Submitted:
13 August 2025
Posted:
14 August 2025
You are already at the latest version
Abstract
Turf-forming Polysiphonia-looking algae were collected from a small (< 1.0 km2 area) Agarophyton tenuistipitata (Gracilariaceae, Gracilariales) farm on the East coast of the Bay of Bengal. DNA was extracted from silica gel-preserved specimens, and plastid-encoded rbcL, nuclear-encoded small subunit SSU, large subunit LSU, and universal plastid amplicon (UPA) were amplified and sequenced. Maximum likelihood (ML) and Bayesian inference were performed for the phylogenetic analysis. Four single-locus species delimitation methods (SDMs), namely the Generalized Mixed Yule-Coalescent (GMYC) method, a Poisson Tree Processes (PTP) model, the Automatic Barcode Gap Discovery (ABGD), and the Assemble Species by Automatic Partitioning (ASAP) method, were performed to segregate the putative species from other taxa in the Polysiphonia sensu lato clades. Our results revealed that rbcL had 1.4% interspecific genetic divergence, whereas LSU, UPA, and SSU had 1.6%, 2.5%, and 5.4% genetic divergence, respectively, from the nearest neighbors. Both comparative genetic and distinct morphological data revealed that the collected Bay of Bengal specimens comprise a species new to science. In addition, the above-mentioned SDMs supported the genetic data and segregated our specimens as Melanothamnus coxsbazarensis sp. nov. as a distinct species.
Keywords:
1. Introduction
2. Materials and Methods
2.1. Study Area and Sample Collections
2.2. DNA Extraction, PCR Amplification, and Sequencing Protocols
2.3. Sequence Alignment and Phylogenetic Analysis
2.4. Assessing the Species Delimitation Methods
2.5. Morphological Observations
3. Results
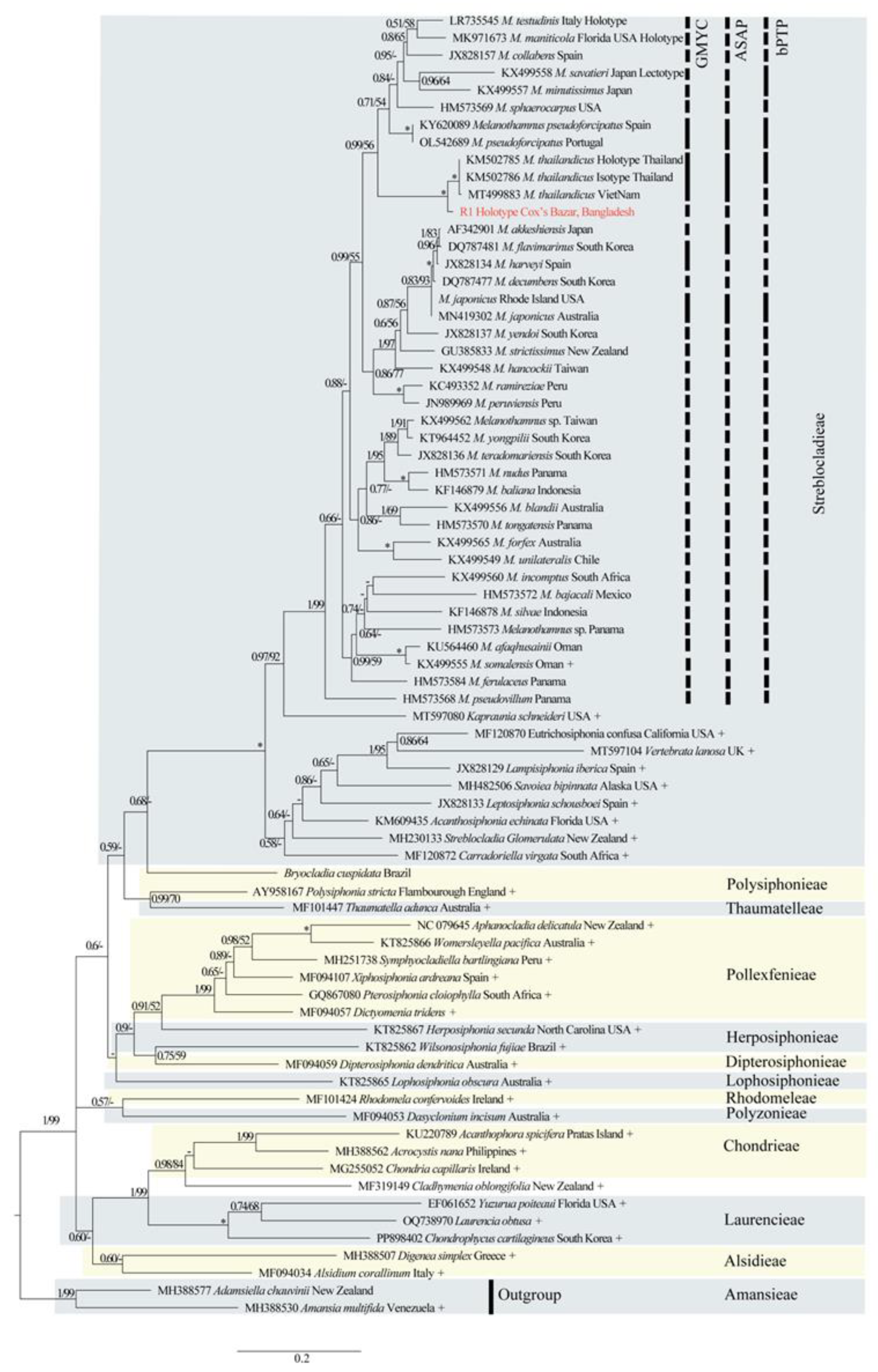
3.1. Phylogenetic Results
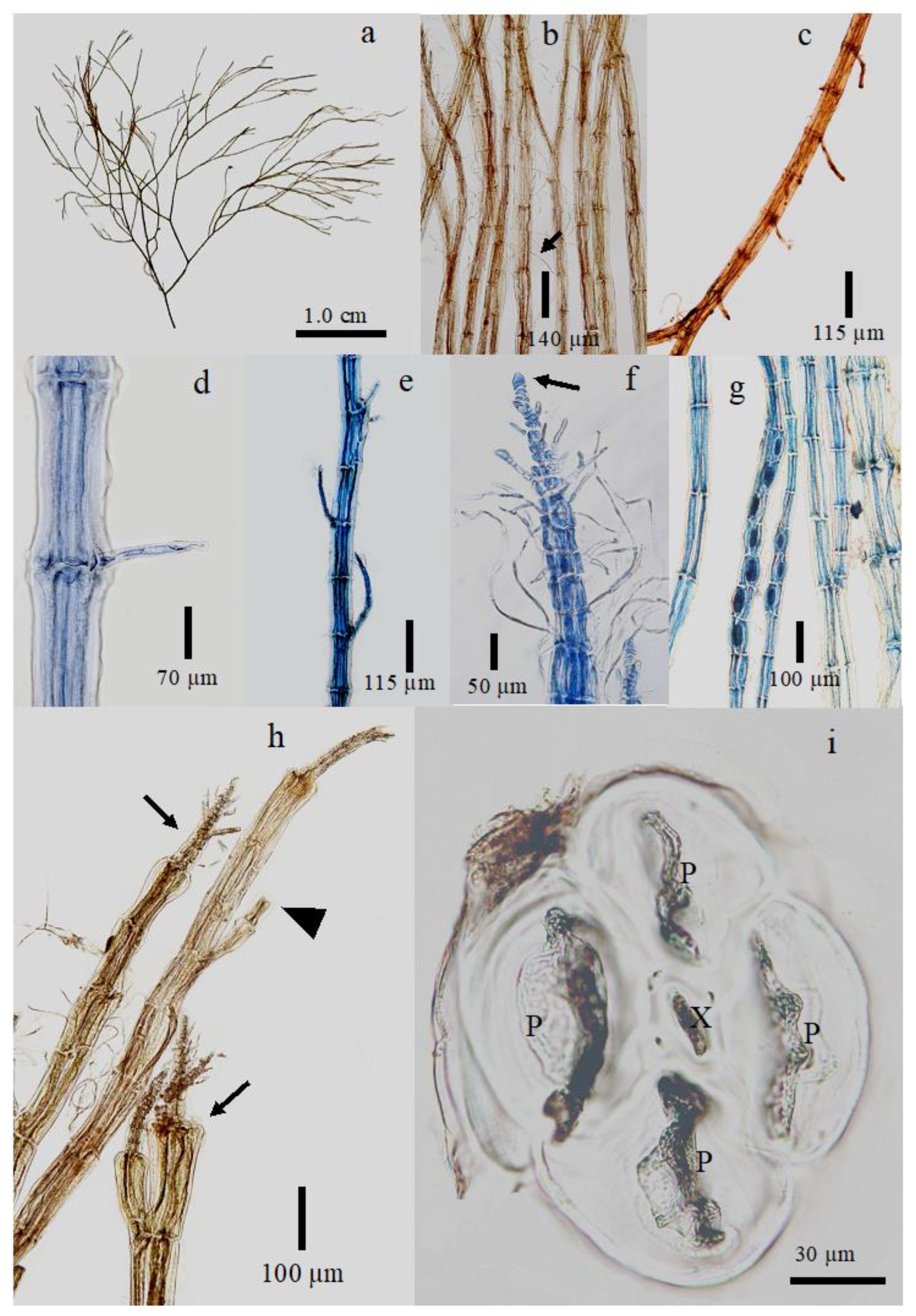
3.2. New Species Descriptions
3.3. Species Delimitation Methods (SDMs)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Falkenberg, P. Die Rhodomelaceen des Golfes von Neapel und der angrenzenden Meeres-Abschnitte. Fauna und Flora des Golfes von Neapel, Monographie 26. Berlin, 1901. pp. i-xvi, 1-754.
- Díaz-Tapia, P.; McIvor, L.; Freshwater, D.W.; Verbruggen, H.; Wynne, M.J.; Maggs, C.A. The genera Melanothamnus Bornet & Falkenberg and Vertebrata S.F. Gray constitute well-defined clades of the red algal tribe Polysiphonieae (Rhodomelaceae, Ceramiales). Eur. J. Phycol. 2017, 52, 1–30. [Google Scholar]
- Hollenberg, G. J. An account of the species of Polysiphonia on the Pacific coast of North America. I. Oligosiphonia. Amer. J. Bot., 1942, 29, 772–85. [Google Scholar] [CrossRef]
- Díaz-Tapia, P.; Ly, M.; Verbruggen, H. Extensive cryptic diversity in the widely distributed Polysiphonia scopulorum (Rhodomelaceae, Rhodophyta): Molecular species delimitation and morphometric analyses. Mol. Phylog. Evol. 2020, 152, 106909. [Google Scholar] [CrossRef]
- Kim, M.S.; Lee, I.K. Neosiphonia flavimarina gen. et sp. nov. with a taxonomic reassessment of the genus Polysiphonia (Rhodomelaceae, Rhodophyta). Phycol. Res. 1999, 47, 271–281. [Google Scholar] [CrossRef]
- Aziz, A.; Rahman, M.T. Marine algae of St. Martin's Island, Bangladesh. IX. New records of green algae (Chlorophyceae). Ban. J. Bot. 2010, 39, 161–168. [Google Scholar] [CrossRef]
- Nam, K.W.; Kang, P.J. Algal flora of Korea. Volume 4, Number 4. Rhodophyta: Ceramiales: Rhodomelaceae: 18 genera including Herposiphonia. pp. [1-6], 1-178, figs 1-102. Incheon: National Institute of Biological Resources, 2012, pp. [1−6], 1−178.
- Díaz-Tapia, P.; Bárbara, I. Seaweeds from sand-covered rocks of the Atlantic Iberian Peninsula. Part 1. The Rhodomelaceae (Ceramiales, Rhodophyta). Cryptog., Algol., 2013, 34, 325–422. [Google Scholar] [CrossRef]
- Diaz-Tapia, P.; Barbara, I.; Cremades, J.; Verbruggen, H.; Maggs, C.A. Three new cryptogenic species in the tribes Polysiphonieae and Streblocladieae (Rhodomelaceae, Rhodophyta). Phycologia, 2017, 56, 605–623. [Google Scholar] [CrossRef]
- Neto, A.I.; Cacabelos, E.; Prestes, A.C.L.; Díaz-Tapia, P.; Moreu, I. New records of marine macroalgae for the Azores. Bot. Mar., 2022, 65, 105–120. [Google Scholar] [CrossRef]
- Guiry, M.D.; Guiry, G.M.E. AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. https://www.algaebase.org; (22 July 2025).
- Fletcher, R.L. Epiphytism and fouling in Gracilaria cultivation: an overview. J. Appl. Phycol. 1995, 7, 325–333. [Google Scholar] [CrossRef]
- Hurtado, A.Q.; Yunque, D.A.; Tibubos, K.; Critchley, A.T. Use of Acadian marine plant extract powder from Ascophyllum nodosum in tissue culture of Kappaphycus varieties, J. Appl. Phycol. 2009, 21, 633–639. [Google Scholar] [CrossRef]
- Muangmai, N.; Maneekat, S.; Petsut, N.; Keawsuralikhit, C. Newly reported marine red alga, Neosiphonia savatieri (Hariot) M.S.Kim et I.K.Lee 1999 (Rhodophyta Rhodomelaceae) from Thailand. Biodiv. J. 2012, 3, 247–250. [Google Scholar]
- Muangmai, N.; Yamagishi, Y.; Maneekat, S.; Kaewsuralikhit, C. The new species Neosiphonia thailandica sp. nov. (Rhodomelaceae, Rhodophyta) from the Gulf of Thailand. Bot. Mar. 2014, 57, 459–467. [Google Scholar] [CrossRef]
- Bình, D.T.; An, K.T.; Cầm, V.H.; Tuấn, T.V. (2020). New record and the molecular phylogeny of epiphyte (Melanothamnus thailandicus) on red algae (Kappaphycus alvarezii) in Khanh Hoa. J Fish. Sci. Technol. 2020, 2, 1–9. [Google Scholar]
- Woodworth, K.A.; Frankovich, T.A.; Freshwater, D.W. Melanothamnus maniticola sp. nov. (Ceramiales, Rhodophyta): an epizoic species evolved for living on the West Indian Manatee. J. Phycol., 2019, 55, 1239–1245. [Google Scholar]
- Serio, D.; Furnari, G.; Moro, I.; Sciuto, K. Molecular and morphological characterisation of Melanothamnus testudinis sp. nov. (Rhodophyta, Rhodomelaceae) and its distinction from Polysiphonia carettia. Phycologia. 2020, 59, 281–291. [Google Scholar] [CrossRef]
- Stuercke, B; Freshwater, W.D. Consistency of Morphological Characters Used to Delimit Polysiphonia sensu lato Species (Ceramiales, Florideophyceae): Analyses of North Carolina, USA Specimens. Phycologia 2008, 47, 541–59. [Google Scholar] [CrossRef]
- Mamoozadeh, N.R.; Freshwater, D.W. Taxonomic notes on Caribbean Neosiphonia and Polysiphonia (Ceramiales, Florideophyceae): five species from Florida, USA and Mexico. Bot. Mar. 2011, 54, 269–92. [Google Scholar] [CrossRef]
- Mamoozadeh, N.R.; Freshwater, D.W. Polysiphonia sensu lato (Ceramiales, Florideophyceae) species of Caribbean Panama including Polysiphonia lobophoralis sp. nov. and Polysiphonia nuda sp. nov. Bot. Mar. 2012, 55, 317–347. [Google Scholar] [CrossRef]
- Savoie, A.M.; Saunders, G.W. A molecular assessment of species diversity and generic boundaries in the red algal tribes Polysiphonieae and Streblocladieae (Rhodomelaceae, Rhodophyta) in Canada. Eur J. Phycol. 2019, 54, 1–25. [Google Scholar] [CrossRef]
- Amos, D.; Aguilar, V.; Barber-Scott, K.; Bustamante, D.E.; Calderon, M.S.; Carrasco, R.; Vang, M.N. Transfer of the marine red alga Erythrocystis saccate (Rhodomelaceae, Rhodophyta) to the tribe Streblocladieae inferred from organellar genome analysis. Phytotaxa, 2021, 507, 266–270. [Google Scholar] [CrossRef]
- Bustamante, D.E.; Yeon, W.B.; Wynne, M.J.; Cho, T.O. Molecular and morphological analyses reveal new taxa additions to the tribe Streblocladieae (Rhodomelaceae, Rhodophyta). J. Phycol. 2021, 57, 817–30. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Tapia, P.; Verbruggen, H. Resolving the taxonomy of the Polysiphonia scopulorum complex and the Bryocladia lineage (Rhodomelaceae, Rhodophyta). J. Phycol. 2024, 60, 49–72. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.A. Mangrove associated macroalgae in the Sundarbans forest, Bangladesh: spatial and temporal changes, MS thesis. Hiroshima University, Hiroshima, Japan. September 20, 2019.
- Islam, M.A.; Mauya, M.Z.; Rafiquzzaman, S.M.; Islam, M.R.; Liao, L.M. First report of the red algal genus Chondria C. Agardh (Rhodomelaceae, Rhodophyta) for the marine flora of Bangladesh. Diversity 2019, 11, 95. [Google Scholar] [CrossRef]
- Islam, M.A.; Islam, M.R.; Aziz, A.; Liao, L.M. Dictyota adnata Zanardini (Phaeophyceae) -A new record from the Sundarbans mangrove forests, Bangladesh. Ban. J Bot. 2020, 49, 407–412. [Google Scholar] [CrossRef]
- Chowdhury, M.S.N.; Hossain, M.S.; AftabUddin, S.; Alamgir, M.; Sharifuzzaman, S.M. Seaweed aquaculture in Bangladesh: Present status, challenges and prospects. Ocean and Coastal Management, 2022, 228. [Google Scholar] [CrossRef]
- Islam, A.K.M.N. Contribution to the Study of the Marine Algae of Bangladesh; Bibliotheca Phycologica; J. Cramer Verlag: Vaduz, Lichtenstein, 1976; p. 276. [Google Scholar]
- Freshwater, D.W.; Rueness, J. Phylogenetic relationships of some European Gelidium (Gelidiales, Rhodophyta) species, based on rbcL nucleotide sequence analysis. Phycologia, 1994, 33, 187–194. [Google Scholar] [CrossRef]
- Sherwood, A.R.; Presting, G.G. Universal primers amplify a 23s rDNA plastid marker in Eukaryotic algae and Cyanobacteria, J. Phycol. 2007, 43, 605–608. [Google Scholar] [CrossRef]
- Harper, J.T.; Saunders, G.W. The Application of Sequences of the Ribosomal Cistron to the Systematics and Classification of the Florideophyte Red Algae (Florideophyceae, Rhodophyta). Cahiers Biol. Mar. 2001, 42, 25–38. [Google Scholar]
- Conklin, K.Y.; Kurihara, A.; Sherwood, A.R. A molecular method for identification of the morphologically plastic invasive algal genera Eucheuma and Kappaphycus (Rhodophyta, Gigartinales) in Hawaii. J. Appl. Phycol., 2009, 21, 691–699. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–4. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML Version 8: A tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van der Mark, P.; Ayres, D.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsen-beck, J.P. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–42. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. Figtree V.1.4.4. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh, 2018.
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 2012, 21, 1864–77. [Google Scholar] [CrossRef]
- Puillandre, N.; Brouillet, S.; Achaz, G. ASAP: assemble species by automatic partitioning. Mol. Ecol. Resources 2021, 21, 609–620. [Google Scholar] [CrossRef]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013, 29, 2869–76. [Google Scholar] [CrossRef]
- Pons, J.; Barraclough, T.G.; Gomez-Zurita, J.; Cardoso, A.; Duran, D.P.; Hazell, S.; Kamoun, S.; Sumlin, W.D.; Vogler, A.P. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Sys. Biol., 2006, 55, 595–609. [Google Scholar] [CrossRef]
- Paradis, E.; Claude, J.; Strimmer, K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics, 2004, 20, 289–290. [Google Scholar] [CrossRef]
- Laboni, H.A.; Sarkar, S.; Islam, M.A.; Islam, M.A.; Islam, M.; Lyzu, C.; Rahman, M.M.; Alam, M.M.; Shohael, A.M.; Karim, M.R. Molecular identification of Gracilaria tenuistipitata isolated from the Bay of Bengal and appraisal of its comprehensive phytochemical profiling, and antioxidant potential. Pharmacol. Res. – Nat. Prod. 2025, 8, 100324. [Google Scholar] [CrossRef]
- Fujisawa, T.; Barraclough, T.G. Delimiting species using single-locus data and the Generalized Mixed Yule Coalescent approach: a revised method and evaluation on simulated data sets, Sys. Biol. 2013, 62, 707–724. [Google Scholar] [CrossRef]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10 Virus Evolution 4,vey016. 2018. [Google Scholar]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. ‘Posterior Summarization in Bayesian Phylogenetics Using Tracer 1. 7’, Sys. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Rojas, R.R.; Fouquet, A.; Ron, S.R.; Hernández-Ruz, E.J.; Melo-Sampaio, P.R.; Chaparro, J.C.; Hrbek, T. (2018). A Pan-Amazonian species delimitation: high species diversity within the genus Amazophrynella (Anura: Bufonidae). Peer J. 2018, 6, e4941. [Google Scholar] [CrossRef] [PubMed]
- Solovyeva, E.N.; Dunayev, E.A.; Nazarov, R.A.; Bondarenko, D.A.; Poyarkov, N.A. COI-Barcoding and Species Delimitation Assessment of Toad-Headed Agamas of the Genus Phrynocephalus (Agamidae, Squamata) Reveal Unrecognized Diversity in Central Eurasia. Diversity 2023, 15, 149. [Google Scholar] [CrossRef]
- Afaq-Husain, S.; Shameel, M. Further investigations on the red alga Melanothamnus afaqhusainii (Ceramiales) from the coast of Pakistan. Pakistan Journal of Botany, 2000, 32, 15–26. [Google Scholar]
- Díaz-Tapia, P.; Kim, M.S.; Secilla, A.; Bárbara, I.; Cremades, J. Taxonomic reassessment of Polysiphonia foetidissima (Rhodomelaceae, Rhodophyta) and similar species, including P. schneideri, a newly introduced species in Europe. Euro. J. Phycol. 2013, 48, 345–362. [Google Scholar]
- Bustamante, D.E.; Won, B.Y.; Miller, K.A.; Cho, T.O. Wilsonosiphonia gen. nov. (Rhodomelaceae, Rhodophyta) based on molecular and morpho-anatomical characters. J. Phycol. 2017, 53, 368–380. [Google Scholar] [CrossRef]
- McIvor, L.; Maggs, C.A.; Provan, J.; Stanhope, M.J. rbcL sequences reveal multiple cryptic introductions of the Japanese red alga Polysiphonia harveyi. Mol. Ecol., 2001, 10, 911–919. [Google Scholar] [CrossRef]
- Sherwood, A.R.; Kurihara, A.; Conklin, K.Y.; Sauvage, T.; Presting, G.G. The Hawaiian Rhodophyta biodiversity survey (2006–2010): a summary of principal findings. BMC Plant Biol. 2010, 10, 258. [Google Scholar] [CrossRef]
- Clarkston, B.E.; Saunders, G.W. (2013). Resolving species diversity in the red algal genus Callophyllis (Kallymeniaceae, Gigartinales) in Canada using molecular assisted alpha taxonomy. Euro. J. Phycol. 2013, 48, 27–46. [Google Scholar] [CrossRef]
- Kim, M.S. Taxonomy of a poorly documented alga, Neosiphonia savatieri Rhodomelaceae, Rhodophyta) from Korea. Nova Hedwigia, 2005, 81, 163–176. [Google Scholar] [CrossRef]
| Morphological features | M. coxsbazarensis | M. thailandicus | M. pseudoforcipatus | M. sphaerocarpus | M. savatieri | M. minutissima | M. collabens |
|---|---|---|---|---|---|---|---|
| Thallus color | Reddish brown | Reddish brown | Dark red to brown or pink | Blackish brown | Dull reddish-brown | Dull red | Red to pale brown |
| Type locality | Cox’s Bazar, Bangladesh | Chon Buri, Thailand |
Galicia, Spain | St. Thomas, Virgin Islands | Kanagawa, Japan | Baja California, Mexico | Cádiz, Spain |
| Plant habit | Prostrate | Erect | Erect | Erect | Erect | Prostrate | Erect |
| Apical cell shape | Domed or acute | Rounded | Rounded | Domed | Rounded | – | Domed |
| Height (cm) | 3–5 | 5–15 | 1.6 | 1–3 | Up to 1 | 0.3–0.6 | Up to 7 cm |
| Number of pericentral cells | 4–5 | 4 | 4 | 4 | 4 | 4 | 6 |
| Branching pattern | Subdichotomous | Dichotomous | Pseudodichotomous | Subdichotomous | Dichotomous to subdichotomous | Subdichotomous | Pseudodichotomous |
| Rhizoidal positions | Throughout the thallus | Discoid holdfast | Basal parts | Basal parts | Basal tuft of rhizoids | Prostrate base | Discoid holdfast |
| Frequency of trichoblasts | Abundant | Scarce | Absent or scarce, at irregular intervals | Abundant | Abundant | Present | Scarce |
| Adventitious endogenous branchlets | Scarce | Abundant | Absent | Absent | Absent | – | Absent |
| Exogenous branches, including cicatrigenous branches | Absent | Absent | Present | Present | Present but no cicatrigenous branches |
– |
Present |
| Cystocarp shape | – | Globular | Globose | Ovate to globose | Globular | Urceolate | Globular |
| References | This study | [15,16] | [2,10] | [7,20] | [14,57] | [3] | [8] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




