
7.43 |
| Dickeya |
wastewater, wastewater treatment plants |
environmental bacterium, frequent plant pathogen |
WWTP |
7.95 |
| Comamonas |
carbon-poor aquitard sediments |
adaptability to nutrient-limited environments, opportunistic pathogen |
Sediment A |
13.51 |
| Massilia |
glacial meltwater |
early colonizers in the rhizosphere |
Snow cannon filter A2 |
5.80 |
| Rhizobium |
agricultural runoff, plants |
biofilm formation, nitrogen fixing |
Snow B |
62.91 |
| Brevundimonas |
oligotrophic environments |
biofilm formation, opportunistic pathogen |
Snow cannon filter A2 |
4.51 |
| Lapillicoccus |
harsh, nutrient-poor, extreme environments |
isolated from stone surfaces; aerobic and mesophilic organisms |
Aged snow A |
2.14 |
| Sphingomonas |
glacier cryoconite, Arctic environments |
psychrophilic, cold-adapted, increases plant resistance to pathogens |
Aged snow A |
5.61 |
| Rudanella |
activated sludge systems |
potentially involved in organic matter degradation |
Outflow reservoir A |
2.74 |
| Pelosinus |
subsurface and sedimentary environments |
anaerobic |
Snowmelt A |
2.02 |
| Xanthomonadaceae |
biofilm, contaminated source water |
biofilm formation |
Snow cannon filter A1 |
4.99 |
| Tepidibacter |
anaerobic conditions |
anaerobic |
Snow cannon filter B |
6.08 |
| Bacteroides |
wildlife feces |
fecal indicator |
Sediment reservoir A |
6.18 |
| Macellibacteroides |
wildlife feces |
fecal indicator |
Sediment reservoir A |
3.88 |
|
Clostridium sensu stricto 1 and 13
|
soils, organic rich sediments, |
spore-forming, can survive disinfection, includes pathogens |
Snowmelt / Sediment reservoir A |
1.21 / 2.4 |
| Methylophilaceae |
stratified water bodies |
methylotrophic bacteria |
Sediment reservoir B |
3.84 |
| Flavobacterium |
environments rich in organic substrates; water |
biofilm formation, fish pathogen, algicidal, biopolymer decomposer, opportunistic pathogen |
Snow A |
41.55 |
| Polymorphobacter |
cold, oligotrophic environments |
adaptability to extreme conditions |
Sediment A |
2.00 |
Table 3.
The samples collected for the analysis.
Table 3.
The samples collected for the analysis.
| No |
Sample characteristics |
Sample code |
| Water sources |
| 1 |
Treated wastewater outflow from a local municipal treatment plant |
WWTP |
| 2 |
River water from the main snowmaking intake |
Intake |
| 3 |
Water from two technical reservoirs used to store water prior to snow production; Reservoirs are equipped with water aeration and UV light disinfection systems |
Reservoir A |
| 4 |
Reservoir B |
| Snow production system |
| 5 |
Biofilm or debris from three snow cannon filters
|
Cannon A1 |
| 6 |
Cannon A2 |
| 7 |
Cannon B |
| 8 |
Two samples of freshly produced technical snow
|
Fresh snow A |
| 9 |
Fresh snow B |
| Post-deposition samples |
| 10 |
Aged snow collected from the ski slope A c.a. 2 months post deposition (February) |
Aged snow A |
| 11 |
Snowmelt water from aged snow patches, collected in April |
Snowmelt |
| 12 |
Reservoir (A) water outflow collected at the end of the snowmaking season (April) |
Outflow A |
| 13 |
Sediments from the bottom of both technical reservoirs, exposed after post-season drainage of water |
Sediment A |
| 14 |
Sediment B |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).