1. Introduction: The Enduring Challenge of Recombinant Protein Production
1.1. The Recombinant Revolution
The advent of recombinant DNA technology in the 1970s marked a watershed moment in molecular biology and biotechnology, fundamentally altering the landscape of medicine, industry, and basic research [
1]. This technology endowed scientists with the remarkable capability to clone a foreign gene and express it in a heterologous host, enabling the production of virtually any protein in abundant quantities [
2]. This breakthrough sparked a revolution, moving proteins from rare, difficult-to-isolate natural products to readily available reagents and therapeutics [
1]. The approval of the first recombinant therapeutic protein, human insulin produced in
Escherichia coli, by the FDA in 1982, was a landmark achievement that paved the way for a multi-billion dollar biopharmaceutical market [
3]. Today, recombinant proteins are indispensable tools used for the treatment of a wide array of diseases, including diabetes, cancer, infectious disorders, and anemia, and serve as critical components in countless diagnostic kits [
4]. Beyond medicine, recombinant enzymes are workhorses in various industrial sectors, including biofuels, food processing, and waste treatment, driving efficiency and sustainability [
5]. This ability to harness cellular machinery for targeted protein synthesis remains one of the most significant achievements of modern biotechnology [
1].
1.2. Persistent Bottlenecks in Biomanufacturing
Despite nearly five decades of advancement, the process of producing recombinant proteins is fraught with challenges that can impede efficiency, escalate costs, and limit the successful production of many “difficult-to-express” targets [
6]. These bottlenecks span the entire biomanufacturing workflow, from the initial expression within the host cell to the final purified product [
7].
A primary challenge originates within the host cell itself. The forced overexpression of a foreign gene imposes a significant metabolic burden, redirecting essential cellular resources such as amino acids, energy (ATP), and redox cofactors (NAD(P)H) away from normal physiological processes like growth and maintenance [
8]. This competition for resources can trigger complex cellular stress responses, including the heat shock response and the unfolded protein response (UPR) in eukaryotes, which can negatively impact cell fitness, reduce growth rates, and ultimately limit the final protein titer [
9]. Furthermore, the expressed protein itself can be toxic to the host, or its aggregation can lead to cellular damage, further constraining productivity . Consequently, the initial focus of the field on simply maximizing transcription and translation through strong promoters and high-copy-number plasmids has evolved [
4]. It is now widely recognized that a successful expression strategy must balance high-level synthesis with the physiological capacity of the host to sustain it, representing a shift from a “brute force” approach to a more nuanced, systems-level understanding of host cell engineering [
4].
Equally significant are challenges related to the quality of the expressed protein. Many expression systems, particularly the rapid and high-yielding bacterial hosts like
E. coli, often produce the target protein in a misfolded and insoluble state [
10]. These aggregates, known as inclusion bodies (IBs), are dense, non-functional particles that accumulate within the cell [
6]. While IBs contain a high concentration of the desired protein and can protect it from proteolytic degradation, recovering bioactive protein from them requires arduous downstream processing steps [
11]. Furthermore, many complex eukaryotic proteins, especially therapeutics, require specific post-translational modifications (PTMs) such as glycosylation, phosphorylation, or the formation of multiple disulfide bonds to achieve their correct three-dimensional structure and biological activity [
7]. Prokaryotic hosts lack the cellular machinery for most of these modifications, necessitating the use of more complex and expensive eukaryotic systems [
7].
These upstream and protein quality issues feed directly into the largest bottleneck in the entire process: downstream processing. The recovery, refolding, and purification of a recombinant protein from the complex milieu of host cell components can account for up to 80% of the total manufacturing cost [
12]. The conventional process for recovering protein from IBs involves solubilization with high concentrations of harsh chaotropic agents like 6–8 M urea or guanidinium chloride (GdnHCl), which completely denature the protein [
13]. The subsequent refolding step, typically achieved by rapidly diluting or dialyzing away the denaturant, is often inefficient, with protein molecules prone to re-aggregating rather than achieving their native conformation, leading to low recovery of active product [
14]. This multi-step, chemically intensive process not only drives up costs but also generates significant chemical waste, creating a “sustainability gap” where the final bioproduct may be green, but the process to make it is not [
15].
1.3. The Emergence of NADES as a Green and Functional Solvent Platform
In the quest for more sustainable and efficient biotechnological processes, a novel class of solvents known as Natural Deep Eutectic Solvents (NADES) has garnered significant attention [
16]. A NADES is a mixture of two or more naturally occurring compounds, typically a hydrogen bond acceptor (HBA) like choline chloride and a hydrogen bond donor (HBD) such as a sugar, amino acid, or organic acid [
17]. When mixed at a specific molar ratio, these components form strong intermolecular hydrogen bonds that disrupt their individual crystal lattices, resulting in a eutectic mixture with a melting point significantly lower than that of any of the starting components [
17].
What makes NADES particularly compelling for biotechnology is their unique combination of properties. They are derived from abundant, low-cost primary metabolites, making them inherently biodegradable, biocompatible, and generally non-toxic [
18]. Their preparation is simple and adheres to the principles of green chemistry, often requiring only gentle heating and stirring with 100% atom economy [
16]. Beyond their green credentials, NADES are “designer solvents” [
18]. Their physicochemical properties—including polarity, viscosity, and solubilizing power—can be precisely tuned by judiciously selecting the HBA and HBD components, altering their molar ratio, or adjusting the water content of the mixture [
16]. This designability allows for the creation of task-specific solvents tailored for particular applications, from the selective extraction of bioactive compounds to providing a stabilizing microenvironment for enzymatic reactions [
18,
19].
1.4. Scope and Thesis of the Review
This review will critically assess the intersection of these two fields. It posits that NADES are not merely “green” replacements but are functional materials that can actively solve long-standing problems in heterologous protein production [
18]. The review will systematically map the potential integration points of NADES across the entire bioprocess workflow, from cell culture to final formulation [
20].
2. The Landscape of Heterologous Protein Expression Platforms
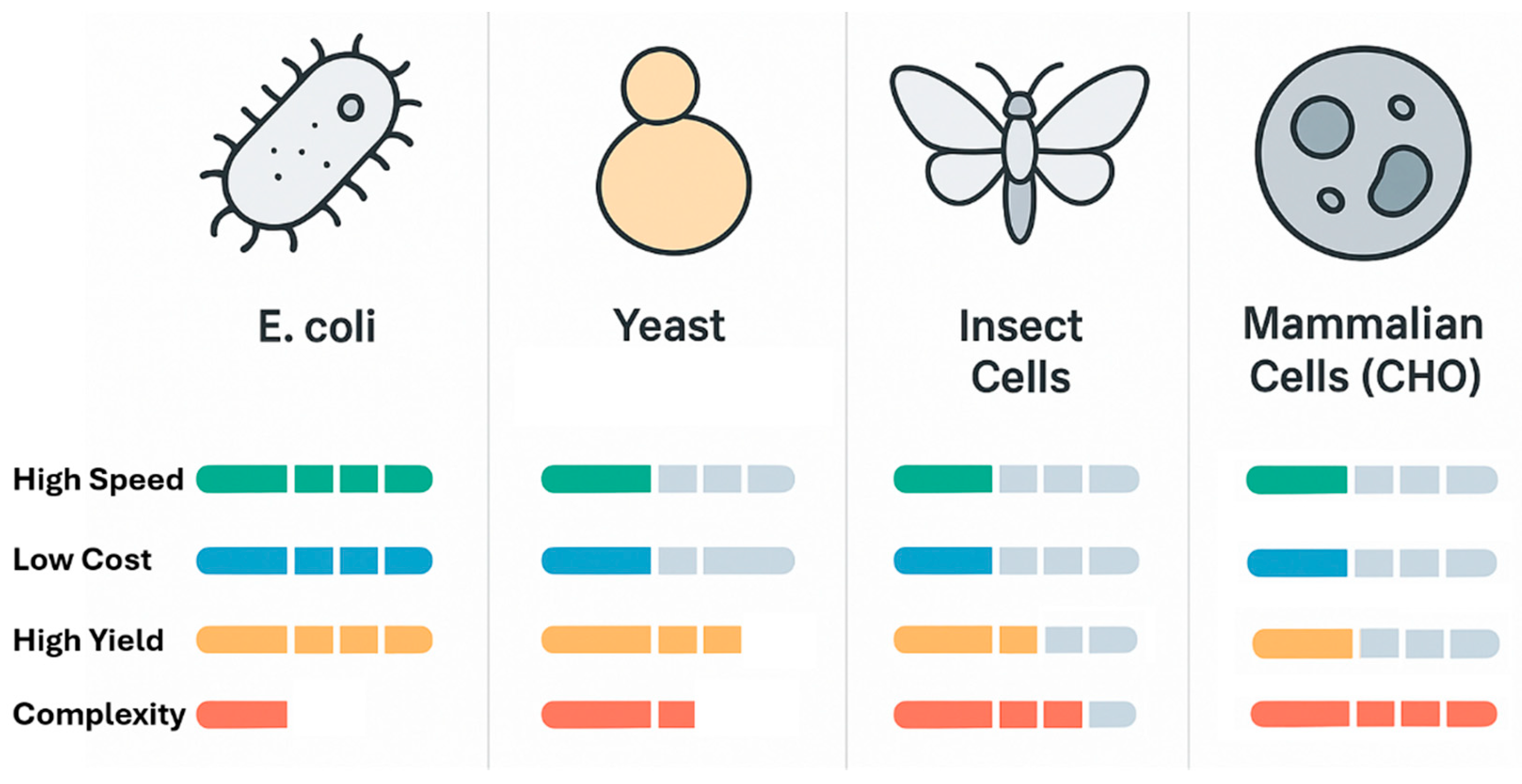
The selection of an appropriate expression system is the first and one of the most critical decisions in any recombinant protein production project. There is no universal host capable of efficiently producing all types of proteins; instead, a diverse array of platforms, spanning from simple bacteria to complex mammalian cells, offers a range of trade-offs between speed, cost, yield, and the ability to perform complex post-translational modifications . The optimal choice depends on the specific characteristics of the target protein and its intended downstream application .
2.1. The Prokaryotic Workhorse: Escherichia coli
Since the dawn of recombinant DNA technology,
Escherichia coli has been the undisputed workhorse for heterologous protein expression [
21]. Its dominance is rooted in a compelling set of advantages: a thoroughly characterized genome and physiology, an extensive and sophisticated genetic toolbox, extremely rapid growth rates (doubling every 20–30 minutes), and the ability to reach very high cell densities in simple, inexpensive media [
22]. These factors combine to offer unparalleled speed and cost-effectiveness, making
E. coli the platform of choice for producing a vast number of proteins for research, industrial, and diagnostic applications [
7]. Under optimal conditions, the expressed protein can accumulate to extraordinary levels, sometimes constituting up to 30–50% of the total cellular protein, resulting in high volumetric productivity [
22].
However, the simplicity and speed of the
E. coli system are also the source of its most significant limitations, particularly for the production of complex eukaryotic proteins [
7]. The primary drawback is its lack of eukaryotic PTM machinery.
E. coli cannot perform glycosylation, a critical modification for the stability and function of many therapeutic proteins[
7]. Furthermore, the highly reducing environment of the bacterial cytoplasm is not conducive to the formation of disulfide bonds, which are essential for the structural integrity of many secreted proteins [
23]. The rapid rate of transcription and translation in
E. coli often outpaces the cell’s protein folding machinery, leading to the misfolding and aggregation of the nascent polypeptide chains into insoluble and inactive inclusion bodies [
7].
Over the decades, significant efforts have been made to overcome these limitations through host strain and process engineering. Specialized
E. coli strains, such as the SHuffle® series, have been engineered with a more oxidizing cytoplasm and co-expression of disulfide bond isomerases to facilitate the correct formation of disulfide bonds [
24]. Other strains, like Lemo21(DE3), allow for precise, tunable control over the expression level of T7 RNA polymerase, thereby slowing down transcription and translation to promote proper folding and improve the yield of soluble protein [
25]. Additional strategies include co-expressing molecular chaperones to assist in the folding process or reducing the cultivation temperature after induction, which slows down cellular processes and often enhances solubility [
7].
2.2. Eukaryotic Microbial Factories: Yeast Systems
Yeast expression systems, particularly
Saccharomyces cerevisiae and
Komagataella phaffii (formerly
Pichia pastoris), represent a powerful compromise, combining the ease of handling and rapid growth of microorganisms with the advanced protein processing capabilities of a eukaryotic cell . As eukaryotes, they possess the machinery for PTMs, including protein folding, disulfide bond formation, and glycosylation, making them suitable for producing complex proteins that fail to express correctly in bacteria [
23].
Saccharomyces cerevisiae, the common baker’s yeast, was the first eukaryotic organism to have its genome sequenced and has been a cornerstone of biological research for decades [
26]. Its Generally Recognized as Safe (GRAS) status, extensive genetic toolkit, and long history of use in industrial fermentations (e.g., brewing, baking) make it an attractive host [
26]. It is capable of secreting proteins into the culture medium, which simplifies downstream purification [
23]. However,
S. cerevisiae has notable limitations, including a tendency to hyperglycosylate proteins with high-mannose structures that can be immunogenic and are different from those found in humans [
27]. Furthermore, expression yields are often lower than those achievable in other systems [
27].
Komagataella phaffii has emerged as one of the most successful and widely used eukaryotic systems for high-level recombinant protein production, especially for industrial enzymes and biopharmaceuticals [
28]. Its key advantages include the ability to grow to extremely high cell densities in simple, defined media, which translates to very high volumetric productivity [
28]. It possesses powerful and tightly regulated promoters, most notably the alcohol oxidase 1 (AOX1) promoter, which is strongly induced by methanol, and the constitutive glyceraldehyde-3-phosphate dehydrogenase (GAP) promoter [
29].
K. phaffii is also an efficient secretor of proteins and performs glycosylation that, while still of the high-mannose type, is generally less extensive and closer to the core structure of mammalian glycans than that of
S. cerevisiae [
28]. Recent advancements in the
K. phaffii platform have focused on overcoming secretion bottlenecks by engineering the protein processing and trafficking pathways, as well as developing sophisticated methods for generating and screening stable, multi-copy integrants of the expression cassette to maximize gene dosage and protein yield [
30].
2.3. Platforms for Complexity: Insect and Mammalian Cell Systems
For the production of the most complex mammalian proteins, particularly those requiring authentic, multi-antennary glycosylation for therapeutic efficacy, higher eukaryotic systems are often indispensable [
31].
Insect Cell Systems, primarily utilizing the Baculovirus Expression Vector System (BEVS), are a robust platform for producing large and complex proteins, including intracellular proteins and multi-protein complexes that are challenging to express in other systems [
32]. The baculovirus vector can accommodate large DNA inserts, allowing for the simultaneous expression of multiple subunits of a protein complex [
32]. Insect cells perform a wide range of PTMs, such as phosphorylation, acetylation, and disulfide bond formation, that are highly similar to those in mammalian cells [
33]. While their N-glycosylation pathway is simpler than that of mammals (paucimannosidic), engineered cell lines are available that can produce more complex, human-like glycan structure [
32]. A significant advantage of BEVS is its inherent safety; the baculoviruses used are insect-specific and cannot replicate in mammalian cells, reducing biosafety concerns [
34]. However, the workflow is more complex and time-consuming than microbial systems, involving the generation of recombinant baculovirus, infection of the insect cell culture, and a lytic production cycle [
33].
Mammalian Cell Systems, with Chinese Hamster Ovary (CHO) cells being the predominant platform, are the gold standard for the production of therapeutic proteins for human use, including the vast majority of monoclonal antibodies [
31]. Their unparalleled advantage is the ability to perform authentic, human-like PTMs, ensuring that the final product has the correct structure, stability, and biological activity, while minimizing the risk of immunogenicity [
35]. However, this fidelity comes at a significant cost. Mammalian cells grow much more slowly than microbes, require complex and expensive culture media, are susceptible to viral contamination, and generally produce lower yields on a per-volume basis [
35]. The process of generating a stable, high-producing cell line is also a lengthy and resource-intensive endeavor [
36].
2.4. A Comparative Outlook and the Rise of Cell-Free Systems
The choice of an expression system involves a critical evaluation of these trade-offs, as summarized in
Table 1 [
37]. A simple, non-glycosylated bacterial enzyme can be produced rapidly and cheaply in
E. coli, whereas a complex human monoclonal antibody for therapeutic use necessitates the fidelity of a mammalian CHO cell line (
Figure 1).
Emerging as a powerful alternative, Cell-Free Protein Synthesis (CFPS) systems bypass the constraints of a living cell altogether [
38]. These systems consist of cellular extracts (typically from
E. coli, wheat germ, or rabbit reticulocytes) containing all the necessary machinery for transcription and translation [
38]. By simply adding an expression plasmid or linear DNA template, protein can be synthesized in a test tube within hours [
38]. This offers unparalleled speed and control over the reaction environment; inhibitors can be omitted, and non-natural amino acids or cofactors can be directly added to the reaction mixture [
38]. However, CFPS systems are generally more expensive and have historically been difficult to scale for large-scale industrial production, though significant progress is being made in this area [
39].
Table 1.
Comparative Analysis of Major Heterologous Expression Systems. This table provides a summary of the key characteristics and trade-offs associated with the primary platforms used for recombinant protein production. The selection of a system is a strategic decision based on the specific requirements of the target protein and the desired scale of production.
Table 1.
Comparative Analysis of Major Heterologous Expression Systems. This table provides a summary of the key characteristics and trade-offs associated with the primary platforms used for recombinant protein production. The selection of a system is a strategic decision based on the specific requirements of the target protein and the desired scale of production.
| Feature |
Escherichia coli |
Saccharomyces cerevisiae |
Komagataella phaffii (Pichia pastoris) |
Insect Cells (BEVS) |
Mammalian Cells (CHO) |
Cell-Free Systems |
| Speed (Time to Protein) |
Very Fast (1–3 days) [7] |
Fast (3–7 days) [26] |
Fast (3–10 days) [29] |
Moderate (7–14 days) [33] |
Slow (Weeks to Months for stable line) [35] |
Extremely Fast (Hours) [38] |
| Cost (Media & Setup) |
Very Low [21] |
Low [27] |
Low [30] |
Moderate [33] |
Very High [35] |
High (per mg protein) |
| Typical Yield |
High (1–10 g/L) [22] |
Moderate (0.1–1 g/L) [26] |
Very High (1–15+ g/L) [29] |
High (0.1–1 g/L) [33] |
Moderate to High (0.5–10+ g/L) [35] |
Low to Moderate (mg/mL scale) [38] |
| Post-Translational Mods. |
None (Limited engineered options) [7] |
Yes (Hyperglycosylation) [27] |
Yes (Less hyperglycosylation) [28] |
Yes (Complex, near-mammalian) [32] |
Yes (Authentic, human-like) [35] |
Limited (Can be supplemented) [38] |
| Disulfide Bond Formation |
Challenging (Requires engineered strains) [23] |
Efficient (Secretory pathway) [26] |
Efficient (Secretory pathway) [30] |
Efficient (Secretory pathway) [33] |
Efficient (Secretory pathway) [40] |
Possible (Requires redox control) [38] |
| Scalability |
Excellent [21] |
Excellent [26] |
Excellent [30] |
Good [34] |
Excellent (Industry standard) [36] |
Challenging for large scale [39] |
| Key Advantage |
Speed, cost, and high yield for simple proteins [7] |
GRAS status, well-characterized genetics [26] |
Very high cell density and secretion levels [28] |
High yield of complex, multi-subunit proteins [32] |
Authentic PTMs for therapeutics [35] |
Speed, open system for modifications [38] |
| Primary Limitation |
Inclusion bodies, lack of PTMs [7] |
Hyperglycosylation, lower yields [27] |
Methanol use (for AOX1), secretion bottlenecks [29] |
More complex workflow than microbes [33] |
Slow, expensive, complex media [35] |
Cost, scalability, lower yields [38] |
3. Natural Deep Eutectic Solvents (NADES): Properties and Biotechnological Promise
The search for sustainable technologies has propelled the exploration of green solvents as alternatives to volatile and often toxic organic compounds [
16]. Among these, Natural Deep Eutectic Solvents (NADES) have emerged as a particularly promising and versatile class of liquids [
18]. Their unique properties, derived from naturally occurring metabolites, make them not just environmentally benign alternatives but also highly functional media for a range of biotechnological applications [
18,
19].
3.1. Fundamentals: From Eutectic Mixtures to Designer Solvents
The term “eutectic” describes a mixture of components that, at a specific molar ratio, melts or solidifies at a single temperature that is lower than the melting points of the individual constituents [
17]. A NADES is a specific type of deep eutectic solvent (DES) where the components are primary metabolites, such as amino acids, organic acids, sugars, and choline derivatives [
18]. These components, often solids at room temperature, form a liquid mixture through the establishment of an extensive hydrogen-bonding network [
17].
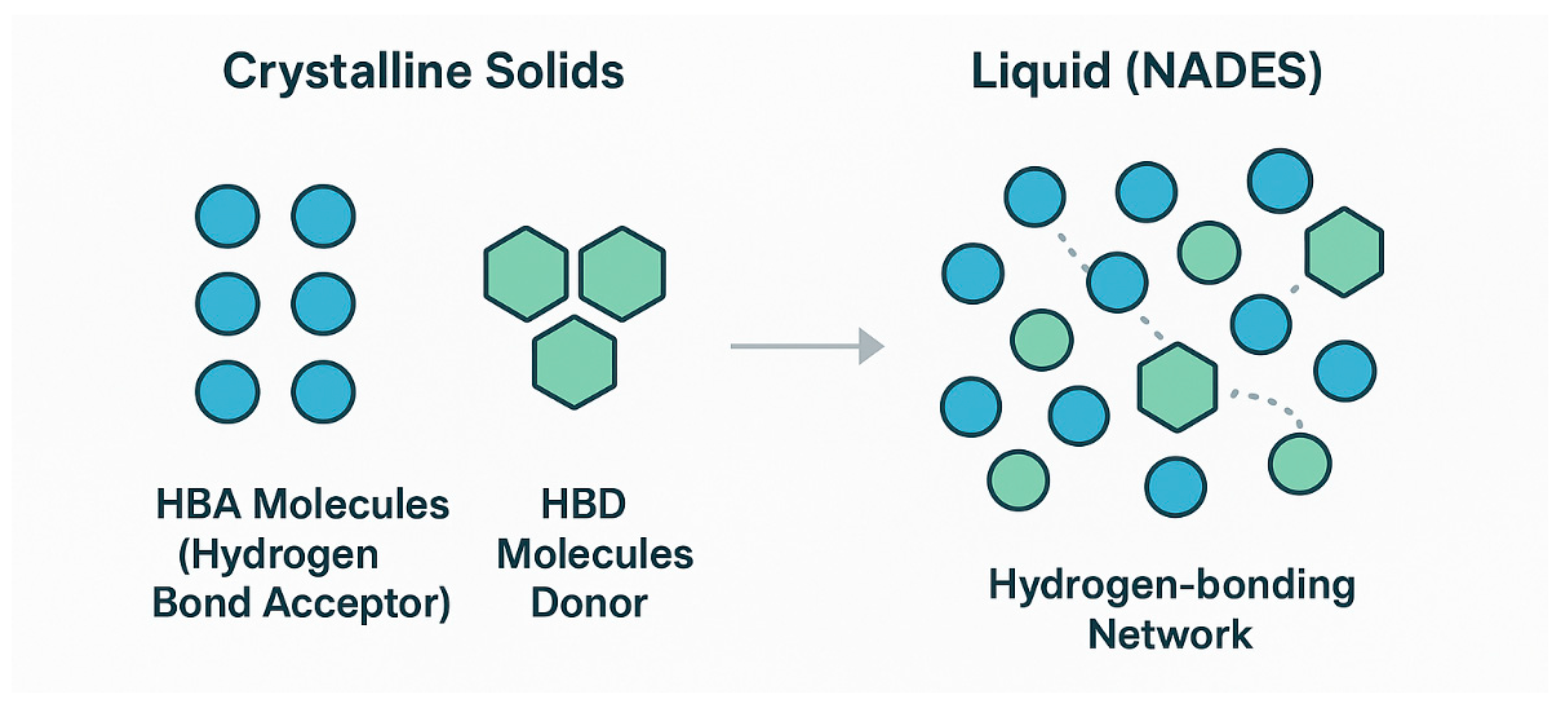
The formation of a NADES is driven by the interaction between a hydrogen bond acceptor (HBA) and a hydrogen bond donor (HBD) (
Figure 2) [
17]. The HBA, typically a quaternary ammonium salt like choline chloride, interacts strongly with the HBD (e.g., urea, glycerol, or glucose) [
17]. This interaction disrupts the crystalline lattice structures of the individual solid components, leading to charge delocalization and a significant depression of the mixture’s freezing point [
18]. The result is a liquid with unique supramolecular organization, often described as a third liquid phase within biological systems, distinct from water and lipids [
41].
The preparation of NADES is remarkably simple and aligns with the principles of green chemistry [
16]. Common methods include:
Heating and Stirring: The most common method, where HBA and HBD components are mixed and heated (typically 50–100 °C) with stirring until a clear, homogeneous liquid is formed [
17].
Evaporation/Freeze-Drying: The components are first dissolved in water, and the water is subsequently removed by rotary evaporation or lyophilization to leave the pure NADES [
17].
Grinding: For some combinations, simply grinding the solid components together in a mortar and pestle at room temperature is sufficient to induce the formation of the liquid eutectic mixture [
42].
Ultrasound- or Microwave-Assisted Synthesis: These methods use energy input to accelerate the formation of the H-bond network, offering faster and more efficient synthesis [
16].
3.2. Key Physicochemical Properties for Bioprocessing
The true power of NADES lies in their “designer” nature; their properties are not fixed but can be precisely tailored for a specific task [
18]. This tunability is achieved by varying the choice of HBA and HBD, their molar ratio, and the water content of the mixture [
16].
Viscosity and Conductivity: NADES are generally characterized by high viscosity, a consequence of their extensive and strong intermolecular hydrogen-bonding network [
16]. This high viscosity can be a significant challenge for industrial applications, as it can impede mass transfer and handling [
43]. However, viscosity is highly sensitive to both temperature and water content; increasing either can dramatically reduce viscosity, making the solvent more tractable [
16]. Conductivity is typically inversely proportional to viscosity, as lower viscosity allows for greater mobility of any charged species within the mixture [
43].
Polarity and Solubilizing Power: NADES can be designed to span a very wide range of polarities, from highly hydrophilic (e.g., sugar-based) to hydrophobic (e.g., menthol- or fatty acid-based) [
16]. This versatility allows them to dissolve a remarkable array of solutes, including many natural and synthetic compounds that are poorly soluble in water [
18]. For example, certain NADES have been shown to increase the solubility of compounds like rutin and quercetin by several orders of magnitude compared to water [
44]. This exceptional solubilizing ability is attributed to the complex H-bond network that can interact favorably with diverse functional groups on a solute molecule [
18].
Biocompatibility and Biodegradability: Because their constituent parts are natural metabolites, NADES are generally considered to be biocompatible, non-toxic, and readily biodegradable [
18]. This is a stark contrast to many conventional organic solvents and even some ionic liquids, which can be toxic and environmentally persistent [
16]. Numerous studies have demonstrated the low cytotoxicity of various NADES formulations against different cell lines, including human cells and microorganisms like
E. coli[
18]. However, it is crucial to note that biocompatibility is not universal and is highly dependent on the specific NADES composition; for instance, some organic acid-based NADES can exhibit higher toxicity than polyol-based ones [
44].
3.3. Established Applications in Biotechnology
The unique combination of green credentials and functional properties has led to the rapid adoption of NADES in various areas of biotechnology [
18].
Green Extraction: NADES are highly effective solvents for the extraction of bioactive compounds from natural sources like plants and microbial biomass [
45]. Their ability to be tailored to match the polarity of a target analyte allows for highly selective and efficient extractions, often surpassing the performance of traditional solvents like methanol or ethanol [
18].
Biocatalysis and Enzyme Stabilization: NADES provide a unique microenvironment that can significantly enhance the stability and activity of enzymes [
46]. The extensive hydrogen-bonding network of the solvent can interact with the enzyme’s surface, preventing unfolding at high temperatures or in the presence of denaturing conditions [
46]. This has led to their use as media for enzymatic reactions, where they can not only stabilize the biocatalyst but also improve the solubility of hydrophobic substrates, leading to enhanced reaction rates and yields [
46].
Table 2.
Representative NADES Formulations and Their Key Properties for Bioprocessing. This table highlights the “designer” nature of NADES by showcasing different combinations of Hydrogen Bond Acceptors (HBAs) and Hydrogen Bond Donors (HBDs) and linking their resulting properties to potential applications within the recombinant protein production workflow.
Table 2.
Representative NADES Formulations and Their Key Properties for Bioprocessing. This table highlights the “designer” nature of NADES by showcasing different combinations of Hydrogen Bond Acceptors (HBAs) and Hydrogen Bond Donors (HBDs) and linking their resulting properties to potential applications within the recombinant protein production workflow.
| HBA |
HBD |
Molar Ratio (HBA:HBD) |
Key Property/Characteristic |
Potential Application in Protein Production |
| Choline Chloride |
Urea |
1:2 |
Strong protein denaturing and solubilizing capacity; high polarity [16] |
Inclusion Body (IB) solubilization; component in refolding buffers [47] |
| Choline Chloride |
Glycerol |
1:2 |
High biocompatibility; cryoprotective properties; forms stable ATPS [44] |
In vivo media additive; cryopreservation of engineered strains; ATPS for purification [44] |
| Choline Chloride |
L-Arginine |
Varies |
Known aggregation suppressor; enhances protein solubility [14] |
Additive in refolding buffers to prevent protein aggregation [14] |
| Betaine |
Sorbitol |
1:1 |
Osmoprotectant; excellent protein stabilizer; high biocompatibility [48] |
In vivo media additive to enhance soluble expression; excipient for final product formulation [43] |
| L-Proline |
Glycerol |
1:2 |
Protein solubilizing agent; osmoprotectant; organocatalytic properties [19] |
In vivo media additive; component in mild solubilization/refolding buffers [19] |
| Choline Chloride |
Lactic Acid |
1:2 |
Acidic pH; high solubilizing power for biomass [44] |
Pre-treatment of biomass for protein extraction (e.g., plant-based systems) [49] |
| Menthol |
Fatty Acid (e.g., Decanoic Acid) |
Varies |
Hydrophobic; low water miscibility; can solubilize apolar molecules [18] |
Extraction and stabilization of membrane proteins; biphasic reaction systems [18] |
4. Integrating NADES into the Recombinant Protein Workflow: A Critical Analysis
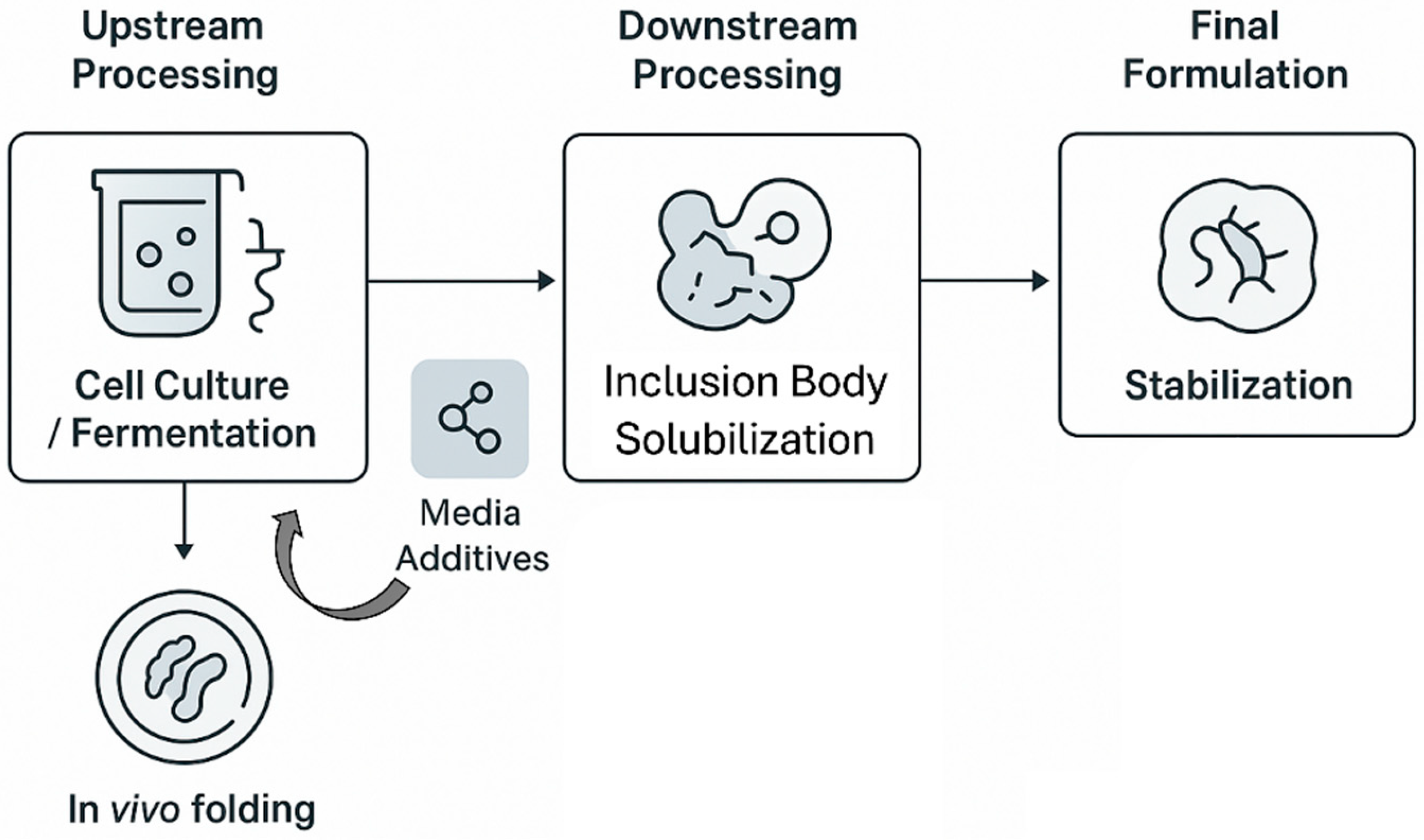
The true transformative potential of NADES in biotechnology lies not just in replacing conventional solvents but in their ability to be functionally integrated at multiple stages of the recombinant protein production workflow. Their unique properties—biocompatibility, tunability, and stabilizing effects—offer novel solutions to challenges in upstream expression, downstream processing, and final product formulation. This section critically analyzes the current and prospective applications of NADES at each of these key stages (
Figure 3).
4.1. Upstream Applications: Enhancing In Vivo Protein Expression and Strain Preservation
The application of NADES directly within the cellular environment (in vivo) is an emerging frontier with significant potential to enhance protein production at its source. This requires a deep understanding of the solvent’s interaction with the living host cell.
4.1.1. Biocompatibility with Production Hosts
A fundamental prerequisite for any in vivo application is that the solvent must be non-toxic and maintain the physiological health of the host microorganism. While NADES are generally considered biocompatible, studies reveal that this property is highly dependent on the specific chemical composition of the solvent [
50]. Research investigating the effect of various NADES on
E. coli has shown that some formulations, particularly those based on choline chloride, can compromise cell membrane integrity [
50]. In contrast, NADES formulated with alternative HBAs like betaine, L-proline, or L-carnitine have demonstrated superior biocompatibility, better preserving the cell membrane and, consequently, enhancing the catalytic efficiency of whole-cell biocatalysis systems [
50]. For example, transmission electron microscopy (TEM) images have shown that
E. coli cells exposed to novel betaine-based NADES maintained better membrane integrity compared to those treated with conventional choline chloride-based NADES [
50]. This composition-dependent biocompatibility is a critical consideration, suggesting that for in vivo applications, NADES based on natural osmolytes like betaine and proline are likely to be the most promising candidates.
4.1.2. Potential of NADES as Media Additives to Improve Soluble Protein Yield
A major challenge in high-level recombinant protein expression is the cellular stress induced by the overexpression process, which often leads to protein misfolding and aggregation [
10]. One established strategy to counteract this is the addition of small molecule “chemical chaperones” or osmolytes to the culture medium [
48]. These compounds, such as glycine betaine, sorbitol, and proline, help stabilize the native conformation of proteins and can improve the yield of soluble, active product [
48].
A compelling area for future research lies in the observation that these very same stabilizing osmolytes are common components of NADES [
51]. This raises the intriguing possibility that using a fully formed NADES as a media additive could provide a synergistic effect that is greater than the sum of its individual components [
45]. The extensive hydrogen-bonding network of a NADES creates a unique microenvironment that could act as a “super-chaperone,” providing a highly structured and protective hydration shell around nascent polypeptide chains, thereby favoring correct folding over aggregation. While studies have shown that adding individual NADES components to cell lysis buffers can increase the recovery of soluble protein , the direct addition of complete NADES formulations to fermentation media to enhance in vivo soluble protein expression remains a largely unexplored but highly promising strategy. The enhanced stability observed for enzymes in NADES solutions in vitro provides a strong rationale for investigating this in vivo application [
46].
4.1.3. NADES as Advanced Cryoprotectants for Engineered Microbial Strains
The long-term preservation of genetically engineered microbial production strains is essential for industrial biotechnology. The standard method involves cryopreservation, typically using glycerol or dimethyl sulfoxide (DMSO) as cryoprotective agents (CPAs) [
52]. However, these conventional CPAs can exhibit cytotoxicity and often result in significant loss of cell viability upon thawing [
52].
NADES have emerged as superior, green alternatives for cryopreservation [
42]. Studies on yeast like
Saccharomyces cerevisiae have demonstrated that NADES composed of natural metabolites (e.g., proline, glucose, sorbitol) can significantly improve post-thaw cell viability (to 75–80%) compared to glycerol or DMSO (around 50%) [
52]. The mechanism behind this enhanced protection is multifaceted. NADES exhibit excellent biocompatibility and maintain cell morphology, preventing the severe cell distortion often seen with conventional CPAs [
52]. More importantly, their primary cryoprotective effect stems from their potent ice recrystallization inhibition activity. During freezing and thawing, the growth of large, sharp ice crystals is a major cause of cell damage [
52]. The extensive hydrogen-bonding network within NADES effectively restrains water molecular motion, preventing the growth of these damaging crystals and resulting in the formation of smaller, less harmful ice structures [
52]. This capability makes NADES ideal for the long-term, stable banking of high-value recombinant strains of
E. coli,
P. pastoris, and other microbial factories, ensuring process consistency and reproducibility.
4.2. Downstream Processing I: A Green Paradigm for Inclusion Body Solubilization and Refolding
The formation of inclusion bodies (IBs) is one of the most common bottlenecks in recombinant protein production, particularly in
E. coli [
11]. While IBs represent a highly concentrated and relatively pure source of the target protein, the process of recovering active protein from these aggregates is notoriously challenging and costly [
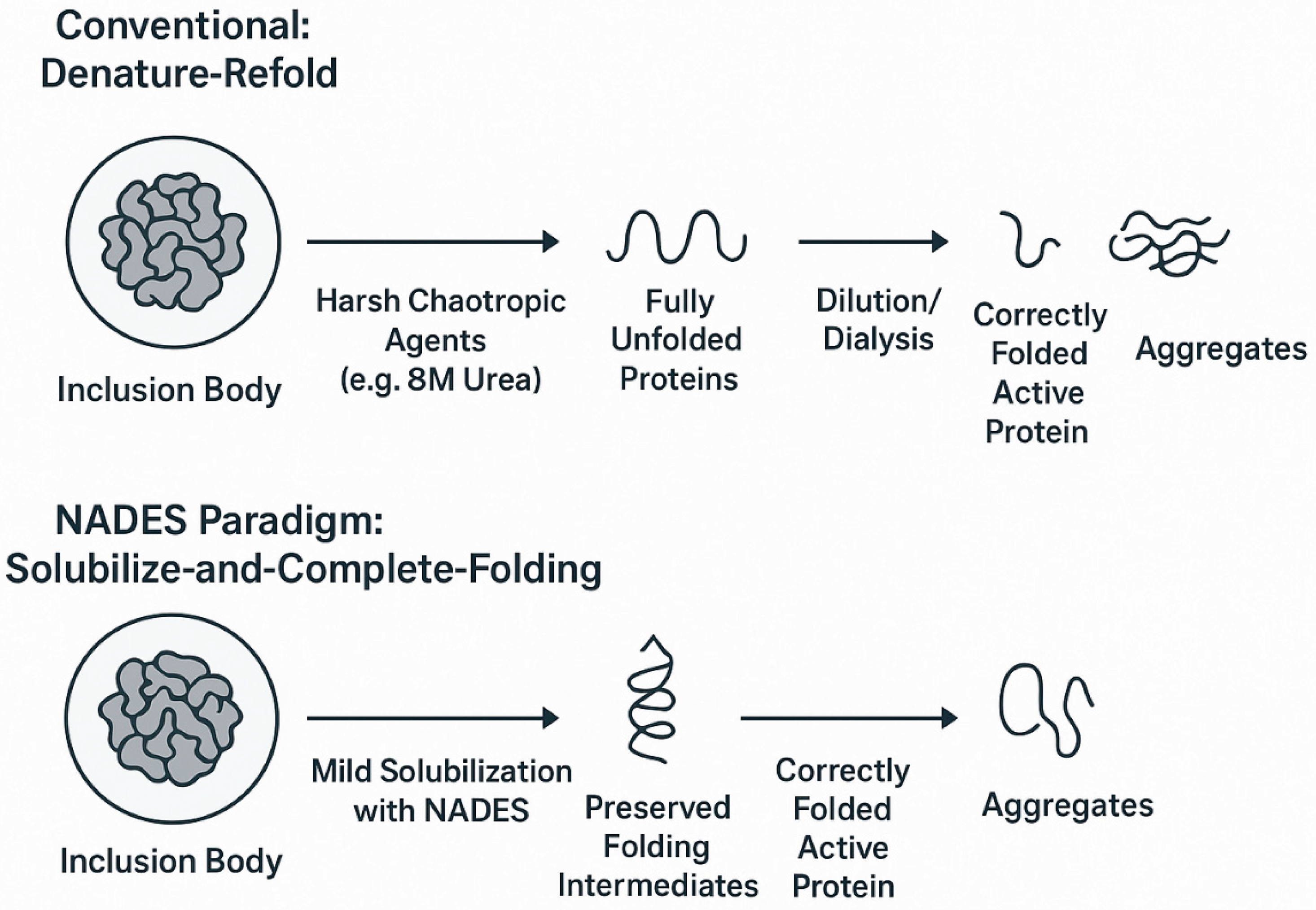
13]. NADES offer a potential paradigm shift in how IBs are processed, moving from harsh, complete denaturation to milder, more efficient refolding strategies.
4.2.1. The Conventional Challenge of “Denature-Refold”
The traditional workflow for IB processing involves two main steps: solubilization and refolding. First, the purified IBs are solubilized using high concentrations (6–8 M) of strong chaotropic agents like urea or guanidinium chloride [
13]. These agents disrupt all non-covalent interactions, completely unfolding the aggregated protein into linear polypeptide chains [
13]. Second, the denaturant is rapidly removed, typically by massive dilution or slow dialysis, to allow the protein to refold into its native, bioactive conformation [
53].
This “denature-refold” approach is fundamentally inefficient. It erases any native-like secondary structures that may have been present within the IBs, forcing the protein to navigate its entire complex folding landscape from a fully unfolded state—a thermodynamically and kinetically challenging process [
54]. Consequently, folding intermediates are highly prone to intermolecular aggregation, competing with the desired intramolecular folding pathway and leading to very low recovery yields of active protein [
55]. This has led to the view of IB processing as a highly empirical, “trial-and-error” field, where optimal refolding conditions must be laboriously screened for each new protein [
53].
4.2.2. A New Strategy: “Solubilize-and-Complete-Folding” with NADES
A conceptual advance in the field has been the recognition that proteins within IBs are not completely misfolded but often exist as a collection of partially folded intermediates with significant native-like secondary structure (α-helices and β-sheets) [
54]. This insight has given rise to the strategy of “mild solubilization,” which aims to dissolve the aggregates while preserving these pre-existing correct structures [
54]. By doing so, the subsequent refolding process is no longer a complete de novo folding event but rather the simpler task of arranging these preserved secondary structural elements into the correct final tertiary structure. This “solubilize-and-complete-folding” paradigm has been shown to dramatically improve refolding yields (
Figure 4) [
54].
NADES, with their highly tunable solubilizing power, are ideal candidates for implementing this strategy. By carefully selecting the HBA, HBD, and molar ratio, it is possible to create a solvent environment that is strong enough to disrupt the intermolecular interactions holding the aggregate together but gentle enough to leave the intramolecular native-like structures intact.
4.2.3. Functional Roles of Specific NADES in Refolding
Urea-based NADES: Urea is a canonical denaturant, and its incorporation into NADES (e.g., choline chloride:urea) creates a powerful solubilizing agent [
47]. The unique aspect of urea-based NADES is the influence of water content on their interaction with proteins. Studies have shown a non-monotonic, “re-entrant” behavior: in neat or low-hydration DES, a protein may remain partially folded. As water content increases to an intermediate level, the hydrogen-bonding network of the DES is disrupted, releasing urea molecules that can act as potent denaturants and increase protein unfolding. However, at very high hydration (>40% water), the protein can begin to refold . This complex behavior suggests that urea-based NADES could be used as a single, tunable system where solubilization and refolding are controlled simply by modulating the water content, potentially eliminating the need for cumbersome dialysis or dilution steps.
Arginine-based NADES: L-arginine is widely used as an additive in refolding buffers. Its primary role is not to actively promote folding but to act as an “aggregation suppressor” [
14]. It is thought to interact with exposed hydrophobic patches and charged residues on folding intermediates, preventing them from forming intermolecular aggregates and giving them more time to fold correctly [
14]. The development of arginine-based NADES is a promising area of research, as such solvents could combine the solubilizing properties of the eutectic mixture with the inherent anti-aggregation activity of arginine, creating a bifunctional refolding medium .
4.3. Downstream Processing II: NADES in Advanced Purification Strategies
Following solubilization and refolding, the target protein must be purified from host cell proteins (HCPs), nucleic acids, and other contaminants. Chromatography is the standard method, but it often requires multiple, sequential column steps. Aqueous Two-Phase Systems (ATPS) offer a scalable and cost-effective alternative for initial capture and purification, and NADES are revolutionizing this technology.
4.3.1. Principles of Aqueous Two-Phase Systems (ATPS)
ATPS is a liquid-liquid extraction technique that occurs when two water-soluble but immiscible polymers (e.g., polyethylene glycol (PEG) and dextran) or a polymer and a salt (e.g., PEG and phosphate) are mixed above a critical concentration, causing them to separate into two distinct aqueous phases [
56]. Biomolecules like proteins will partition between the two phases based on a combination of factors, including their size, surface charge, hydrophobicity, and specific affinity for the phase-forming components [
56]. This allows for the selective separation of the target protein from contaminants in a single extraction step, which is gentle, rapid, and easily scalable [
56].
4.3.2. NADES-based ATPS for Green and Efficient Purification
Recently, NADES have been successfully used as a novel phase-forming component in ATPS, typically in combination with a salting-out agent like potassium phosphate [
56]. These NADES-based ATPS offer several advantages over traditional polymer-based systems. They replace synthetic polymers with biodegradable, natural components, creating a fully green and sustainable purification platform [
56].
Case studies have demonstrated the remarkable efficiency of these systems. For example, an ATPS composed of a choline chloride:glycerol NADES and potassium phosphate was able to extract 98.16% of bovine serum albumin (BSA) and 94.36% of trypsin into the NADES-rich phase in a single step [
44]. Spectroscopic analysis confirmed that the protein’s native conformation was preserved during the extraction process [
44]. The mechanism for this highly efficient partitioning appears to involve the formation of NADES-protein aggregates, which drives the selective migration of the target protein into the NADES-rich phase [
56]. The tunability of NADES allows for the optimization of the ATPS for specific proteins by adjusting factors like pH, salt concentration, and the NADES composition itself, enabling high recovery and purification factors [
56].
4.4. Final Formulation: NADES for Long-Term Protein Stabilization
The final step in protein production is to formulate the purified protein in a buffer that ensures its long-term stability and activity during storage and transport. Many therapeutic proteins are prone to aggregation or degradation over time, requiring cold-chain storage and complex formulations with multiple excipients [
46].
4.4.1. Mechanisms of Protein Stabilization in NADES
NADES have demonstrated exceptional capabilities as protein-stabilizing media, often surpassing traditional aqueous buffers [
46]. The primary mechanism is believed to be kinetic stabilization. The dense and highly structured hydrogen-bonding network of the NADES creates a “cage” around the protein, physically constraining its conformational flexibility and suppressing the large-scale structural fluctuations that precede unfolding and aggregation . Molecular dynamics simulations have supported this “solvent slaving” mechanism, showing a significant slowdown of protein backbone dynamics in NADES compared to water . This effect can dramatically increase the thermal stability of proteins; for example, the half-denaturation temperature of bovine serum albumin was significantly enhanced even in dilute NADES solutions [
46].
4.4.2. Prospects for Integrated and Stable Formulations
The excellent stabilizing properties and inherent biocompatibility of NADES open up the prospect of significant process intensification. In a conventional workflow, the protein is purified and then transferred into a final formulation buffer through an additional step like dialysis or diafiltration. In a NADES-based process, the solvent used for the final purification step (e.g., in an ATPS) could potentially serve as the final formulation buffer itself . This would eliminate an entire unit operation, saving time, reducing protein loss, and lowering costs. Studies have shown that proteins and even delicate macromolecules like mRNA can remain stable and functional for months at room temperature when stored in NADES, highlighting their potential to create highly stable, long-shelf-life biopharmaceutical products that may not require a continuous cold chain [
50].
5. Future Perspectives and Concluding Remarks
5.1. Synthesis and Outlook: Towards a Fully Integrated Bioprocess
The convergence of heterologous protein expression and Natural Deep Eutectic Solvents (NADES) technology heralds a significant opportunity to redefine the landscape of biomanufacturing. This review has systematically detailed the potential for NADES to be integrated at every critical stage of the protein production pipeline, acting not merely as passive “green” replacements for conventional solvents but as active, functional materials that address specific, long-standing challenges [
18]. The overarching vision that emerges is that of a “NADES-native” bioprocess—a fully integrated workflow where a biocompatible and functional solvent system, chosen at the outset, is carried through from cell culture to final formulation [
20]. Such a process could simultaneously enhance soluble protein expression in vivo, simplify inclusion body processing, streamline purification through integrated extraction systems, and provide a final formulation with exceptional long-term stability. This holistic approach promises to yield a process that is not only more sustainable and environmentally benign but also more efficient and economically viable than current paradigms.
5.2. Addressing the Critical Knowledge Gaps
While the potential is immense, the realization of this vision is contingent upon addressing several critical knowledge gaps that currently limit the widespread adoption of NADES in industrial bioprocessing.
Paucity of In Vivo Data: The most significant gap in the current literature is the lack of systematic studies on the in vivo effects of NADES when used as additives in microbial fermentations. While the biocompatibility of certain NADES has been established [
50], and the benefits of their individual components as osmolytes are known [
57], there is a pressing need for research that directly evaluates the impact of complete NADES formulations on host cell physiology, metabolic burden, and the yield of soluble recombinant protein. Understanding how these complex solvents interact with the cellular machinery during active growth and protein synthesis is paramount.
Challenges of Scale-Up and Economic Viability: The transition from laboratory-scale experiments to industrial-scale production presents substantial engineering challenges [
58]. The high viscosity of many NADES formulations is a primary concern, as it can negatively impact mixing, oxygen transfer in bioreactors, and pumping efficiency in downstream equipment [
16]. Furthermore, the economic feasibility of large-scale bioprocesses hinges on the efficient recovery and recycling of the solvent. While recycling of NADES is possible, current methods can be energy-intensive. Comprehensive techno-economic analyses (TEA) are urgently needed to compare the overall cost-benefit of NADES-based processes against well-established conventional methods, factoring in capital expenditure, operating costs, and solvent recycling efficiency [
58].
Deepening Mechanistic Understanding: While empirical evidence for the beneficial effects of NADES is accumulating, a deeper, molecular-level understanding of the underlying mechanisms is often lacking. How exactly do different NADES compositions interact with the protein folding machinery in vivo? What are the precise thermodynamic and kinetic drivers of protein partitioning in NADES-based ATPS? Answering these questions will be crucial for moving beyond empirical screening to the rational design of optimal solvent systems.
5.3. Charting Future Research Directions
To bridge these gaps and unlock the full potential of NADES in recombinant protein production, future research should be directed along several key interdisciplinary avenues:
High-Throughput Screening (HTS) of NADES Libraries: The “designer” nature of NADES means that a vast combinatorial space of potential solvents exists. HTS platforms should be developed to rapidly screen large libraries of NADES compositions for specific functionalities. This could involve using fluorescent reporter proteins to screen for NADES that enhance soluble expression in vivo, or employing light-scattering techniques to identify formulations that are most effective at solubilizing specific inclusion bodies or preventing aggregation during refolding.
Computational Modeling and Rational Solvent Design: Empirical screening should be complemented by computational approaches. Molecular dynamics (MD) simulations are a powerful tool for investigating protein-NADES interactions at the atomic level . These simulations can provide crucial insights into how NADES affect protein stability, conformation, and folding pathways, thereby enabling the rational, in silico design of NADES tailored for specific target proteins or process steps . This approach could significantly accelerate the development cycle and reduce the experimental burden of HTS.
Bioprocess Engineering and Integration: Research is needed to develop novel bioprocess equipment and workflows optimized for handling NADES. This could include the design of bioreactors that can efficiently mix and aerate viscous cultures, or the development of advanced separation technologies, such as membrane-based systems or centrifugal extractors, for efficient protein recovery and solvent recycling from NADES-based ATPS. The goal should be to design integrated, continuous processes that maximize the benefits of process intensification offered by NADES.
5.4. Conclusions
The production of recombinant proteins remains a vital engine of the biotechnology industry. However, to meet the growing demands for complex therapeutics and sustainable industrial products, the field must evolve beyond its current paradigms. Natural Deep Eutectic Solvents are poised to be a key enabler of this evolution. They are not merely green alternatives but are highly functional, tunable materials that offer elegant solutions to some of the most persistent and vexing problems in protein biomanufacturing—from cellular stress and protein aggregation to the complexities and costs of purification. The synergistic integration of NADES with heterologous expression platforms has the potential to create a new generation of bioprocesses that are more efficient, economical, and fundamentally sustainable. Realizing this potential will require a concerted and interdisciplinary effort from microbiologists, biochemists, chemists, and bioengineers to explore this exciting and promising scientific frontier.
Author Contributions
Conceptualization, V.C.-R.; methodology, V.C.-R.; formal analysis, V.C.-R.; investigation, V.C.-R. and J.A.M.-O.; resources, V.C.-R.; data curation, V.C.-R.; writing—original draft preparation, V.C.-R. and J.A.M.-O; writing—review and editing, V.C.-R.; visualization, V.C.-R.; supervision, V.C.-R.; project administration, V.C.-R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
All the research data can be found in the text.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Berg, P.; Mertz, J.E. Personal Reflections on the Origins and Emergence of Recombinant DNA Technology. Genetics 2010, 184, 9–17. [Google Scholar] [CrossRef]
- Cohen, S.N.; Chang, A.C.Y.; Boyer, H.W.; Helling, R.B. Construction of Biologically Functional Bacterial Plasmids In Vitro. Proc. Natl. Acad. Sci. 1973, 70, 3240–3244. [Google Scholar] [CrossRef]
- Quianzon, C.C.; Cheikh, I. History of insulin. J. Community Hosp. Intern. Med. Perspect. 2012, 2, 18701. [Google Scholar] [CrossRef]
- Jayakrishnan, A.; Wan Rosli, W.R.; Tahir, A.R.M.; Razak, F.S.A.; Kee, P.E.; Ng, H.S.; Chew, Y.-L.; Lee, S.-K.; Ramasamy, M.; Tan, C.S.; et al. Evolving Paradigms of Recombinant Protein Production in Pharmaceutical Industry: A Rigorous Review. Sci 2024, 6, 9. [Google Scholar] [CrossRef]
- Chapman, J.; Ismail, A.E.; Dinu, C.Z. Industrial Applications of Enzymes: Recent Advances, Techniques, and Outlooks. Catalysts 2018, 8, 238. [Google Scholar] [CrossRef]
- Bhatwa, A.; Wang, W.; Hassan, Y.I.; Abraham, N.; Li, X.-Z.; Zhou, T. Challenges Associated With the Formation of Recombinant Protein Inclusion Bodies in Escherichia coli and Strategies to Address Them for Industrial Applications. Front. Bioeng. Biotechnol. 2021, 9. [Google Scholar] [CrossRef] [PubMed]
- Francis, D.M.; Page, R. Strategies to Optimize Protein Expression in E. coli. Curr. Protoc. Protein Sci. 2010, 61. [Google Scholar] [CrossRef]
- Snoeck, S.; Guidi, C.; De Mey, M. “Metabolic burden” explained: stress symptoms and its related responses induced by (over)expression of (heterologous) proteins in Escherichia coli. Microb. Cell Fact. 2024, 23, 96. [Google Scholar] [CrossRef] [PubMed]
- Raschmanová, H.; Zamora, I.; Borčinová, M.; Meier, P.; Weninger, A.; Mächler, D.; Glieder, A.; Melzoch, K.; Knejzlík, Z.; Kovar, K. Single-Cell Approach to Monitor the Unfolded Protein Response During Biotechnological Processes With Pichia pastoris. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Schramm, F.D.; Schroeder, K.; Jonas, K. Protein aggregation in bacteria. FEMS Microbiol. Rev. 2020, 44, 54–72. [Google Scholar] [CrossRef]
- García-Fruitós, E. Inclusion bodies: a new concept. Microb. Cell Fact. 2010, 9, 80. [Google Scholar] [CrossRef]
- Spalding, B.J. Downstream Processing: Key To Slashing Production Costs 100 Fold. Nat. Biotechnol. 1991, 9, 229–232. [Google Scholar] [CrossRef]
- Singh, A.; Upadhyay, V.; Panda, A.K. Solubilization and Refolding of Inclusion Body Proteins. In; 2015; pp. 283–291.
- Zhao, D.; Liu, Y.; Zhang, G.; Zhang, C.; Li, X.; Wang, Q.; Shi, H.; Su, Z. Interaction of arginine with protein during refolding process probed by amide H/D exchange mass spectrometry and isothermal titration calorimetry. Biochim. Biophys. Acta - Proteins Proteomics 2015, 1854, 39–45. [Google Scholar] [CrossRef]
- Burnett, M.J.B.; Burnett, A.C. Therapeutic recombinant protein production in plants: Challenges and opportunities. PLANTS, PEOPLE, PLANET 2020, 2, 121–132. [Google Scholar] [CrossRef]
- Ferreira, C.; Sarraguça, M. A Comprehensive Review on Deep Eutectic Solvents and Its Use to Extract Bioactive Compounds of Pharmaceutical Interest. Pharmaceuticals 2024, 17, 124. [Google Scholar] [CrossRef]
- Li, D. Natural deep eutectic solvents in phytonutrient extraction and other applications. Front. Plant Sci. 2022, 13. [Google Scholar] [CrossRef]
- Liu, Y.; Friesen, J.B.; McAlpine, J.B.; Lankin, D.C.; Chen, S.-N.; Pauli, G.F. Natural Deep Eutectic Solvents: Properties, Applications, and Perspectives. J. Nat. Prod. 2018, 81, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Karadendrou, M.-A.; Kostopoulou, I.; Kakokefalou, V.; Tzani, A.; Detsi, A. L-Proline-Based Natural Deep Eutectic Solvents as Efficient Solvents and Catalysts for the Ultrasound-Assisted Synthesis of Aurones via Knoevenagel Condensation. Catalysts 2022, 12, 249. [Google Scholar] [CrossRef]
- Tripathi, N.K.; Shrivastava, A. Recent Developments in Bioprocessing of Recombinant Proteins: Expression Hosts and Process Development. Front. Bioeng. Biotechnol. 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Cook, J.D.; Ye, W.; Lee, J.E.; Sidhu, S.S. Optimization of peptidic HIV-1 fusion inhibitor T20 by phage display. Protein Sci. 2019, 28, 1501–1512. [Google Scholar] [CrossRef] [PubMed]
- İncir, İ.; Kaplan, Ö. Escherichia coli as a versatile cell factory: Advances and challenges in recombinant protein production. Protein Expr. Purif. 2024, 219, 106463. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Tsang, J.S.H. Use of ribosomal promoters from Burkholderia cenocepacia and Burkholderia cepacia for improved expression of transporter protein in Escherichia coli. Protein Expr. Purif. 2006, 49, 219–227. [Google Scholar] [CrossRef]
- Lobstein, J.; Emrich, C.A.; Jeans, C.; Faulkner, M.; Riggs, P.; Berkmen, M. SHuffle, a novel Escherichia coli protein expression strain capable of correctly folding disulfide bonded proteins in its cytoplasm. Microb. Cell Fact. 2012, 11, 753. [Google Scholar] [CrossRef]
- Schlegel, S.; Löfblom, J.; Lee, C.; Hjelm, A.; Klepsch, M.; Strous, M.; Drew, D.; Slotboom, D.J.; de Gier, J.-W. Optimizing Membrane Protein Overexpression in the Escherichia coli strain Lemo21(DE3). J. Mol. Biol. 2012, 423, 648–659. [Google Scholar] [CrossRef]
- Tripodi, F.; Nicastro, R.; Reghellin, V.; Coccetti, P. Post-translational modifications on yeast carbon metabolism: Regulatory mechanisms beyond transcriptional control. Biochim. Biophys. Acta - Gen. Subj. 2015, 1850, 620–627. [Google Scholar] [CrossRef]
- Bonander, N.; Bill, R.M. Optimising Yeast as a Host for Recombinant Protein Production (Review). In; 2012; pp. 1–9.
- Kastberg, L.L.B.; Jacobsen, I.H.; Özdemir, E.; Workman, C.T.; Jensen, M.K.; Förster, J. Characterizing heterologous protein burden in Komagataella phaffii. FEMS Yeast Res. 2025, 25. [Google Scholar] [CrossRef] [PubMed]
- Türkanoğlu Özçelik, A.; Yılmaz, S.; Inan, M. Pichia pastoris Promoters. In; 2019; pp. 97–112.
- Karbalaei, M.; Rezaee, S.A.; Farsiani, H. Pichia pastoris : A highly successful expression system for optimal synthesis of heterologous proteins. J. Cell. Physiol. 2020, 235, 5867–5881. [Google Scholar] [CrossRef] [PubMed]
- Sharker, S.M.; Rahman, A. A Review on the Current Methods of Chinese Hamster Ovary (CHO) Cells Cultivation for the Production of Therapeutic Protein. Curr. Drug Discov. Technol. 2021, 18, 354–364. [Google Scholar] [CrossRef] [PubMed]
- Sari-Ak, D.; Alomari, O.; Shomali, R.; Lim, J.; Thimiri Govinda Raj, D. Advances in CRISPR-Cas9 for the Baculovirus Vector System: A Systematic Review. Viruses 2022, 15, 54. [Google Scholar] [CrossRef]
- Possee, R.D.; Chambers, A.C.; Graves, L.P.; Aksular, M.; King, L.A. Recent Developments in the Use of Baculovirus Expression Vectors. Curr. Issues Mol. Biol. 2020, 215–230. [Google Scholar] [CrossRef]
- Hong, Q.; Liu, J.; Wei, Y.; Wei, X. Application of Baculovirus Expression Vector System (BEVS) in Vaccine Development. Vaccines 2023, 11, 1218. [Google Scholar] [CrossRef]
- Li, Z.-M.; Fan, Z.-L.; Wang, X.-Y.; Wang, T.-Y. Factors Affecting the Expression of Recombinant Protein and Improvement Strategies in Chinese Hamster Ovary Cells. Front. Bioeng. Biotechnol. 2022, 10. [Google Scholar] [CrossRef]
- O’Flaherty, R.; Bergin, A.; Flampouri, E.; Mota, L.M.; Obaidi, I.; Quigley, A.; Xie, Y.; Butler, M. Mammalian cell culture for production of recombinant proteins: A review of the critical steps in their biomanufacturing. Biotechnol. Adv. 2020, 43, 107552. [Google Scholar] [CrossRef]
- Gecchele, E.; Merlin, M.; Brozzetti, A.; Falorni, A.; Pezzotti, M.; Avesani, L. A Comparative Analysis of Recombinant Protein Expression in Different Biofactories: Bacteria, Insect Cells and Plant Systems. J. Vis. Exp. 2015. [Google Scholar] [CrossRef]
- Khambhati, K.; Bhattacharjee, G.; Gohil, N.; Braddick, D.; Kulkarni, V.; Singh, V. Exploring the Potential of Cell-Free Protein Synthesis for Extending the Abilities of Biological Systems. Front. Bioeng. Biotechnol. 2019, 7. [Google Scholar] [CrossRef]
- Maharjan, A.; Park, J. Cell-free protein synthesis system: A new frontier for sustainable biotechnology-based products. Biotechnol. Appl. Biochem. 2023, 70, 2136–2149. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, N.; Murphy, L.; Tyther, R. Post-translational Modifications of Recombinant Proteins: Significance for Biopharmaceuticals. Mol. Biotechnol. 2008, 39, 113–118. [Google Scholar] [CrossRef]
- VanderWeide, J.; Tombesi, S.; Castellarin, S.D.; Sabbatini, P. Canopy architecture and fruit microclimate, not ripening-related phytohormones, control phenylpropanoid accumulation in response to early leaf removal in ‘Merlot’ (Vitis vinifera L.) grapevines. Plant Physiol. Biochem. 2020, 157, 291–302. [Google Scholar] [CrossRef]
- Cajnko, M.M.; Vicente, F.A.; Novak, U.; Likozar, B. Natural deep eutectic solvents (NaDES): translating cell biology to processing. Green Chem. 2023, 25, 9045–9062. [Google Scholar] [CrossRef]
- Mohd Fuad, F.; Mohd Nadzir, M. The formulation and physicochemical properties of betaine-based natural deep eutectic solvent. J. Mol. Liq. 2022, 360, 119392. [Google Scholar] [CrossRef]
- Che Zain, M.S.; Yeoh, J.X.; Lee, S.Y.; Shaari, K. Physicochemical Properties of Choline Chloride-Based Natural Deep Eutectic Solvents (NaDES) and Their Applicability for Extracting Oil Palm Flavonoids. Sustainability 2021, 13, 12981. [Google Scholar] [CrossRef]
- di Lorenzo, R.; Bernardi, A.; Grumetto, L.; Sacchi, A.; Avagliano, C.; Coppola, S.; de Giovanni di Santa Severina, A.F.; Bruno, C.; Paparo, L.; Laneri, S.; et al. Phenylalanine Butyramide Is a New Cosmetic Ingredient with Soothing and Anti-Reddening Potential. Molecules 2021, 26, 6611. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Zhu, M.; Hu, T.; Liu, C. Natural deep eutectic solvent—A novel green solvent for protein stabilization. Int. J. Biol. Macromol. 2023, 247, 125477. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, Z.; Lou, C. Thermodynamic Irreversibility Analysis of Thermal Radiation in Coal-Fired Furnace: Effect of Coal Ash Deposits. Materials (Basel). 2023, 16, 799. [Google Scholar] [CrossRef] [PubMed]
- Garbe, M.; Lehmann, L.T.; Berger, R.G.; Ersoy, F. Improvement in the Stability and Enzymatic Activity of Pleurotus sapidus Lipoxygenase Dissolved in Natural Deep Eutectic Solvents (NADESs). Life 2024, 14, 271. [Google Scholar] [CrossRef]
- Smink, D.; Juan, A.; Schuur, B.; Kersten, S.R.A. Understanding the Role of Choline Chloride in Deep Eutectic Solvents Used for Biomass Delignification. Ind. Eng. Chem. Res. 2019, 58, 16348–16357. [Google Scholar] [CrossRef]
- Al Fuhaid, L.; Khattak, S.; Alghuneim, A.; Gallouzi, I.; Choi, Y.H.; Verpoorte, R.; Witkamp, G.-J.; Farinha, A. NADES as Biocompatible Media for Thermally Stable RNA Molecules 2025.
- Wang, L.; Li, T.; Liu, N.; Liu, X. Identification of tomato AHL gene families and functional analysis their roles in fruit development and abiotic stress response. Plant Physiol. Biochem. 2023, 202, 107931. [Google Scholar] [CrossRef]
- Jesus, A.R.; Meneses, L.; Duarte, A.R.C.; Paiva, A. Natural deep eutectic systems, an emerging class of cryoprotectant agents. Cryobiology 2021, 101, 95–104. [Google Scholar] [CrossRef]
- Basu, A.; Li, X.; Leong, S.S.J. Refolding of proteins from inclusion bodies: rational design and recipes. Appl. Microbiol. Biotechnol. 2011, 92, 241–251. [Google Scholar] [CrossRef]
- Singh, A.; Upadhyay, V.; Upadhyay, A.K.; Singh, S.M.; Panda, A.K. Protein recovery from inclusion bodies of Escherichia coli using mild solubilization process. Microb. Cell Fact. 2015, 14, 41. [Google Scholar] [CrossRef]
- Wang, H. .; Ng, T.. A ribonuclease from Chinese ginseng (Panax ginseng) flowers. Protein Expr. Purif. 2004, 33, 195–199. [Google Scholar] [CrossRef]
- Buarque, F.; Gautério, G.; Coelho, M.; Lemes, A.; Ribeiro, B. Aqueous Two-Phase Systems Based on Ionic Liquids and Deep Eutectic Solvents as a Tool for the Recovery of Non-Protein Bioactive Compounds—A Review. Processes 2022, 11, 31. [Google Scholar] [CrossRef]
- Xu, J.; Zhu, Z.; Duan, P.; Li, W.; Zhang, Y.; Wu, J.; Hu, Z.; Roque, R.S.; Liu, J. Cloning, expression, and purification of a highly immunogenic recombinant gonadotropin-releasing hormone (GnRH) chimeric peptide. Protein Expr. Purif. 2006, 50, 163–170. [Google Scholar] [CrossRef]
- De Brabander, P.; Uitterhaegen, E.; Delmulle, T.; De Winter, K.; Soetaert, W. Challenges and progress towards industrial recombinant protein production in yeasts: A review. Biotechnol. Adv. 2023, 64, 108121. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).