1. Introduction
1.1. Genomics and Protein Structure Prediction: A Unified Frontier Enabled by Deep Learning
Computational biology combines advanced computing with biological research to explore complex living systems, particularly in genomics and protein structure prediction [1]. Within this interdisciplinary realm, genomics and protein structure prediction represent two pivotal, yet intrinsically linked, areas of research. The journey from genetic information encoded in DNA to the functional machinery of proteins is a central dogma of molecular biology: DNA is transcribed into RNA, which is then translated into protein sequences. The linear sequence of amino acids in a protein subsequently folds into a unique 3D structure, which in turn determines its biological activity. Understanding this intricate flow of information from gene to protein structure to function is paramount for advancing our knowledge of biological systems and developing novel therapeutic interventions. This review combines genomics and protein structure prediction into a single, cohesive narrative due to their inherent biological interconnectedness and the synergistic role AI and ML plays in bridging these domains. The rationale is rooted in the central dogma of molecular biology: genomic information (DNA/RNA sequences) directly encodes the amino acid sequences of proteins, and these sequences, in turn, determine the protein's three-dimensional structure, which is crucial for its function. AI and ML provide the computational framework to traverse this biological pathway, enabling a holistic understanding of biological systems from the genetic blueprint to the functional molecular machinery. By treating these fields jointly, we can better illustrate how advancements in one area, driven by deep learning, often directly impact and accelerate progress in the other, leading to a more comprehensive and integrated view of biological processes and disease mechanisms. Since the 1990s, machine learning has evolved from basic neural networks analysing gene expression data to sophisticated deep learning algorithms. Models such as convolutional neural networks (CNNs), recurrent neural networks (RNNs), and transformers now detect complex patterns in genomic and proteomic datasets, enabling accurate predictions [2].
In 2015, researchers from Harvard and MIT developed DeepBind, a groundbreaking deep learning algorithm that identifies RNA-binding protein sites, revealing previously unknown regulatory elements in the genome [3]. Scientists increasingly rely on such algorithms to address biological challenges, from predicting protein structures to identifying disease-causing mutations. For example, DeepMind’s AlphaFold uses advanced neural networks to accurately predict proteins’ three-dimensional structures, opening new frontiers in structural biology [38]. These advancements have driven significant progress in genomics, medical diagnosis, and drug discovery. The use of AI and ML in computational biology has resulted in noteworthy breakthroughs spanning diverse niches, like genomics, medical diagnosis, and drug discovery. AI enables precise analysis of genomic data, identifying disease-causing mutations and supporting the development of personalized treatments. It also predicts functional pathways for new drugs, streamlining target identification and reducing reliance on trial-and-error experiments. By analyzing vast genomic, proteomic, and other biological datasets, deep learning uncovers subtle patterns often missed by traditional statistical methods, enhancing our understanding of biological systems.
The growing demand for personalized medicine and efficient drug discovery drives the adoption of AI in life sciences [4]. However, challenges remain. AI algorithms require large, high-quality datasets, which can be scarce in some biological fields [3]. Additionally, interpreting their results is complex, as they detect subtle patterns that may not align with traditional biological models.
Despite these challenges, AI has the potential to transform computational biology by deepening our understanding of biological systems and improving healthcare outcomes. This review explores its applications, addresses associated challenges, and highlights key advancements, such as AlphaFold and DeepBind, and their potential impact on personalized medicine and drug discovery in the coming years.
1.2. Brief History and Evolution of Deep Learning
The journey of deep learning, from its theoretical origins to its current state as a transformative technology, is marked by periods of intense research and significant breakthroughs. Rina Dechter introduced the term "deep learning" to the machine learning community in 1986, and Igor Aizenberg and colleagues applied it to artificial neural networks in 2000, focusing on Boolean threshold neurons [5]. The concept originated in 1943, when Warren McCulloch and Walter Pitts developed a computer model based on human neural networks, using "threshold logic" to simulate cognitive processes [6]. Since then, deep learning has evolved continuously, with brief setbacks during the "AI Winters" (periods of reduced funding and interest in AI research) [5].
Table 1 outlines the history and evolution of deep learning. In 1943, Warren McCulloch and Walter Pitts pioneered neural networks with a computational model called threshold logic, using mathematical algorithms to mimic cognitive processes [6]. In 1958, Frank Rosenblatt developed the perceptron, a two-layer neural network for pattern recognition based on simple arithmetic operations. He also proposed adding more layers, though practical implementation was delayed until 1975.
In 1980, Kunihiko Fukushima introduced the Neocognitron, a hierarchical, multilayered neural network that excelled in handwriting and pattern recognition tasks. By 1989, researchers developed algorithms for deep neural networks, though their lengthy training times (often days) limited practicality. In 1992, Juyang Weng’s Cresceptron enabled automated 3D object recognition in complex scenes, advancing neural network applications.
In the mid-2000s, Geoffrey Hinton and Ruslan Salakhutdinov’s seminal paper popularized deep learning by demonstrating the effectiveness of layer-by-layer neural network training [5]. In 2009, the NIPS Workshop on Deep Learning for Speech Recognition showed that pre-training could be skipped with large datasets, significantly reducing error rates. By 2012, deep learning algorithms achieved human-level performance in pattern recognition tasks, marking a major milestone in the field.
In 2014, Google acquired DeepMind, a UK-based AI startup, for £400 million, accelerating AI research advancements. In 2015, Facebook implemented DeepFace, a deep learning system with 120 million parameters, enabling accurate automatic tagging and identification in photographs. In 2016, DeepMind’s AlphaGo defeated professional Go player Lee Sedol in a highly publicized Seoul tournament, showcasing deep learning’s capabilities. By 2024, transformer-based models like AlphaFold3 predicted protein complexes and ligand interactions, while genomic language models (gLMs) forecasted gene co-regulation in single-cell data, advancing precision medicine [38,99]. These developments, driven by large datasets and enhanced computational power, highlight deep learning’s transformative impact on biological research (
Table 2).
Deep learning uses artificial neural networks (ANNs) to perform complex computations on large datasets. These networks consist of interconnected neuron layers that process and extract patterns from input data. Deep learning processes data through multiple layers of neural networks, with each layer extracting and transforming features before passing them to the next. A fully connected deep neural network includes an input layer, several hidden layers, and an output layer. Neurons in each layer receive inputs from the previous layer, process them, and pass outputs forward, ultimately producing the final result. Through nonlinear transformations, these layers learn complex patterns and representations from the input data [7].
Deep learning employs various algorithms, each suited to specific tasks. These include radial basis function networks, multilayer perceptron, self-organizing maps, convolutional neural networks (CNNs), recurrent neural networks (RNNs), long short-term memory networks (LSTMs), and transformers. CNNs excel in genomics, as demonstrated by DeepBind for RNA-binding protein site prediction and DeepCpG for DNA methylation analysis [32]. RNNs and LSTMs handle sequential data effectively, while transformers, used in AlphaFold3, model complex protein interactions and genomic sequences [38]. These algorithms drive advancements in precision medicine and drug discovery by detecting subtle patterns in large biological datasets (
Table 2). More recently, by 2024, transformer-based models like AlphaFold3 have advanced to predict protein complexes and ligand interactions with unprecedented accuracy, while genomic language models (gLMs) have emerged to forecast gene coregulation in single-cell data, significantly advancing precision medicine. These continuous developments, driven by the availability of massive datasets and enhanced computational power, underscore deep learning’s transformative influence across diverse scientific disciplines, including biological research.
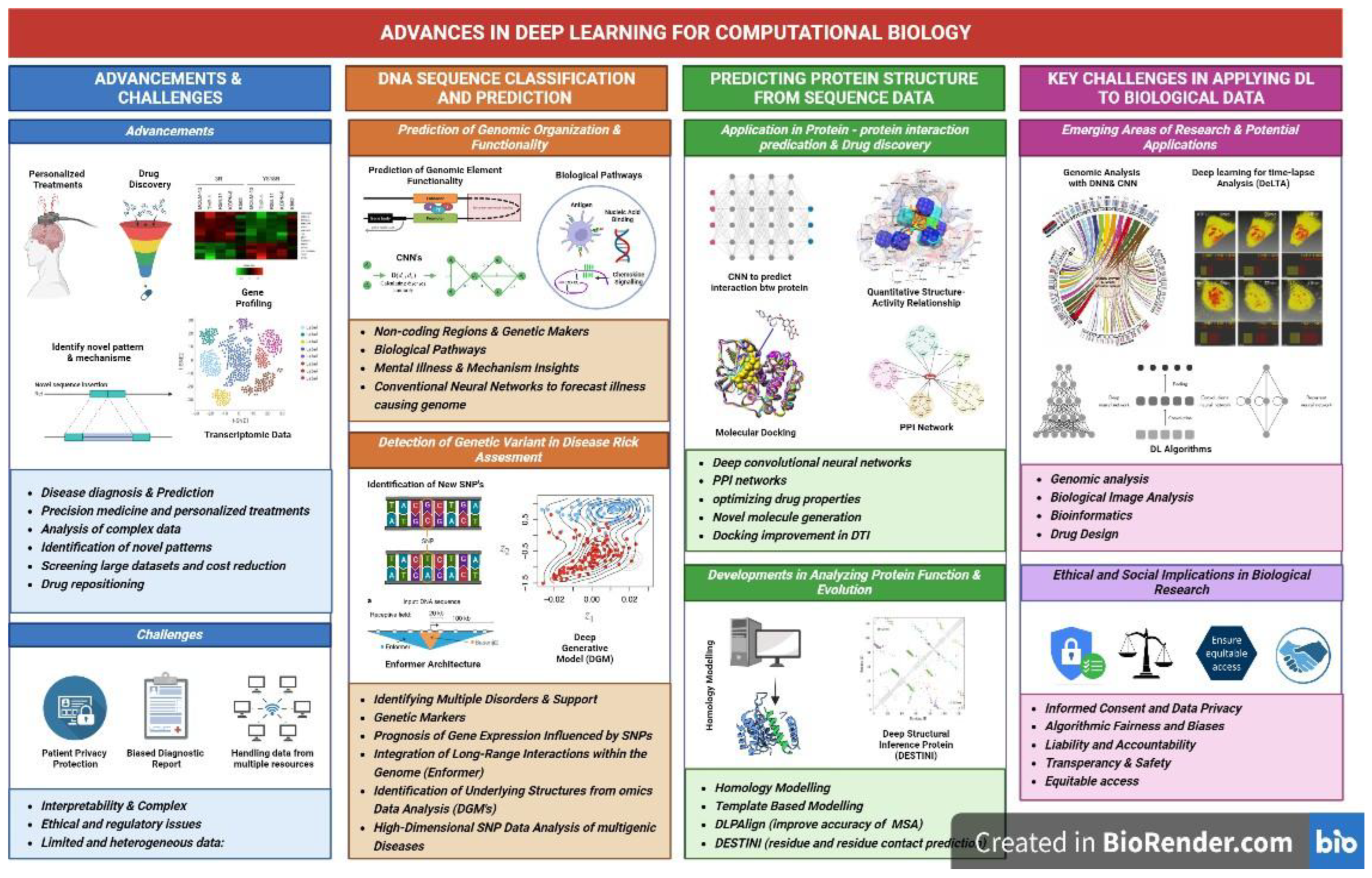
2. Advantages and Challenges of Using Deep Learning in Computational Biology
Advancements in genomics and imaging technologies have generated vast molecular and cellular profiling data from numerous global sources. This data surge challenges traditional analysis methods [8]. Deep learning, a subset of machine learning, has emerged as a powerful tool for bioinformatics, extracting insights from large datasets by identifying patterns and making accurate predictions [10]. For instance, DeepBind uses convolutional neural networks (CNNs) to predict RNA-binding protein sites, while AlphaFold employs transformers for precise protein structure prediction [32,38]. These applications demonstrate deep learning’s transformative potential in biology and medicine, though challenges persist (
Figure 1).
2.1. Advantages of Using Deep Learning
Deep learning enhances disease diagnosis and prediction. Ching et al. [11] highlights its ability to develop accurate, data-driven diagnostic tools that identify pathological samples. It also rapidly screens large datasets, reducing drug discovery costs by identifying targets and predicting responses [11]. Furthermore, deep learning supports drug repositioning by analyzing transcriptomic data to identify new therapeutic targets [12]. Deep learning supports precision medicine by developing personalized treatments [13]. It integrates patient-specific data, including genomic profiles, clinical records, and lifestyle factors, to tailor therapies [14]. By analyzing large datasets with high accuracy, deep learning identifies genetic markers, variations, drug efficacy, protein interactions, and clinical prognoses, optimizing treatment selection and disease monitoring [15]. For example, Dinov et al. [16] developed a deep learning protocol for Parkinson’s disease diagnosis, achieving high accuracy and demonstrating potential for drug discovery and personalized medicine.
Deep learning models efficiently handle large, complex biological datasets [8]. These algorithms extract intricate patterns, improving the accuracy of predictions and data classification. By learning relevant features from vast datasets, they minimize the need for human intervention [17]. This is particularly valuable in biomedicine and molecular biology, where complex, heterogeneous data often pose analytical challenges. The scalability and transferability of deep learning models enable efficient handling of large, complex datasets [18]. These models can be trained in specific biological tasks with minimal modifications, reducing resource demands and improving generalization of new data. Additionally, deep learning identifies novel patterns that conventional methods may miss. For example, Liu et al. [25] used deep learning to predict functional implications of non-coding genomic variations with greater accuracy than traditional approaches.
2.2. Challenges of Using Deep Learning
A key challenge in applying deep learning to computational biology is interpretability [11]. Complex model architecture often functions as "black boxes," making it difficult for researchers to understand how predictions reflect biological mechanisms. Interpretability is critical for building trust among clinicians and stakeholders, particularly in medical diagnostics, where decisions must rely on reliable factors rather than data artefacts. Ongoing efforts aim to develop techniques that clarify deep learning’s decision-making processes [8]. Deep learning enhances diagnostic accuracy in medicine but raises ethical and regulatory concerns, particularly regarding patient privacy [20]. Robust guidelines on informed consent and data protection can mitigate these issues. Additionally, biased diagnostic reports risk discriminating between patient groups, potentially leading to incorrect diagnoses or unequal treatment access [20,21]. Transparent and ethical use of deep learning models promotes accountability in biomedical research and healthcare.
Although deep learning models handle large datasets effectively, they require high-quality, labelled data for training [22]. In healthcare and biomedicine, obtaining such data is challenging due to privacy regulations and data heterogeneity. Moreover, biological data from sources like electronic health records and pathological reports often vary in format and standards, reducing model performance and generalization. Deep learning models, despite their advanced capabilities, demand significant computational resources and specialized hardware for training and deployment [23]. High-performance computing infrastructure is essential, posing challenges for small non-profit organizations and research institutions with limited resources.
3. Interconnecting Genomics and Protein Structure Prediction through Deep Learning
The central dogma of molecular biology—DNA to RNA to protein—establishes a direct link: genomic information dictates protein sequences, and these sequences, in turn, determine protein structures and functions. Deep learning provides the computational framework to traverse this biological pathway, enabling a holistic understanding of biological systems from the genetic blueprint to the functional molecular machinery. This section review combines genomics and protein structure prediction into a single narrative due to their biological interdependence and the synergistic advancements driven by deep learning.
3.1. Role of Deep Learning in Genomic Variant Detection and Precision Medicine
Deep learning has transformed genomic variant detection and gene expression analysis. Genomics, encompassing an organism’s entire genetic makeup, provides critical insights into biological processes, diseases, and individual differences. Deep neural networks enable researchers to analyze gene expression profiles and genetic variations, advancing personalized medicine, drug discovery, and disease mechanism understanding [24]. Specifically, these algorithms accurately classify variants to identify disease-causing mutations and support gene expression studies, such as splicing-code analysis and long noncoding RNA identification [24].
The use of deep learning in genomic variant detection has enabled the prediction of the organization and functionality of various genomic elements such as promoters, enhancers, and gene expression levels [25]. Deep learning detects gene variants to predict their effects on disease risk and gene expression. To accomplish this, a genome is split into optimal, non-overlapping fragments using fragmentation and windowing techniques [26]. A three-step procedure—fragmenting, model training for forecasting variant effects, and evaluating with test data—constitutes deep learning-based identification of genetic variations [26]. A deep learning model demonstrated favorable precision in distinguishing patients from controls and the ability to identify individuals with multiple disorders during research on genetic variants in non-coding areas [27]. These regions were enriched with pathways related to immune responses, antigen binding, chemokine signaling, and G-protein receptor activities, offering insights into mental illness mechanisms [27]. By utilizing deep neural networks, researchers have gained insights into gene expression profiles, genetic variations, and single-cell RNA sequencing data, advancing personalized medicine and drug discovery [18]. For genomic variant detection, algorithms precisely classify variants to identify disease-causing mutations [24]. In single-cell transcriptomics, graph neural networks (GNNs) like scGNN model cell-type interactions and gene regulation [84]. Additionally, genome language models (gLMs), leveraging transformer-based architectures, have emerged in 2024 to predict gene co-regulation in single-cell data, enhancing precision medicine applications [118].
Deep learning methods, such as convolutional neural networks (CNNs), predict genetic variations that may cause diseases [25]. A CNN-based model outperformed traditional methods in forecasting the functional impacts of non-coding genomic variants, achieving high accuracy in variant classification but requiring large datasets to prevent overfitting (
Table 2, [79]). Recurrent neural networks (RNNs) model sequential dependencies for gene expression prediction, though they struggle with long-range interactions [80]. Deep learning also identifies single-nucleotide polymorphisms (SNPs) affecting gene expression levels, revealing new variants linked to expression changes [28].
Gene expression relies on transcriptional regulators, such as pre-mRNA splicing, polyadenylation, and transcription, to produce functional proteins. While high-throughput screening provides quantitative data on gene expression, traditional experimental and computational methods struggle to analyze large genomic regions. Deep learning overcomes this limitation, accurately predicting gene expression levels and identifying enhancer-promoter interactions. For example, the Enformer model, described in Nature Genetics, improved gene expression predictions by integrating long-range genomic interactions (up to 100 kb) using massive parallel assays [29].
Deep generative models (DGMs) enhance gene expression analysis by identifying underlying structures, such as pathways or gene programmers, from omics data [30]. These models provide a framework to account for latent and observable variables, effectively analyzing high-dimensional SNP data to understand multigenic diseases. DGMs also predict how nucleotide changes affect DNA beyond gene expression datasets, offering new insights into genetic regulation [30]. Deep learning has transformed our understanding of genetics by identifying genomic variants and analyzing gene expression, accelerating the discovery of disease-related genes, drug targets, and therapies [24]. It enables clinicians to make precise decisions based on individual genomic profiles. Despite challenges like overfitting and interpretability, deep learning often outperforms traditional methods, supported by robust computational pipelines for genomics research.
3.2. Advancements in Deep Learning for Epigenetic Data Analysis
Recent advancements in deep learning have enhanced the analysis of epigenetic data, deepening our understanding of gene expression and chromatin dynamics regulation [31]. These methods extract critical insights into how genetic and environmental factors, such as nutrition and lifestyle, influence epigenetic modifications, particularly in obesity and metabolic diseases [31]. Convolutional neural networks (CNNs) have advanced epigenetic analysis by capturing spatial dependencies in DNA methylation patterns. For example, DeepCpG, developed by Angermueller et al. [2017], uses CNNs to predict methylation states across genomes, outperforming traditional methods but requiring high-quality, well-annotated data [32]. Similarly, transformers model long-range interactions in chromatin dynamics, though they are computationally intensive (
Table 2, [99]).
Epigenetic alterations significantly impact health, influenced by environmental factors like exercise, stress, and diet [31]. Deep learning enables rapid analysis of large epigenetic datasets, with applications like DNA methylation ageing clocks. For instance, DeepMAge, trained on 4,930 blood DNA profiles, predicts age with a median error of 2.77 years, outperforming linear regression-based clocks [34]. Deep generative models (DGMs) have also advanced epigenetic analysis in 2024, identifying latent structures in DNA methylation data to uncover regulatory mechanisms [122]. Additionally, the analysis of histone modification data has been explored using deep learning techniques. Key markers for gene activity and chromatin structure include various modifications such as acetylation and methylation. To unravel the intricate connection between patterns in these modifications and gene expression, neural networks like attention-based ones or those based on deep belief have proven effective. In particular, Yin [2019] introduced their model called DeepHistone, which leverages multiple profiles from different histones to predict levels of gene expression with high precision, leading to new insights into epigenetic mechanisms previously unknown [33].
Moreover, studies conducted on animals have shown that epigenetic modifications are linked to metabolic health outcomes in humans. Animal models provide ideal opportunities for rigorously controlled studies that can offer insight into the roles of specific epigenetic marks in indicating present metabolic conditions and predicting future risks of obesity and metabolic diseases [31]. Examples include maternal nutritional supplementation, undernutrition, or overnutrition during pregnancy, resulting in altered fat deposition and energy homeostasis among offspring. Corresponding changes in DNA methylation, histone post-translational alterations, and gene expression were observed, primarily affecting genes regulating insulin signaling and fatty acid metabolism [31]. Recent studies indicate paternal nutrition levels also affect their children's fat disposition, with corresponding detrimental effects on their bodies' epigenetic characterizations [31].
Although deep learning-based techniques demonstrate potential in epigenetic data analysis, they possess constraints. Substantial amounts of top-notch data are necessary for these models to train adequately. Additionally, interpreting results from deep learning can be challenging; thus, understanding biological mechanisms leading to model predictions is difficult. Thus, evaluating input quality and model performance is critical before endorsing results. The latest advancements underscore the promise of deep learning methods for scrutinizing epigenetic data. Neural networks' potency allows scientists to discern concealed patterns, grasp far-reaching relationships, and make precise forecasts from extensive epigenomic datasets. These progressions offer significant enlightenment into gene expression's regulatory mechanisms, which can aid in comprehending diseases and designing specific treatments. The initiatives undertaken by these experts are merely a few illustrations of the thrilling headway attained within this domain, sparking further innovations in research on epigenetics.
3.3. Applications of Deep Learning in Protein Structure Prediction
Deep learning has transformed protein structure prediction by accurately determining proteins’ three-dimensional shapes. This capability is critical for understanding protein functions, advancing drug discovery, and designing therapeutics. Deep learning models effectively capture complex patterns in protein sequences, enabling precise structure predictions [38].
Predicting the structure of a protein with precision, based solely on its sequence, proves to be challenging, but deep learning presents itself as a viable solution. Recent applications employing this approach have successfully predicted both three-state and eight-state secondary structures in proteins [35]. Protein secondary structure prediction serves as an intermediate process, linking the primary sequence and tertiary structure predictions. The three traditional classifications of secondary structures include helix, strand, and coil. However, predicting 8-state secondary structures from protein sequences is a much more intricate task referred to as the Q8 problem- which offers greater precision in providing structural information for varied applications. Thus, several techniques of deep learning such as SC-GSN network, bidirectional long short-term memory (BLSTM) approach, a conditional neural field with multiple layers, and DCRNN have been employed to forecast the eight-state secondary structures [35]. In addition, a next step conditioned convolutional neural network (CNN) was utilized to identify sequence motifs linked with particular secondary structure elements by analyzing the amino acid sequences. For instance, in 2019, AlQuraishi's research introduced "Alphafold," a CNN-powered model that accurately forecasted protein secondary structure. Its competence in capturing sequence-structure connections resulted in better forecasts when weighed against conventional means [36].
Deep learning significantly impacts protein-protein interactions and binding site prediction. Convolutional neural networks (CNNs) and transformers analyze protein sequences and structures, detecting intricate interactions (e.g., DeepPPI, AlphaFold) [37,38]. CNNs excel in capturing local structural patterns, ideal for binding site prediction, but require extensive training data (
Table 2, [95]). Transformers model long-range dependencies, enabling accurate protein complex predictions, though computationally demanding [116]. DeepPPI predicts interactions from sequence data, enhancing understanding of protein networks [37].
Significant advancements have been made in the tertiary structure prediction of proteins using deep learning. Abriata et al., employed a deep learning contact-map approach to achieve a notable breakthrough in the 13th Critical Assessment of Techniques for Protein Structure Prediction (CASP13) [38]. To determine protein folding accurately, predicting residue-residue contacts is crucial. Deep learning approaches leverage vast protein databases to capture intricate patterns and dependencies between residues. This aids in long-range contact prediction by developing deep-learning models that guide the assembly of protein structures with greater precision. Wang et al.'s [2021] method utilized a deep residual network which proved effective in anticipating residue-residue interactions for precise folding predictions through their model's accuracy improvement [39]. Meanwhile, the "AlphaFold 2" model created by Senior et al. [2020] is another significant illustration worth noting. Through the integration of RNNs and attention mechanisms, AlphaFold 2 achieved extraordinary precision in prognosticating protein tertiary structures, surpassing other techniques in the Critical Assessment of Structure Prediction (CASP) competition as well. In 2024, AlphaFold 3 extended these capabilities by predicting protein complexes and ligand interactions with high accuracy, further advancing its utility in drug discovery and structural biology [117]. Such success can be attributed to how RNNs effortlessly capture long-range dependencies within protein sequences without issue. [40].
These applications showcased the extensive range and influence of deep learning in predicting protein structure. With its adeptness at identifying complex patterns and connections within protein sequences and structures, deep learning has enabled significant progress in comprehending aspects such as folding, function, and interactions related to proteins. Although there may be upcoming challenges and opportunities, the extensive implications of deep learning's capability to reveal fresh insights regarding proteins are immense in terms of comprehending basic life processes, personalized medicine, as well as drug discovery.
4. Deep Learning Models for Prediction of Protein Structure from Sequence Data
Deep learning, a subset of machine learning, has significantly advanced computational biology, particularly in protein structure and interaction prediction [41]. These algorithms process large, complex datasets, learning abstract features for tasks like data augmentation in bioinformatics [42,43]. Deep learning architectures accept diverse inputs, including protein sequences, 3D structures, and network topologies, for applications like structure prediction and text mining. Key neural network components include fully connected, convolutional, and recurrent layers [44,45].
Deep learning architectures, including convolutional neural networks (CNNs), recurrent neural networks (RNNs), and transformers, accept diverse inputs like protein sequences and 3D structures [44,45]. CNNs extract local features for secondary structure prediction, offering high accuracy but needing large datasets (
Table 2, [79]). RNNs model sequential dependencies, suitable for residue contact prediction, but struggle with long sequences [80]. Transformers, used in AlphaFold, capture global interactions for tertiary structure prediction, though resource-intensive [38,116]. These methods drive advancements in protein modeling.
4.1. Applications of Deep Learning in Protein-Protein Interaction Prediction and Drug Discovery
The latest deep learning techniques that are employed in PPI models may include Deep convolutional neural networks. This technique is widely used due to its potential to extract features from structural data. For instance, based on Torrisi et al [44], the structural network information along with the sequence-based features predicts the interactions between proteins. Besides that, in order to extract structural information from 2D volumetric representations of proteins, the pre- trained ResNet50 model was used. The results indicate that methodologies for image-related tasks can be extended to work on protein structures [45]. However, these techniques of analyzing molecular structure have drawbacks such as elevated computational expenses and as well as interpretability [45].
There are various deep learning methods that could be utilized for protein-protein interaction networks. First, the DeepPPI is a multilayer perception learning structure that requires protein sequences as its source of input features [46]. The encoding method utilized by this method is the seven sequence-based features which use concatenation as its combining method. Moving on to the second method which is DPPI, is a convolutional neural network structure that also uses protein sequences as its source of input features [47]. protein-positioning specific scoring matrices, PSSM which is derived by PSI-BLAST is used as the encoding method for this deep earning method. Next, the DeePFE-PPI is also a method that was created in 2019 using multilayer prescription which uses protein sequences as an input. The encoding method that is utilized in this method is pre-trained model embedding (Word2vec) [48]. Besides that, S-VGAE is also an example of graph convolutional Neural networks which utilize protein sequences and topology information of protein-protein interaction networks. The encoding method employed in this technique is a conjoint method and it is combined via the concatenation method [49].
Besides protein-protein interaction, Deep learning is also utilized in drug discovery for optimizing the properties of drugs, determining new drugs as well as predicting drug-target interactions. In addition, deep learning is also employed in predicting the molecular properties of drugs such as solubility, bioactivity, toxicity, and many more [50]. In addition, it is also used to produce novel molecules that have preferred properties. Next, in QSAR studies for drug discovery, the deep neural network is used to predict the bioactivity of the drugs and their chemical structures [50].
Moreover, deep learning methods are also applied to lead to optimized integration of traditional in silico drug discovery methods." This clarifies the intent and improves flow. Based on the research, which is entitled, (AtomNet from Atomwise company, the first major application of deep learning into DTI prediction) clearly shows the application of convolutional neural networks which is a type of deep learning technique to predict the molecular bioactivity in proteins [51]. In addition, in terms of docking, deep learning techniques have been employed to improve the accuracy of both traditional docking modules and scoring functions. For instance, the docking proved that the application of deep learning had improved the binding mode prediction accuracy over the baseline docking process. Besides that, this paper had also proven the fact that Deep learning could be successfully utilized in the rational docking process [52].
4.2. Recent Developments in Deep Learning-Based Techniques for Analyzing Protein Function and Evolution
Recently, there were a few developments that are used in protein analysis by incorporating deep learning algorithms. An example of it would be combining deep learning with homology modelling. Furthermore, Homology modelling is the most popular protein structure prediction method that is utilized to generate the 3D structure of a protein. This is based on two principles which are the amino acid that is used to determine the 3D structure, and the 3D structure that is preserved regarding the primary structure [53]. Therefore, it is convenient and an effective way to build a 3D model using known structures of homologous proteins that have a certain sequence similarity. However, it does have some challenges when using this method such as weak sequence structures, modelling of the rigid body shifts and many more [53]. However, incorporating deep learning models has resulted in great improvement in the protein’s model accuracy.
The deep learning-based methods are employed to improve accuracy in each step of template- based modelling of protein. For instance, DLPAlign is an example of a deep learning technique that is combined with sequence alignment [53]. This straightforward and beneficial approach may aid to increase the accuracy of the progressive multiple sequence analysis method by basically providing training to the model based on convolutional neural networks CNNs [53]. Besides that, DESTINI is also a recent method which applies deep learning techniques algorithm, for protein residue and residue contact prediction along with template-based structure modelling [55].
In short, Deep Learning techniques have provided various achievements in collaborative sectors, namely model quality assessment (QA), a subsequent stage in protein structure prediction. Basically, QA is followed by structure predictions to quantify the deviation from the natively folded protein structures in both template-based and template-free techniques.
4.3. Challenges and Future Directions
There are various challenges when using Deep learning techniques when analyzing biological- related specimens such as protein structure prediction. First, deep learning requires a large amount of high-quality data. Hence, only biological analysis could be done if only a large amount of data is gathered [56]. Next, the deep learning model is incapable of multitasking when it is applied in an analysis procedure. Deep learning models are capable of handling one issue at a time. Furthermore, the interpretability of deep learning models is also a challenge of interest for many researchers to overcome. This is because it is difficult to understand and identify how they obtain their predictions. New techniques are being developed by researchers to overcome this problem. The future direction of deep learning is to create hybrid models by incorporating other machine learning techniques to improve performance and interpretability [56].
5. Key Challenges and Future Directions
As mentioned in the previous section, high-quality data, the inability to multi-task and data interpretability are some of the key challenges experienced in the application of AI systems such as deep learning into biological data. There are several other challenges, especially in terms of ethics and social implications which are addressed in the sub-sections below. Addressing these challenges of deep learning requires specific and innovative approaches specific to the types of biological data used. Thus, overcoming these challenges would ultimately pave a path to improvement in biological research.
5.1. Emerging Areas of Research and Potential Applications
Computational biology is defined as an interdisciplinary field which involves the use of techniques from various other fields such as biology, mathematics, statistics, computer science and more. Applications of deep learning in computational biology can be seen in various areas including in the study of genomics and proteomics. There are many major achievements that are obtained specifically in areas such as protein structure prediction, and rapid advancement in other areas of research from the traditional approaches including genomic engineering, multi-omics, and phylogenetics can also be seen [57].
The study of genomes and their interaction with other genes and external factors is commonly known as genomics. One of the primary studies conducted in genomics is the study of regulatory mechanisms and non-coding transcription factors [58]. One of the major current applications of deep learning research of genomics and transcriptomics is one of the emerging areas of research in deep learning. Deep learning is used to identify variations in genomic data, this includes DNA sequencing and gene expression. For example, it is used to predict the functions of genes, discover gene regulatory networks, and identify biomarkers in diseases. As a result of this application, the metabolic pathways can also be optimized. A study identified several challenges in genomics including mapping the effects of mutation within a population and the DNA sequence prediction in a genome which has complex interactions and variations. To combat these challenges, deep learning methods are employed in genomic studies. Deep learning is used to identify variations in genomic data, including DNA sequencing, gene expression, and drug perturbation effects. For example, it predicts gene functions, discovers regulatory networks, and identifies disease biomarkers, optimizing metabolic pathways [59].
In single-cell transcriptomics, graph neural networks (GNNs) like scGNN analyze cell-type classification and gene co-regulation [84]. In 2024, scGNN has further advanced, modelling cell-type interactions and gene regulation with high precision, driving progress in precision medicine [119]. In drug perturbation analysis, deep learning models predict molecular responses to drug treatments, aiding drug discovery [18]. Deep Neural Networks (DNNs) and Convolutional Neural Networks (CNNs) address challenges like mapping mutation effects and predicting DNA sequence functions [59]. DNNs, trained on DNA sequence datasets, identify protein-binding sites and predict splicing outcomes, while CNNs analyze mutation effects in single nucleotide variants [58]. Deep Neural Networks (DNN) and Convolutional Neural Networks (CNN) are algorithms of deep learning that are employed in genomic studies [59]. Deep Neural Networks (DNNs) solve DNA sequence prediction by training on sequence datasets and the corresponding protein structures. This enables the identification of the proteins which are specific and binds to a particular DNA sequence.
The DNN models are also able to predict splicing outcomes for new DNA sequences based on the training of splicing patterns. CNN, on the other hand, addresses the remaining issue; the prediction of mutation effects [58]. This model can analyze and identify the potential causes of mutation in a DNA sequence and determine the mutation or disease on the single nucleotide variant that is affected. Both CNN and DNN are powerful tools of deep learning which can provide valuable information on the complex structures of genomes. The application of the algorithm in the field of genomics would greatly improve the analysis of complex structures, functions and interactions of genomes.
In the field of biological image analysis, the deep learning algorithm CNN is found to be an efficient tool that is able to undertake several tasks such as classification, feature detection, pattern recognition and feature extraction (58). Since the CNN models are effective in processing grid-like data such as images, it is commonly utilized in image analysis [59]. Staking more convolutional layers in the model aids in detecting complex and abstract features in biological images. The CNN model is able to learn and identify delicate patterns and subtle differences in biological images which improve the accuracy of a diagnosis. DeLTA is an example of a deep learning tool used to analyze biological images, specifically, time-lapse microscopy images [60]. The Deep Learning for Time-lapse Analysis (DeLTA) is able to analyze the growth of a single cell and the gene expressions in microscopy images. It was found to be able to process and capture microscopy images with high accuracy and without the need for human interventions. Furthermore, deep learning is incorporated into healthcare, specifically radiology. Tasks such as classifying patients based on chest X-rays diagnosis and nodule detection in computed tomography images are done using deep learning [61]. The analysis of a large number of radiology data depends on the efficiency of the powerful deep learning algorithms. Thus, deep learning algorithms have the potential to revolutionize biological image analysis by providing automated and accurate analysis.
Deep learning in the proteomics field can mainly be shown in the protein-protein interactions predictions. Protein complexes can also be identified through deep learning. Protein structure and function can then be predicted from the data obtained and be used for various activities such as the identification of targeted proteins for drug development. Deep learning is applied in the study of phylogenetics where the limitation in the classification methods where the branch lengths of the phylogeny cannot be inferred is to be overcome [57].
While there are many applications of deep learning that bring significant advancements, there are other potential applications of deep learning in the biological field that can be further discussed and implied. For instance, deep learning is applied in the identification of protein-protein interactions. Therefore, a similar technique can be applied in drug design. In terms of drug design, the application and incorporation of deep learning have made the process more time and cost-effective as compared to traditional drug design methods [62]. The use of deep learning in drug design is identified to be more flexible due to the neural network architecture of the algorithm [63]. Especially in the current era, with the combat against COVID-19, deep learning has shown great potential in accelerating the drug design process. The deep learning models are able to identify antimicrobial compounds against a disease or a virus by training the model with the ability to identify molecules against the virus or bacteria. Similarly, another study showed the use of deep learning models in the use of de novo drug design where the model was trained to identify the physical and chemical properties of the drugs, classifying them based on their features and allowing automated extraction to create a novel ligand against the target protein [65]. Drug repurposing, which is a quicker method to achieve and complete drug designing for a disease or illness, is found to incorporate deep learning approaches. An article reported the use of network- based approaches in drug repurposing to identify the target molecule for known drugs to speed up the process [66]. Another study in relation to COVID-19, used the Molecule Transformer-Drug Target Interaction (MT-DTI), a deep learning model trained with chemical sequences and amino acids sequences to identify the commercially available antiviral drugs with similar properties of interaction with the SARS-CoV-2 virus [64]. These are just some examples of the emerging use of deep learning models in drug design. The appearance of the COVID-19 disease has boosted the application of AI systems in the field to improve the speed and efficiency of the process.
To summarize, the deep learning algorithm is a powerful artificial intelligence tool that is widely used in the field of computational biology. The application of the tool is just in its beginning phase as there are more fields and complex challenges that are to be explored and tackled in the upcoming future. The application of this AI technology will help to shape the future of computational biology by improving the predictions and understanding biological processes.
5.2. Ethical and Social Implications
The advancements in digital technology allow the incorporation of Artificial Intelligence tools such as Machine Learning and Deep Learning in various fields of research. These techniques use various algorithms to identify complex and non-linear correlations in massive datasets and could improve prediction accuracy by learning from minor algorithmic errors encountered. Despite the use of a powerful machine learning tool, such as deep learning algorithms, in the field of research and healthcare is found to be revolutionary, it inevitably raises ethical concerns and social implications that require careful consideration [67].
Four major ethical issues were identified regarding the use of AI tools in the healthcare system; informed consent of data, safety and transparency, algorithmic fairness and biases and data privacy [68]. These concerns may be identified in the healthcare sector, but these concerns are also integrated in the usage of deep learning techniques in biological research which involves the use of deep neural networks to analyze and interpret volumes of biological data.
The field of biological research involves the use of various biological data which includes genomic sequences, protein structures, medical images and scans and more. The deep learning algorithm has access to this data, containing sensitive biological data, to aid in research. As medical records and genomic information of individuals are involved and used, it is critical to ensure the privacy of the individual, as well as the informed consent for data usage, is obtained [68]. As highlighted in 2024 reviews, these challenges persist, particularly in rare disease genomics, where data scarcity and ethical concerns around privacy and bias necessitate robust regulatory frameworks [120]. Privacy violations and mishandling of personal information are examples of invasion of data privacy without individual consent [70].
The potential bias in the algorithms of the deep learning tool is one of the main ethical concerns surrounding AI systems. The algorithms utilized in these systems can perpetuate biases and negatively impact marginalized groups [68]. This is because the training data is not representative of the diverse populations leading to biased results and disparities in research outcomes. The biases can be found in different stages of biological research, including data collection and annotation, if they are not addressed [71]. Therefore, efforts should be taken to address the bias in data collection to promote the inclusivity of all data regardless of population type, disease groups, diversity and other factors involved in research.
It is also crucial to promote transparency and safety in the AI tools used in biological research. Deep learning models are opaque making it difficult to understand the process of prediction and decision in research. As deep learning models are made up of multiple layers of artificial neurons, where each layer corresponds to a different learning pattern, it poses a challenge to accurately identify the pattern learned by each layer functions to make a prediction. Lacking transparency in the algorithm decision-making process begins the questioning of the ability to scrutinize the AI results as a reasonable explanation leading to the data being unprovable and uninterpretable by humans [68,71]. Thus, transparent models are required to make sure the researchers can observe the prediction pattern to validate and understand the results obtained. This ensures the accountability of the scientific research process. However, it was discussed that full transparency may cause friction against certain ethical concerns as it may leak private and sensitive data into the open [69]. Hence, there should be limitations to the disclosure of the algorithms.
Security in biological research not only involves maintaining the data and privacy of personal data, but it also involves the responsible use of technology. The technology at hand, deep learning, must be used responsibly and ethically in research. Scientists must incorporate ethical frameworks to avoid potential misuse and unintended consequences or risks that may occur [68]. Furthermore, risk assessments are to be conducted to aid in decision-making and reduce the possible negative impacts. The deployment and implementation of this technology must be considered well with proper safety measures, regular monitoring and evaluations.
Moreover, there is the concern of liability and accountability where questions would arise to who would be the person to be held accountable towards any form of mistakes or errors caused by deep learning algorithms used in research [72]. As the algorithms are continually learning and evolving, it is difficult and complex to determine liability. Thus, legal frameworks are to be adapted with clear lines of responsibility to address the challenges faced by the AI system in biological research [73].
Ensuring equitable access to the deep learning tool is one of the social implications of AI systems in biological research. Promoting equitable access to scientists and researchers would be able to participate in the advancement of deep learning algorithms in biological research [74]. It would also prevent exacerbating disparities in biological research while promoting an inclusive and collaborative research environment. Fostering the exchange of ideas and knowledge of experts would further aid deep learning to be integrated into the biological research community.
In a nutshell, these are some of the social implications and ethical concerns revolving around the use of AI systems such as Deep Learning in the field of biological research. Deep learning is the future of more efficient and advanced research; however, the ethical concern mentioned above should be addressed to ensure the responsibility and accuracy of the algorithm in biological research
6. Future Prospects and Potential Impact of Deep Learning on Biological Research and Clinical Practice
Deep learning has significantly advanced computational biology, particularly in genomics and protein structure prediction, with breakthroughs like AlphaFold and DeepBind driving progress in precision medicine and drug discovery. The following sections summarize these advancements and their potential impact.
Table 3 compares key deep learning models from 2020 to 2025, highlighting recent advancements in genomics and protein structure prediction, including AlphaFold 3 and genome language models.
The application of deep learning models is poised to have a substantial influence in the field of biological research and clinical practices. With their ability to analyze large and intricate data, deep learning models could be beneficial in assisting with pathological diagnosis, drug discovery, genomic data identification or even personalized treatments. As noted in 2025, hybrid CNN-transformer models are gaining traction for single-cell omics analysis, balancing local and global feature detection to improve predictions [121]. By harnessing deep learning algorithms researchers can examine biological data consisting of gene expression or protein structure to identify new patterns or molecules which could yield an insight into the biological structure mechanisms. By harnessing deep learning algorithms researchers can examine biological data consisting of gene expression or protein structure to identify new patterns or molecules which could yield an insight into the biological structure mechanisms. In addition to that, deep learning models are also being used to facilitate accelerating new drug target development and research, developing new accurate diagnostic tests, and aiding in improving clinical trial designs [75]. The future prospect of deep learning models in both biological research and clinical practice are promising given that this technology and its algorithms continue to be developed which could potentially catapult humanity into a new era of making diagnosis for diseases or illnesses in addition to providing a more practical way for providing better patient care with precise treatments and prevention of diseases [76].
On top of that, deep learning models have the potential to be used to improve the efficiency and effectiveness of healthcare delivery systems by automating menial tasks and accelerated diagnostics tests [77]. Deep learning models possess the ability to integrate and analyze a variety of data types, including genomics, proteomics, imaging, and clinical data. This enables the exploration of concealed patterns and relationships within these datasets, empowering machine learning to offer a holistic comprehension of diseases and provide guidance for translational research endeavors. Besides that, deep learning has broad applicability in addressing diverse challenges. By training on extensive datasets, deep learning models excel at navigating tasks such as image classification, object detection, speech recognition, and machine translation aside from that, deep learning is a rapidly growing field, and it is being used in a variety of domains, including healthcare, computer vision, natural language processing, and robotics [78]. The summary of the recent advancements of deep learning in computational biology can be referred to
Table 2. LLMs are emerging as powerful tools in computational biology. Models like Evo and other transformer-based architectures, originally developed for natural language processing, are being adapted to understand and generate biological sequences (DNA, RNA, protein). These models can learn complex patterns and relationships within biological data, enabling tasks such as de novo protein design, predicting the effects of genetic mutations, and even simulating\ biological processes. Their ability to handle multi-task processing addresses a limitation of earlier deep learning models and represents a significant hot spot and future development direction in the field. LLMs can bridge the gap between sequence information and functional outcomes, offering new avenues for discovery in both genomics and protein science. For instance, genomic language models (gLMs) trained on DNA sequences are advancing our understanding of genomes and can generalize across a plethora of genomic tasks.
In protein science, protein language models (PLMs) are revolutionizing protein structure prediction, function annotation, and design by learning the probability distribution of amino acids within proteins. Evo, a genomic foundation model, exemplifies this trend, capable of both prediction and generative design from molecular to whole-genome scale, and can predict the effects of gene mutations with unparalleled accuracy. This type of model not only has outstanding performance but also can be applied to multiple downstream tasks, solving the problem of multi-task processing mentioned in the article to a certain extent. Continued advancements in integrating diverse multiomics datasets (genomics, transcriptomics, proteomics, metabolomics, epigenomics) will provide a more comprehensive understanding of biological systems. Deep learning models capable of effectively processing and synthesizing these heterogeneous data types will be crucial for uncovering complex disease mechanisms and developing truly personalized medicine. Beyond prediction, generative deep learning models (e.g., GANs, VAEs) are increasingly being used for de novo design of biological molecules, including proteins with desired functions or novel drug compounds. This shift from analysis to design holds immense potential for accelerating therapeutic development and synthetic biology Continued research into explainable AI (XAI) will be vital to make deep learning models more transparent and trustworthy for biological and clinical applications. This includes developing methods to visualize learned features, identify influential input elements, and provide human-understandable explanations for model predictions.
Author Contributions
Zaw Myo Hein: Writing – review & editing, Writing – original draft, Funding acquisition, Data curation. Dhanyashri Guruparan: Writing – review & editing, Writing – original draft, Visualization, Resources, Investigation. Blaire Okunsai: Writing – review & editing. Che Mohd Nasril Che Mohd Nassir: Writing – review & editing, Writing – original draft. Muhammad Danial Che Ramli: Writing – review & editing, Writing – original draft, Data curation. Suresh Kumar: Writing – review & editing, Writing – original draft, Visualization, Resources, Project administration, Investigation, Data curation, Conceptualization.
Funding
This work was supported by Ajman University
Institutional Review Board Statement
Not applicable.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors express their gratitude to Ajman University, UAE, for their support for the article processing charge (APC). The authors would also like to acknowledge that the assistance of ChatGPT 4.0, an AI language model developed by OpenAI, was used to help in the refinement of certain and limited sections of this manuscript, particularly for the language editing. Further, all AI-assisted content has been duly reviewed and thoroughly edited by the authors to ensure accuracy, scientific rigor, and adherence to the standards of academic writing. Authors are fully responsible for the content of this manuscript, even those parts improved by an AI tool, and are thus liable for any breach of publication ethics.
Abbreviations
The following abbreviations are used in this manuscript:
| LLMs |
Large Language Models |
| scGNN |
single-cell analysis using graph neural networks |
| CNNs |
Convolutional neural networks |
| RNNs |
Recurrent neural networks |
| DNA |
Deoxyribonucleic Acid |
| RNA |
Ribonucleic Acid. |
| ANNs |
Artificial neural networks |
| gLMs |
genomic language models |
| LSTMs |
Long short-term memory networks |
| GAN |
Generative adversarial network |
| GNNs |
Graph neural networks |
| SNPs |
Single-nucleotide polymorphisms |
| DGMs |
Deep generative models |
| DBN |
Deep Belief Networks |
| RL |
Reinforcement Learning |
| VAE |
Variational Autoencoders |
| DBM |
Deep Boltzmann Machines |
| DQN |
Deep Q- Networks |
| GCN |
Graph Convolutional Networks |
| VGAE |
Variational Graph Autoencoders |
| DNN |
Deep Neural Networks |
| CRNN |
Convolutional Recurrent Neural Networks |
| BLSTM |
Bidirectional long short-term memory |
| CASP |
Critical Assessment of Structure Prediction |
| PPI |
Protein-protein interaction |
| PSSM |
Protein-positioning specific scoring matrices |
| QA |
Quality assessment |
| AI |
Artificial Intelligence |
| PLMs |
Protein language models |
References
- Way, G.P.; Greene, C.S.; Carninci, P.; Carvalho, B.S.; de Hoon, M.; Finley, S.D.; Gosline, S.J.C.; Lȇ Cao, K.-A.; Lee, J.S.H.; et al. A field guide to cultivating computational biology. PLoS Biology 2021, 19, e3001419. [Google Scholar] [CrossRef] [PubMed]
- Faustine, A.; Pereira, L.; Bousbiat, H.; Kulkarni, S. UNet-NILM: A Deep Neural Network for Multi-tasks Appliances State Detection and Power Estimation in NILM. Proceedings of the 5th International Workshop on Non-Intrusive Load Monitoring 2020. [Google Scholar] [CrossRef]
- Kindel, W.F.; Christensen, E.D.; Zylberberg, J. Using deep learning to reveal the neural code for images in primary visual cortex.
- Machine Learning Industry Trends Report Data Book, 2022-2030 [Internet]. Available online: https://www.grandviewresearch.com/sector-report/machine-learning-industry-data-book (accessed on 9 May 2023).
- Wang, H.; Raj, B. On the Origin of Deep Learning. arXiv 2017. [Google Scholar] [CrossRef]
- Wang, H.; Raj, B. On the origin of deep learning. arXiv 2017, arXiv:1702.07800. [Google Scholar] [CrossRef]
- Sarker, I.H. Deep Learning: A Comprehensive Overview on Techniques, Taxonomy, Applications and Research Directions. SN Computer Science 2021, 2. [Google Scholar] [CrossRef]
- Stegle, O.; Pärnamaa, T.; Parts, L. Deep learning for computational biology. Molecular Systems Biology 2016, 12, 878. [Google Scholar] [CrossRef]
- Ferrucci, D.A.; Brown, E.D.; Chu-Carroll, J.; Fan, J.Z.; Gondek, D.C.; Kalyanpur, A.; Lally, A.; Murdock, J.W.; Nyberg, E.; Prager, J.M.; et al. Building Watson: An Overview of the DeepQA Project. Ai Magazine 2010, 31, 59. [Google Scholar] [CrossRef]
- Larrañaga, P.; Calvo, B.; Santana, R.; Bielza, C.; Galdiano, J.; Inza, I.; Lozano, J.A.; Armañanzas, R.; Santafé, G.; Pérez, A.; Robles, V. Machine learning in bioinformatics. Briefings in Bioinformatics 2006, 7, 86–112. [Google Scholar] [CrossRef] [PubMed]
- Ching, T.; Himmelstein, D.; Beaulieu-Jones, B.K.; Kalinin, A.A.; Do, B.T.; Way, G.P.; Ferrero, E.; Agapow, P.; Zietz, M.; Hoffman, M.M.; Xie, W.; et al. Opportunities and obstacles for deep learning in biology and medicine. Journal of the Royal Society Interface 2018, 15, 20170387. [Google Scholar] [CrossRef]
- Litjens, G.; Sánchez, C.I.; Timofeeva, N.; Hermsen, M.; Nagtegaal, I.D.; Kovacs, I.E.; De Kaa, C.a.H.; Bult, P.; Van Ginneken, B.; Van Der Laak, J. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Scientific Reports 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Mamoshina, P.; Vieira, A.; Putin, E.; Zhavoronkov, A. Applications of Deep Learning in Biomedicine. Molecular Pharmaceutics 2016, 13, 1445–1454. [Google Scholar] [CrossRef] [PubMed]
- Carracedo-Reboredo, P.; Liñares-Blanco, J.; Rodriguez-Fernandez, N.; Cedrón, F.; Novoa, F.J.; Carballal, A.; Maojo, V.; Pazos, A.; Fernandez-Lozano, C. A review on machine learning approaches and trends in drug discovery. Computational and Structural Biotechnology Journal 2021, 19, 4538–4558. [Google Scholar] [CrossRef]
- Zhao, L.; Ciallella, H.L.; Aleksunes, L.M.; Zhu, H. Advancing computer-aided drug discovery (CADD) by big data and data-driven machine learning modeling. Drug Discovery Today 2020, 25, 1624–1638. [Google Scholar] [CrossRef]
- Dinov, I.D.; Heavner, B.; Tang, M.; Glusman, G.; Chard, K.; D’Arcy, M.; Madduri, R.; Pa, J.; Spino, C.; Kesselman, C.; et al. Predictive Big Data Analytics: A Study of Parkinson’s Disease Using Large, Complex, Heterogeneous, Incongruent, Multi-Source and Incomplete Observations. PLOS ONE 2016, 11, e0157077. [Google Scholar] [CrossRef]
- Min, S.; Lee, B.; Yoon, S. Deep learning in bioinformatics. Briefings in Bioinformatics, bbw068.
- Ma, Q.; Xu, D. Deep learning shapes single-cell data analysis. Nat Rev Mol Cell Biol 2016, 23, 303–304. [Google Scholar] [CrossRef] [PubMed]
- Romão, M.C.; Castro, N.F.; Pedro, R.; Vale, T. Transferability of deep learning models in searches for new physics at colliders. Physical Review D 2020, 101. [Google Scholar] [CrossRef]
- Prabhu, S.P. Ethical challenges of machine learning and deep learning algorithms. Lancet Oncology 2019. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.F.; Singh, A.; Otaki, Y.; Tamarappoo, B.; Kavanagh, P.; Parekh, T.; Hu, L.; Gransar, H.; Sharir, T.; Einstein, A.J.; et al. Mitigating bias in deep learning for diagnosis of coronary artery disease from myocardial perfusion SPECT images. European Journal of Nuclear Medicine and Molecular Imaging 2022, 50, 387–397. [Google Scholar] [CrossRef]
- Data sharing in the age of deep learning. Nature Biotechnology 2023, 41, 433. [CrossRef]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.Q.; Duan, Y.; Al-Shamma, O.; Santamaría, J.V.G.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. Journal of Big Data 2021, 8. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Huss, M.; Abid, A.; Mohammadi, P.; Torkamani, A.; Telenti, A. A primer on deep learning in genomics. Nature Genetics 2018, 51, 12–18. [Google Scholar] [CrossRef]
- Liu, J.; Li, J.; Wang, H.; Yan, J. Application of deep learning in genomics. Science China. Life Sciences 2020, 63, 1860–1878. [Google Scholar] [CrossRef]
- Jo, T.; Nho, K.; Bice, P.; Saykin, A.J. Deep learning-based identification of genetic variants: application to Alzheimer’s disease classification. Briefings in Bioinformatics 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Qu, H.-Q.; Mentch, F.D.; Qu, J.; Chang, X.; Nguyen, K.; Tian, L.; Glessner, J.; Sleiman, P.M.A.; Hakonarson, H. Application of deep learning algorithm on whole genome sequencing data uncovers structural variants associated with multiple mental disorders in African American patients. Molecular Psychiatry 2022. [Google Scholar] [CrossRef] [PubMed]
- Montesinos-López, O.A.; Montesinos-López, A.; Pérez-Rodríguez, P.; Barrón-López, J.A.; Martini, J.W.R.; Fajardo-Flores, S.B.; Gaytan-Lugo, L.S.; Santana-Mancilla, P.C.; Crossa, J. A review of deep learning applications for genomic selection. BMC Genomics 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Avsec, Ž.; Agarwal, V.; Visentin, D.; Ledsam, J.R.; Grabska-Barwinska, A.; Taylor, K.R.; Assael, Y.; Jumper, J.; Kohli, P.; Kelley, D.R. Effective gene expression prediction from sequence by integrating long-range interactions. Nature Methods 2021, 18, 1196–1203. [Google Scholar] [CrossRef]
- Treppner, M.; Binder, H.; Hess, M. Interpretable generative deep learning: an illustration with single cell gene expression data. Human Genetics 2022. [Google Scholar] [CrossRef]
- Van Dijk, S.J.; Tellam, R.L.; Morrison, J.L.; Muhlhausler, B.S.; Molloy, P.L. Recent developments on the role of epigenetics in obesity and metabolic disease. Clinical Epigenetics 2015, 7. [Google Scholar] [CrossRef] [PubMed]
- Angermueller, C.; Lee, H.J.; Reik, W.; Stegle, O. DeepCpG: accurate prediction of single-cell DNA methylation states using deep learning. Genome Biology 2017, 18. [Google Scholar]
- Yin, Q.; Wu, M.; Liu, Q.; Lv, H.; Jiang, R. DeepHistone: a deep learning approach to predicting histone modifications. BMC Genomics 2019. [Google Scholar] [CrossRef]
- Galkin, F.; Mamoshina, P.; Kochetov, K.; Sidorenko, D.; Zhavoronkov, A. DeepMAge: A Methylation Aging Clock Developed with Deep Learning. Aging and Disease 2021, 12, 1252. [Google Scholar] [CrossRef]
- Zhang, B.; Li, J.; Lü, Q. Prediction of 8-state protein secondary structures by a novel deep learning architecture. BMC Bioinformatics 2018, 19. [Google Scholar] [CrossRef]
- AlQuraishi, M. AlphaFold at CASP13. Bioinformatics 2019, 35, 4862–4865. [Google Scholar] [CrossRef]
- Du, X.; Sun, S.; Hu, C.; Yao, Y.; Yan, Y.; Zhang, Y. DeepPPI: Boosting Prediction of Protein–Protein Interactions with Deep Neural Networks. Journal of Chemical Information Modeling 2017, 57, 1499–1510. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Mo, B.; Zhao, J.; Zhao, J.; Zhao, J. Theory-based residual neural networks: A synergy of discrete choice models and deep neural networks. Transportation Research Part B 2021. [Google Scholar] [CrossRef]
- Cramer, P. AlphaFold2 and the future of structural biology. Nature Structural & Molecular Biology 2021, 28, 704–705. [Google Scholar] [CrossRef]
- Baek, M.; Baker, D. Deep learning and protein structure modeling. Nature methods 2022, 19, 13–14. [Google Scholar] [CrossRef]
- Zeng, H.; Edwards, M.D.; Liu, G.; Gifford, D.K. Convolutional neural network architectures for predicting DNA–protein binding. Bioinformatics 2016, 32, i121–i127. [Google Scholar] [CrossRef]
- Zhang, S.; Zhou, J.; Hu, H.; Gong, H.; Chen, L.; Cheng, C.; Zeng, J. A deep learning framework for modeling structural features of RNA-binding protein targets. Nucleic acids research 2016, 44, e32–e32. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Torrisi, M.; Pollastri, G.; Le, Q. Deep learning methods in protein structure prediction. Computational and Structural Biotechnology Journal 2020, 18. [Google Scholar] [CrossRef]
- Jha, K.; Saha, S.; Singh, H. Prediction of protein-protein interaction using graph neural networks. Scientific reports 2022, 12, 8360. [Google Scholar] [CrossRef]
- Du, X.; Sun, S.; Hu, C.; Yao, Y.; Yan, Y.; Zhang, Y. DeepPPI: Boosting Prediction of Protein- Protein Interactions with Deep Neural Networks. J Chem Inf Model. 2017, 57, 1499–1510. [Google Scholar] [CrossRef]
- Hashemifar, S.; Neyshabur, B.; Khan, A.A.; Xu, J. Predicting protein-protein interactions through sequence-based deep learning. Bioinformatics 2018, 34, i802–i810. [Google Scholar] [CrossRef]
- Yao, Y.; Du, X.; Diao, Y.; Zhu, H. An integration of deep learning with feature embedding for protein–protein interaction prediction. PeerJ 2019, 2019, e7126. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Fan, K.; Song, D.; Lin, H. Graph-based prediction of Protein-protein interactions with attributed signed graph embedding. BMC Bioinformatics 2020, 21, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Dara, S.; Dhamercherla, S.; Jadav, S.S.; Babu, C.M.; Ahsan, M.J. Machine Learning in Drug Discovery: A Review. Artificial intelligence review 2022, 55, 1947–1999. [Google Scholar] [CrossRef]
- Wen, M.; Zhang, Z.; Niu, S.; Sha, H.; Yang, R.; Yun, Y.; Lu, H. Deep-Learning- Based Drug-Target Interaction Prediction. Journal of proteome research 2017, 16, 1401–1409. [Google Scholar] [CrossRef]
- Suh, D.; Lee, J.W.; Choi, S.; Lee, Y. Recent applications of deep learning methods on evolution-and contact-based protein structure prediction. International Journal of Molecular Sciences 2021, 22, 6032. [Google Scholar] [CrossRef]
- Kuang, M.; Liu, Y.; Gao, L. DLPAlign: a deep learning based progressive alignment method for multiple protein sequences. In CSBio'20: Proceedings of the Eleventh International Conference on Computational Systems-Biology and Bioinformatics; 2020; pp. 83–92. [Google Scholar]
- Gao, M.; Zhou, H.; Skolnick, J. DESTINI: A deep-learning approach to contact- driven protein structure prediction. Scientific reports 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Yang, Y.; Xia, C.Q.; Mirza, A.H.; Shen, H.B. Recent methodology progress of deep learning for RNA–protein interaction prediction. Wiley Interdisciplinary Reviews: RNA 2019, 10, e1544. [Google Scholar] [CrossRef] [PubMed]
- Sapoval, N.; Aghazadeh, A.; Nute, M.G.; et al. Current progress and open challenges for applying deep learning across the biosciences. Nat Commun 2022, 13, 1728. [Google Scholar] [CrossRef]
- Akay, A.; Hess, H. Deep Learning: Current and Emerging Applications in Medicine and Technology. IEEE Journal of Biomedical and Health Informatics 2019, 23, 906–920. [Google Scholar] [CrossRef]
- Choudhary, K.; DeCost, B.L.; Low, W.C.; Jain, A.; Tavazza, F.; Cohn, R.; Wolverton, C. Recent advances and applications of deep learning methods in materials science. Npj Computational Materials 2022, 8. [Google Scholar] [CrossRef]
- O’Connor, O.M.; Alnahhas, R.N.; Lugagne, J.; Dunlop, M.J. DeLTA 2.0: A deep learning pipeline for quantifying single-cell spatial and temporal dynamics. PLOS Computational Biology 2022, 18, e1009797. [Google Scholar] [CrossRef]
- Fernandez-Quilez, A. Deep learning in radiology: ethics of data and on the value of algorithm transparency, interpretability and explainability. AI Ethics 2023, 3, 257–265. [Google Scholar] [CrossRef]
- Zhang, Y.; Ye, T.; Xi, H.; Juhas, M.; Li, J.J. Deep Learning Driven Drug Discovery: Tackling Severe Acute Respiratory Syndrome Coronavirus 2. Frontiers in Microbiology 2021, 12. [Google Scholar] [CrossRef]
- Chen, H.; Engkvist, O.; Wang, Y.; Olivecrona, M.; Blaschke, T. The rise of deep learning in drug discovery. Drug Discovery Today 2018, 23, 1241–1250. [Google Scholar] [CrossRef] [PubMed]
- Beck, B.R.; Shin, B.; Choi, Y.; Park, S.; Kang, K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug- target interaction deep learning model. Computational and Structural Biotechnology Journal 2020, 18, 784–790. [Google Scholar] [CrossRef] [PubMed]
- Bai, Q.; Liu, S.; Tian, Y.; Xu, T.; Banegas-Luna, A.J.; Pérez-Sánchez, H.; Yao, X. Application advances of deep learning methods for de novo drug design and molecular dynamics simulation. Wiley Interdisciplinary Reviews: Computational Molecular Science 2021, 12. [Google Scholar] [CrossRef]
- Pan, X.; Lin, X.; Cao, D.; Zeng, X.; Yu, P.S.; He, L.; Cheng, F. Deep learning for drug repurposing: methods, databases, and applications. arXiv 2022, arXiv:10.48550/arxiv.2202.05145. [Google Scholar] [CrossRef]
- Jobin, A.; Vayena, E. The global landscape of AI ethics guidelines. Nature Machine Intelligence 2019, 1, 389–399. [Google Scholar] [CrossRef]
- Naik, N.; Tokas, T.; Shetty, D.K.; Swain, D.; Shah, M.; Paul, R.; Somani, B.K. Legal and Ethical Consideration in Artificial Intelligence in Healthcare: Who Takes Responsibility? Frontiers in Surgery 2022, 9. [Google Scholar] [CrossRef] [PubMed]
- Lo Piano, S. Ethical principles in machine learning and artificial intelligence: cases from the field and possible ways forward. Humanities & Social Sciences Communications 2020. [Google Scholar] [CrossRef]
- Ching, T.; Himmelstein, D.; Beaulieu-Jones, B.K.; Kalinin, A.A.; Do, B.T.; Way, G.P.; Greene, C.S. Opportunities and obstacles for deep learning in biology and medicine. Journal of the Royal Society Interface 2018, 15, 20170387. [Google Scholar] [CrossRef]
- Sharma, P.B.; Ramteke, P. Recommendation for Selecting Smart Village in India through Opinion Mining Using Big Data Analytics. Indian Scientific Journal of Research in Engineering and Management 2023, 07. [Google Scholar] [CrossRef]
- Naik, N.; Tokas, T.; Shetty, D.K.; Swain, D.; Shah, M.; Paul, R.; Somani, B.K. Legal and Ethical Consideration in Artificial Intelligence in Healthcare: Who Takes Responsibility? Frontiers in Surgery 2022, 9. [Google Scholar] [CrossRef]
- Braun, M.; Hummel, P.; Beck, S.C.; Dabrock, P. Primer on an ethics of AI-based decision support systems in the clinic. Journal of Medical Ethics 2021, 47, e3. [Google Scholar] [CrossRef] [PubMed]
- Jarrahi, M.H.; Askay, D.A.; Eshraghi, A.; Smith, P. Artificial intelligence and knowledge management: A partnership between human and AI. Business Horizons 2022, 66, 87–99. [Google Scholar] [CrossRef]
- Terranova, N.; Venkatakrishnan, K.; Benincosa, L.J. Application of Machine Learning in Translational Medicine: Current Status and Future Opportunities. Aaps Journal 2021, 23. [Google Scholar] [CrossRef]
- Hartl, D.; De Luca, V.; Kostikova, A.; Laramie, J.M.; Kennedy, S.D.; Ferrero, E.; Siegel, R.M.; Fink, M.; Ahmed, S.; Millholland, J.; et al. Translational precision medicine: an industry perspective. Journal of Translational Medicine 2021, 19. [Google Scholar] [CrossRef] [PubMed]
- Zitnik, M.; Nguyen, F.; Wang, B.; Leskovec, J.; Goldenberg, A.; Hoffman, M.M. Machine learning for integrating data in biology and medicine: Principles, practice, and opportunities. Information Fusion 2018, 50, 71–91. [Google Scholar] [CrossRef] [PubMed]
- Sarker, I.H. Deep Learning: A Comprehensive Overview on Techniques, Taxonomy, Applications and Research Directions. SN Computer Science 2021, 2. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–44. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural computation 1997, 9, 1735–80. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.; Lin, C.H.; Lane, H.Y. De novo peptide and protein design using generative adversarial networks: an update. Journal of Chemical Information and Modeling 2022, 62, 761–74. [Google Scholar] [CrossRef]
- Xu, J.; Wang, S. Analysis of distance-based protein structure prediction by deep learning in CASP13. Proteins: Structure, Function, and Bioinformatics 2019, 87, 1069–81. [Google Scholar] [CrossRef]
- Tan, R.K.; Liu, Y.; Xie, L. Reinforcement learning for systems pharmacology-oriented and personalized drug design. Expert Opinion on Drug Discovery 2022, 17, 849–63. [Google Scholar] [CrossRef]
- Feng, C.; Wang, W.; Han, R.; Wang, Z.; Ye, L.; Du, Z.; Wei, H.; Zhang, F.; Peng, Z.; Yang, J. Accurate de novo prediction of RNA 3D structure with transformer network. bioRxiv 2022. [Google Scholar]
- Wang, J.; Xie, X.; Shi, J.; He, W.; Chen, Q.; Chen, L.; Gu, W.; Zhou, T. Denoising autoencoder, a deep learning algorithm, aids the identification of a novel molecular signature of lung adenocarcinoma. Genomics, proteomics & bioinformatics 2020, 18, 468–80. [Google Scholar]
- Jha, K.; Saha, S.; Singh, H. Prediction of protein–protein interaction using graph neural networks. Scientific Reports 2022, 12, 8360. [Google Scholar] [CrossRef] [PubMed]
- Svensson, V.; Gayoso, A.; Yosef, N.; Pachter, L. Interpretable factor models of single-cell RNA- seq via variational autoencoders. Bioinformatics 2020, 36, 3418–21. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, C.; Das, B.; Rawat, V.S.; Wahlang, J.B.; Nongpiur, A.; Tiewsoh, I.; Lyngdoh, N.M.; Das, D.; Bidarolli, M.; Sony, H.T. Artificial Intelligence and Machine Learning Technology Driven Modern Drug Discovery and Development. International Journal of Molecular Sciences 2023, 24, 2026. [Google Scholar] [CrossRef]
- Hinton, G.E.; Sabour, S.; Frosst, N. Matrix capsules with EM routing. InInternational conference on learning representations 2018. [Google Scholar]
- Das, D.; Chakrabarty, B.; Srinivasan, R.; Roy, A. Gex2SGen: Designing Drug-like Molecules from Desired Gene Expression Signatures. Journal of Chemical Information and Modeling 2023, 63, 1882–93. [Google Scholar] [CrossRef]
- Li, Z.; Gao, E.; Zhou, J.; Han, W.; Xu, X.; Gao, X. Applications of deep learning in understanding gene regulation. Cell Reports Methods 2023, 100384. [Google Scholar] [CrossRef]
- Bansal, H.; Luthra, H.; Chaurasia, A. Impact of Machine Learning Practices on Biomedical Informatics, Its Challenges and Future Benefits. In Artificial Intelligence Technologies for Computational Biology; CRC Press, 2023; pp. 273–294. [Google Scholar]
- Feng, X.; Zhang, H.; Lin, H.; Long, H. Single-cell RNA-seq data analysis based on directed graph neural network. Methods 2023, 211, 48–60. [Google Scholar] [CrossRef]
- Dou, H.; Tan, J.; Wei, H.; Wang, F.; Yang, J.; Ma, X.G.; Wang, J.; Zhou, T. Transfer inhibitory potency prediction to binary classification: A model only needs a small training set. Computer Methods and Programs in Biomedicine 2022, 215, 106633. [Google Scholar] [CrossRef]
- Hou, Z.; Yang, Y.; Ma, Z.; Wong, K.C.; Li, X. Learning the protein language of proteome-wide protein-protein binding sites via explainable ensemble deep learning. Communications Biology 2023, 6, 73. [Google Scholar] [CrossRef] [PubMed]
- Yelmen, B.; Jay, F. An Overview of Deep Generative Models in Functional and Evolutionary Genomics. Annual Review of Biomedical Data Science 2023, 6. [Google Scholar] [CrossRef]
- Chen, P.; Zheng, H. Drug-target interaction prediction based on spatial consistency constraint and graph convolutional autoencoder. BMC bioinformatics 2023, 24, 1–21. [Google Scholar] [CrossRef]
- Zheng, S.; Guo, J.; Langendijk, J.A.; Both, S.; Veldhuis, R.N.; Oudkerk, M.; van Ooijen, P.M.; Wijsman, R.; Sijtsema, N.M. Survival prediction for stage I-IIIA non-small cell lung cancer using deep learning. Radiotherapy and Oncology 2023, 109483. [Google Scholar] [CrossRef]
- Li, Z.; Gao, E.; Zhou, J.; Han, W.; Xu, X.; Gao, X. Applications of deep learning in understanding gene regulation. Cell Rep Methods 2023, 3, 100384. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Cai, X.; Xu, S.; Ji, H. Metapath-aggregated heterogeneous graph neural network for drug– target interaction prediction. Briefings in Bioinformatics 2023, 24, bbac578. [Google Scholar] [CrossRef] [PubMed]
- Veturi, Y.A.; Woof, W.; Lazebnik, T.; Moghul, I.; Woodward-Court, P.; Wagner, S.K.; de Guimarães, T.A.; Varela, M.D.; Liefers, B.; Patel, P.J.; et al. SynthEye: Investigating the Impact of Synthetic Data on Artificial Intelligence-assisted Gene Diagnosis of Inherited Retinal Disease. Ophthalmology Science 2023, 3, 100258. [Google Scholar] [CrossRef]
- Suhartono, D.; Majiid, M.R.; Handoyo, A.T.; Wicaksono, P.; Lucky, H. Towards a more general drug target interaction prediction model using transfer learning. Procedia Computer Science 2023, 216, 370–6. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Sinha, D. Synthetic attack data generation model applying generative adversarial network for intrusion detection. Computers & Security 2023, 125, 103054. [Google Scholar]
- Yang, K.; Huang, H.; Vandans, O.; Murali, A.; Tian, F.; Yap, R.H.; Dai, L. Applying deep reinforcement learning to the HP model for protein structure prediction. Physica A: Statistical Mechanics and its Applications 2023, 609, 128395. [Google Scholar] [CrossRef]
- Bhatti, U.A.; Tang, H.; Wu, G.; Marjan, S.; Hussain, A. Deep learning with graph convolutional networks: An overview and latest applications in computational intelligence. International Journal of Intelligent Systems 2023, 2023, 1–28. [Google Scholar] [CrossRef]
- Mreyoud, Y.; Song, M.; Lim, J.; Ahn, T.H. MegaD: deep learning for rapid and accurate disease status prediction of metagenomic samples. Life 2022, 12, 669. [Google Scholar] [CrossRef]
- Hamdy, R.; Maghraby, F.A.; Omar, Y.M. Convchrome: Predicting gene expression based on histone modifications using deep learning techniques. Current Bioinformatics 2022, 17, 273–83. [Google Scholar] [CrossRef]
- Cai, J.; Wang, S.; Xu, C.; Guo, W. Unsupervised deep clustering via contractive feature representation and focal loss. Pattern Recognition 2022, 123, 108386. [Google Scholar] [CrossRef]
- Stepniewska-Dziubinska, M.M.; Zielenkiewicz, P.; Siedlecki, P. Development and evaluation of a deep learning model for protein–ligand binding affinity prediction. Bioinformatics 2018, 34, 3666–74. [Google Scholar] [CrossRef]
- Jukič, M.; Bren, U. Machine learning in antibacterial drug design. Frontiers in Pharmacology 2022, 13, 1284. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, T.; Sayed, S.; Salah, A.; Houssein, E.H. Long short-term memory neural networks for RNA viruses mutations prediction. Mathematical Problems in Engineering 2021, 2021, 1–9. [Google Scholar] [CrossRef]
- Sharma, P.; Dahiya, S.; Kaur, P.; Kapil, A. Computational biology: Role and scope in taming antimicrobial resistance. Indian Journal of Medical Microbiology 2023, 41, 33–8. [Google Scholar] [CrossRef] [PubMed]
- Giri, S.J.; Dutta, P.; Halani, P.; Saha, S. MultiPredGO: deep multi-modal protein function prediction by amalgamating protein structure, sequence, and interaction information. IEEE Journal of Biomedical and Health Informatics 2020, 25, 1832–8. [Google Scholar] [CrossRef]
- Zhao, J.P.; Hou, T.S.; Su, Y.; Zheng, C.H. scSSA: A clustering method for single cell RNA-seq data based on semi-supervised autoencoder. Methods 2022, 208, 66–74. [Google Scholar] [CrossRef]
- Mamoshina, P.; Vieira, A.; Putin, E.; Zhavoronkov, A. Applications of deep learning in biomedicine. Molecular pharmaceutics 2016, 13, 1445–54. [Google Scholar] [CrossRef]
- Ieremie, I.; Ewing, R.M.; Niranjan, M. TransformerGO: predicting protein–protein interactions by modelling the attention between sets of gene ontology terms. Bioinformatics 2022, 38, 2269–77. [Google Scholar] [CrossRef]
- Sapoval, N.; Aghazadeh, A.; Nute, M.G.; et al. Current progress and open challenges for applying deep learning across the biosciences. Nat Commun 2022, 13, 1728. [Google Scholar] [CrossRef]
- Feng, X.; Zhang, H.; Lin, H.; Long, H. Single-cell RNA-seq data analysis based on directed graph neural network. Methods 2023, 211, 48–60. [Google Scholar] [CrossRef]
- Yelmen, B.; Jay, F. An overview of deep generative models in functional and evolutionary genomics. Annu Rev Biomed Data Sci 2023, 6, 173–189. [Google Scholar] [CrossRef]
- Hou, Z.; Yang, Y.; Ma, Z.; Wong, K.C.; Li, X. Learning the protein language of proteome-wide protein-protein binding sites via explainable ensemble deep learning. Commun Biol 2023, 6, 73. [Google Scholar] [CrossRef]
- de Lima, L.M.; de Assis, M.C.; Soares, J.P.; Grão-Velloso, T.R.; de Barros, L.A.; Camisasca, D.R.; Krohling, R.A. On the importance of complementary data to histopathological image analysis of oral leukoplakia and carcinoma using deep neural networks. Intelligent Medicine 2023. [Google Scholar] [CrossRef]
- Gupta, A.; Zou, J. Feedback GAN for DNA optimizes protein functions. Nature Machine Intelligence 2019, 1, 105–11. [Google Scholar] [CrossRef]
- Lin, P.; Yan, Y.; Huang, S.Y. DeepHomo2. 0: improved protein–protein contact prediction of homodimers by transformer-enhanced deep learning. Briefings in Bioinformatics 2023, 24, bbac499. [Google Scholar] [CrossRef]
- Xavier, R.; de Souza, K.P.; Chateau, A.; Alves, R. Genome assembly using reinforcement learning. In Advances in Bioinformatics and Computational Biology: 12th Brazilian Symposium on Bioinformatics, BSB 2019, Fortaleza, Brazil, October 7–10, 2019, Revised Selected Papers 1; Springer International Publishing, 7 October 2020; pp. 16–28. [Google Scholar]
- Wang, J.; Ma, A.; Chang, Y.; Gong, J.; Jiang, Y.; Qi, R.; Wang, C.; Fu, H.; Ma, Q.; Xu, D. scGNN is a novel graph neural network framework for single-cell RNA-Seq analyses. Nature communications 2021, 12, 1882. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.K.; Azizan, F.L.; Wong, Y.S.; Omar, N. DeepFinder: An integration of feature-based and deep learning approach for DNA motif discovery. Biotechnology & Biotechnological Equipment 2018, 32, 759–68. [Google Scholar]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).