1. Introduction
Every year, spinal cord injuries (SCI) affect approximately 250,000 to 500,000 individuals globally, with an estimated two to three million people living with SCI-related disabilities [
44]. SCI arises from damage to the spinal cord or surrounding structures, disrupting communication between the brain and body [
37]. Causes include traumatic incidents such as vehicular accidents, falls, and sports injuries, as well as non-traumatic factors. Clinical manifestations vary depending on the injury’s severity and location, commonly resulting in sensory and motor impairments, muscular weakness, and complications in physiological functions [
8]. While complete injuries typically lead to permanent deficits, partial injuries may permit some functional recovery.
Technological advancements have significantly improved rehabilitation approaches and patient quality of life. Among these, brain-computer interfaces (BCIs) that leverage electroencephalography (EEG) have emerged as promising tools. EEG-based BCIs enable direct communication between the brain and external devices, offering a non-invasive, portable, and cost-effective solution for individuals with limited motor control [
3,
29]. EEG signals, which capture oscillatory neural activity, are acquired via electrodes placed on the scalp. These signals can be decoded in real time using machine learning algorithms to infer user intentions [
6].
Despite their potential, EEG signals present analytical challenges due to their high dimensionality, low signal-to-noise ratio, and variability across sessions and subjects [
50]. Effective dimensionality reduction is thus essential for improving signal decoding accuracy and computational efficiency. Traditional techniques like Principal Component Analysis (PCA) have been widely used, but recent research focuses on nonlinear methods better suited to the intrinsic geometry of EEG data [
36].
Manifold learning is a powerful class of nonlinear dimensionality reduction methods that seeks low-dimensional representations while preserving local or global data structures [
26,
46,
56]. These methods are particularly advantageous in processing EEG data due to their ability to retain discriminative features critical for classification [
10]. Prominent manifold learning algorithms include ISOMAP [
56], Locally Linear Embedding (LLE) [
46], t-Distributed Stochastic Neighbor Embedding (t-SNE) [
35], Spectral Embedding [
7], and Multidimensional Scaling (MDS) [
55].
In recent years, the integration of manifold learning techniques with shallow classifiers such as k-Nearest Neighbors (k-NN), Support Vector Machines (SVM), and Naive Bayes has shown promise in decoding motor imagery (MI) tasks from EEG [
24,
25]. These combinations enable efficient real-time EEG decoding with reduced computational burden. Moreover, comparative studies suggest that manifold learning can improve classification accuracy in EEG-based BCIs, particularly for applications in neurorehabilitation and assistive technology [
31].
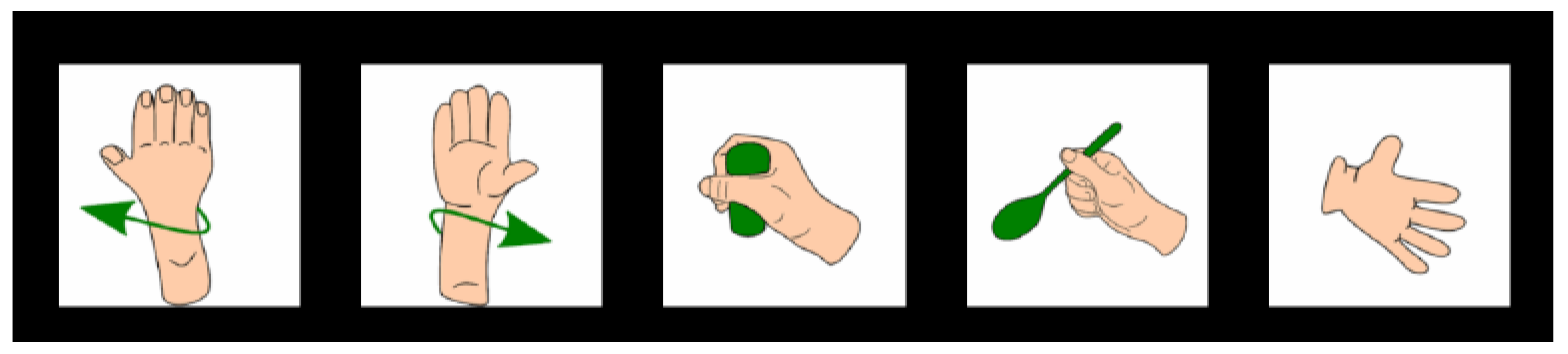
This study aims to explore the effectiveness of manifold learning techniques paired with shallow classifiers for classifying EEG data collected from six healthy participants performing five wrist and hand motor imagery tasks. The performance of various dimensionality reduction-classifier pairs is evaluated across binary, ternary, and five-class scenarios to identify robust, low-complexity pipelines suitable for real-time BCI applications.
1.1. State of the Art
Recent studies have highlighted the potential of manifold learning and advanced feature extraction techniques in enhancing the classification performance of EEG-based BCIs, particularly in motor imagery tasks.
Li et al. [
34] introduced an adaptive feature extraction framework combining wavelet packet decomposition (WPD) and semidefinite embedding ISOMAP (SE-ISOMAP). This approach utilized subject-specific optimal wavelet packets to extract time-frequency and manifold features, achieving 100% accuracy in binary classification tasks and significantly outperforming conventional dimensionality reduction methods.
Yamamoto et al. [
70] proposed a novel method called Riemann Spectral Clustering (RiSC), which maps EEG covariance matrices as graphs on the Riemannian manifold using a geodesic-based similarity measure. They further extended this framework with odenRiSC for outlier detection and mcRiSC for multimodal classification, where mcRiSC reached 73.1% accuracy and outperformed standard single-modal classifiers in heterogeneous datasets.
Krivov and Belyaev [
28] incorporated Riemannian geometry and Isomap to reveal the manifold structure of EEG covariance matrices in a low-dimensional space. Their method, evaluated with Linear Discriminant Analysis (LDA), reported classification accuracies of 0.58 (CSP), 0.61 (PGA), and 0.58 (Isomap) in a four-class task, underlining the potential of manifold methods in representing EEG data structures.
Tyagi and Nehra [
57] compared LDA, PCA, FA, MDS, and ISOMAP for motor imagery feature extraction using BCI Competition IV datasets. A feedforward artificial neural network (ANN) trained with the Levenberg-Marquardt algorithm yielded the lowest mean square error (MSE) with LDA (0.1143), followed by ISOMAP (0.2156), while other linear methods showed relatively higher errors.
Xu et al. [
64] designed an EEG-based attention classification method utilizing Riemannian manifold representation of symmetric positive definite (SPD) matrices. By integrating amplitude and phase information using a filter bank and applying SVM, their approach reached a classification accuracy of 88.06% in a binary scenario without requiring spatial filters.
Lee et al. [
32] assessed the efficacy of PCA, LLE, and ISOMAP in binary EEG classification using LDA. The classification errors were reported as 28.4% for PCA, 25.8% for LLE, and 27.7% for ISOMAP, suggesting LLE’s slight edge in capturing intrinsic EEG data structures.
Li, Luo, and Yang [
33] further evaluated the performance of linear and nonlinear dimensionality reduction techniques in motor imagery EEG classification. Nonlinear methods such as LLE (91.4%) and parametric t-SNE (94.1%) outperformed PCA (70.7%) and MDS (75.7%), demonstrating the importance of preserving local neighborhood structures for robust feature representation.
Sayılgan [
51] investigated EEG-based classification of imagined hand movements in spinal cord injury patients using Independent Component Analysis (ICA) for feature extraction and machine learning classifiers including SVM, k-NN, AdaBoost, and Decision Trees. The highest accuracy was achieved with SVM (90.24%), while k-NN demonstrated the lowest processing time, with the lateral grasp showing the highest classification accuracy among motor tasks.
These studies collectively underline the critical role of dimensionality reduction, particularly manifold learning, in effectively decoding motor intentions from EEG data, thereby improving the performance and applicability of BCI systems in neurorehabilitation contexts.
1.2. Contributions
Electroencephalographic (EEG) recordings, collected from multiple scalp locations, are inherently high-dimensional, often containing redundant information and being susceptible to various noise sources and artifacts. Such properties can hinder the accuracy and robustness of motor intention decoding in Brain–Computer Interface (BCI) systems. While conventional linear dimensionality reduction approaches, such as Principal Component Analysis (PCA) and Linear Discriminant Analysis (LDA), are widely used, they are often inadequate for capturing the complex, nonlinear temporal–spatial relationships embedded in EEG activity patterns. In contrast, manifold learning techniques offer a promising alternative by projecting data into a lower-dimensional space while preserving its intrinsic local geometry.
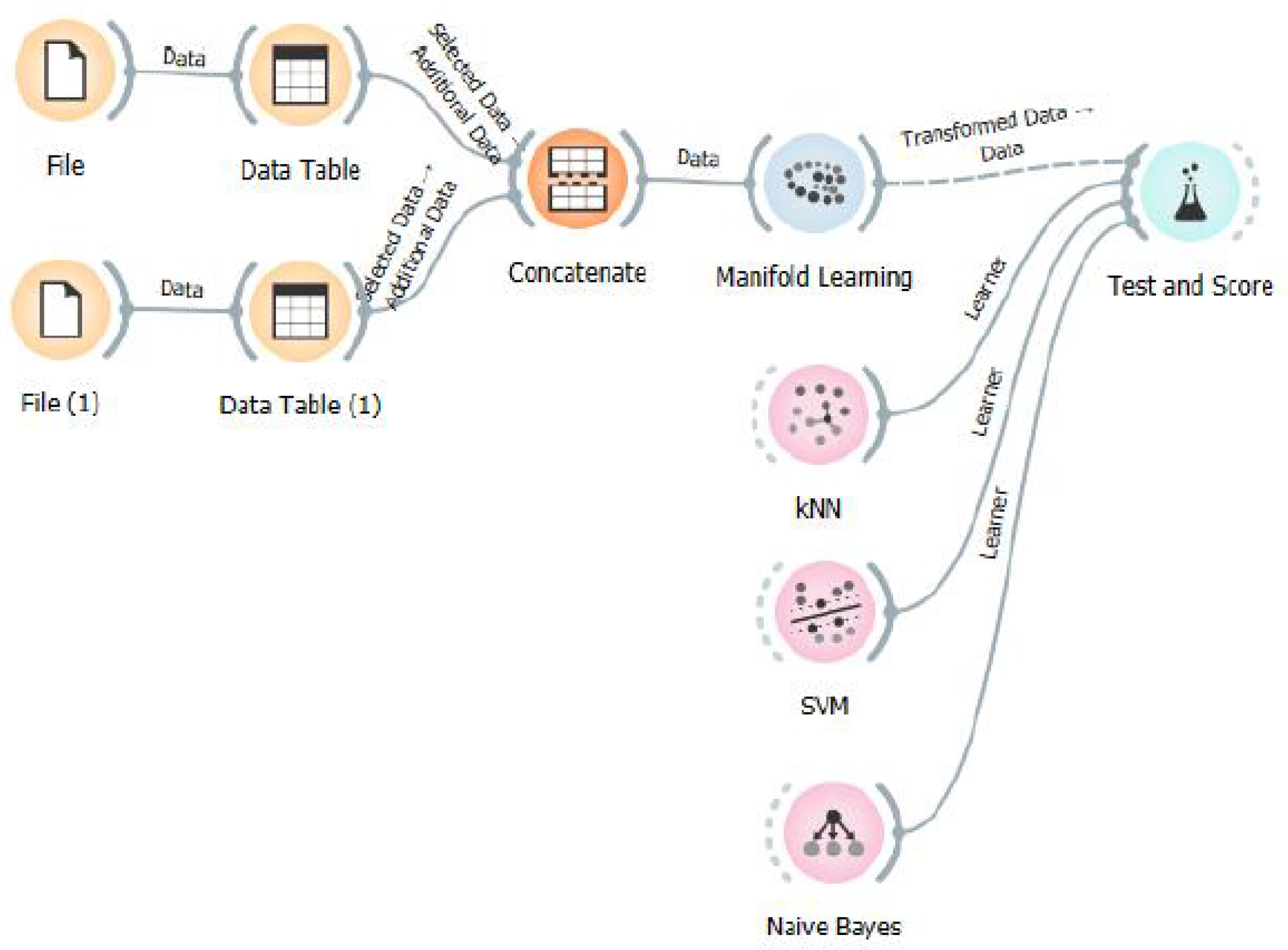
In this work, we present a comprehensive and adaptable manifold learning-based processing framework for EEG analysis, designed to support the development of rehabilitation-oriented BCIs. The proposed pipeline integrates multiple nonlinear dimensionality reduction algorithms with shallow classifiers to alleviate overfitting, enhance inter-class separability, and improve overall decoding performance.
The main contributions of this study can be outlined as follows:
Introduction of a unified manifold learning framework for the classification of motor imagery EEG signals into binary (2-class), ternary (3-class), and multi-class (5-class) categories, using real-time data acquired from healthy participants.
Systematic comparison of five widely recognized manifold learning algorithms: Spectral Embedding, Locally Linear Embedding (LLE), Multidimensional Scaling (MDS), Isometric Mapping (ISOMAP), and t-distributed Stochastic Neighbor Embedding (t-SNE) for their effectiveness as feature transformation tools in motor intent recognition.
Alignment of the proposed methodology with practical rehabilitation needs, specifically for integration into a cost-efficient, 2-degree-of-freedom robotic platform employing a straightforward control strategy.
Emphasis on building a sustainable machine learning model capable of accurately detecting motor intentions in healthy users while ensuring high classification performance, thereby enabling scalability to clinical scenarios.
Addressing a notable gap in the literature by exploring high-accuracy 3-class and 5-class EEG-based BCI paradigms for spinal cord injury (SCI) rehabilitation, and benchmarking binary classification results against state-of-the-art systems.
Analysis of task combination compatibility across different classification schemes, with performance metrics aggregated over all participants to support model generalizability.
Comprehensive evaluation using multiple performance indicators—accuracy, precision, recall, and F1-score to assess the robustness of each manifold–classifier pairing under varying task complexities.
4. Results
As a result of the real-time data processing process, the binary, 3- and 5-classification results of manifold learning methods and classification algorithms are presented in the tables below according to their performance metrics. In this study, AUC, CA (Accuracy), F1 score, and precision were preferred as evaluation criteria. High values obtained in the relevant metrics show that the classification success of the model is high.
According to the results shown in
Table 2, the most effective classification method in the ISOMAP method is k-NN. The values of AUC, CA, F1 score, and precision are between 0.995 and 0.993, indicating that the combination of ISOMAP and k-NN is quite successful. As the number of classes increases, the performances of all classifiers decrease, but this is expected. Naive Bayes shows the best performance after k-NN. However, a serious decrease is seen in the 5-class case. SVM is the model with the lowest performance in all classes.
Table 3 compares the performance of the classification algorithms (SVM, k-NN, Naive Bayes) applied after dimensionality reduction with the Local Linear Embedding (LLE) method in 2, 3, and 5-class classifications through AUC, CA, F1, and Precision metrics. k-NN is the model with the highest metric value in all classes (2-, 3-, 5-). According to the metrics, k-NN is between 0.963-0.815 values. SVM is the model with the lowest performance. According to the SVM results, some values remained around or below 0.5. This shows that the model has weak discrimination power between classes. Naive Bayes method is the method with the best results after k-NN.
Table 4 shows the performances of the classification algorithms (SVM, k-NN, Naive Bayes) with Spectral Embedding method in 2-, 3- and 5-class classifications. k-NN showed the highest performance in all metrics, staying between 0.960-0.721. Although SVM gave a very high AUC (0.960) value, especially in the 3-class case, it showed low performance in other metrics. Naive Bayes gave worse results than k-NN but better than SVM. Although SVM reached the highest value in AUC with 0.960 in 3-class values, it failed in other metrics (CA = 0.391, F1 = 0.365). This shows that the model cannot provide balance while trying to distinguish the classes.
An examination of the results presented in
Table 5 reveals that k-NN emerged as the most successful model, achieving the highest values (0.999–0.890) across all metrics, when compared to t-SNE. The SVM model demonstrates superior performance in this table in comparison to other methods, particularly in the 2- and 3-class problems. The AUC value of SVM is 0.950–0.954 in the 2- and 3-class problems, indicating high distinguishability between classes. The observation that the precision and F1 scores are high and close to each other indicates that there are no unbalanced predictions between classes and that the model maintains its general balance.
However, it is observed that the performance of SVM declines considerably in the 5-class problem. The decline of the Area Under the Curve (AUC) to 0.619 indicates a reduction in the capacity to differentiate between the designated classes. The CA, F1 score, and precision values are all low, at approximately 33%, suggesting that random predictions were made from five classes. SVM was inadequate in more complex and multi-class scenarios. This finding suggests that the high-dimensional discrimination capacity of SVM may be impaired after t-SNE.
Naive Bayes demonstrated average performance in the 2-class problem. An AUC of approximately 0.795 suggests that, while its capacity to differentiate between classes is marginally superior to random guessing, it is not significantly superior. The CA and F1 values ranging from 0.728 to 0.730 suggest the presence of errors in classification, yet the model demonstrates the capacity to discern certain patterns. In the 3-class problem, the performance of Naive Bayes has decreased significantly. While the AUC value (0.768) remains at a moderate level, CA (0.590), precision (0.589), and F1 score (0.586) have all shown significant decline. This decline can be attributed to the assumption of unconditional independence between classes, which may not be adequate for complex, structured data.
In the 5-class problem, the CA, F1, and precision values are below 40%, indicating a weak model success. The AUC value (0.733) is relatively high, but this does not guarantee that the model can reliably differentiate between the classes. It is important to note that if the dataset is imbalanced, the AUC value may be elevated while the F1 score may decline.
Table 6.
MDS-Based Classification Results Across 2-, 3- and 5-Class
Table 6.
MDS-Based Classification Results Across 2-, 3- and 5-Class
| |
2-class |
3-class |
5-class |
| |
AUC |
CA |
F1 |
Prec |
AUC |
CA |
F1 |
Prec |
AUC |
CA |
F1 |
Prec |
| SVM |
0.772 |
0.738 |
0.740 |
0.742 |
0.754 |
0.556 |
0.549 |
0.569 |
0.709 |
0.367 |
0.361 |
0.380 |
| k-NN |
0.974 |
0.971 |
0.971 |
0.969 |
0.976 |
0.943 |
0.943 |
0.928 |
0.971 |
0.884 |
0.819 |
0.835 |
| Naive Bayes |
0.768 |
0.727 |
0.727 |
0.728 |
0.737 |
0.565 |
0.559 |
0.562 |
0.712 |
0.404 |
0.384 |
0.396 |
In the classification analyses performed after the MDS-based dimensionality reduction method, the k-NN algorithm demonstrated the highest level of success in all class combinations (2-, 3-, and 5-class). The values obtained in AUC, accuracy (CA), F1 score and precision metrics reveal that the k-NN model is superior in terms of both discrimination power and prediction balance. Conversely, the SVM model exhibited moderate performance for 2-class problems, but as the number of classes increased, a substantial decline in performance was observed. The Naive Bayes model demonstrated inferior performance in comparison to the k-NN model across all class combinations, failing to exhibit adequate performance within the context of this particular data set and dimensionality reduction method. These findings show that the most appropriate classification algorithm after MDS is k-NN and that SVM and Naive Bayes need to be carefully evaluated for complex multi-class problems. In the classifications made with MDS method, the performance of SVM, Naive Bayes, and k NN models changed according to the number of classes. In the two-class case, SVM gave better results than Naive Bayes, while k-NN outperformed all three models. While SVM and Naive Bayes showed similar performance in three classes, k-NN again provided the highest accuracy and balance. In the 5-class scenario, the success of SVM decreased significantly, and Naive Bayes produced more balanced results. However, k NN was by far the most successful model even in this complex case. These results show that k-NN is a stable and powerful model in every class structure, SVM is successful in simpler tasks and Naive Bayes offers limited but balanced performance for complex structures.
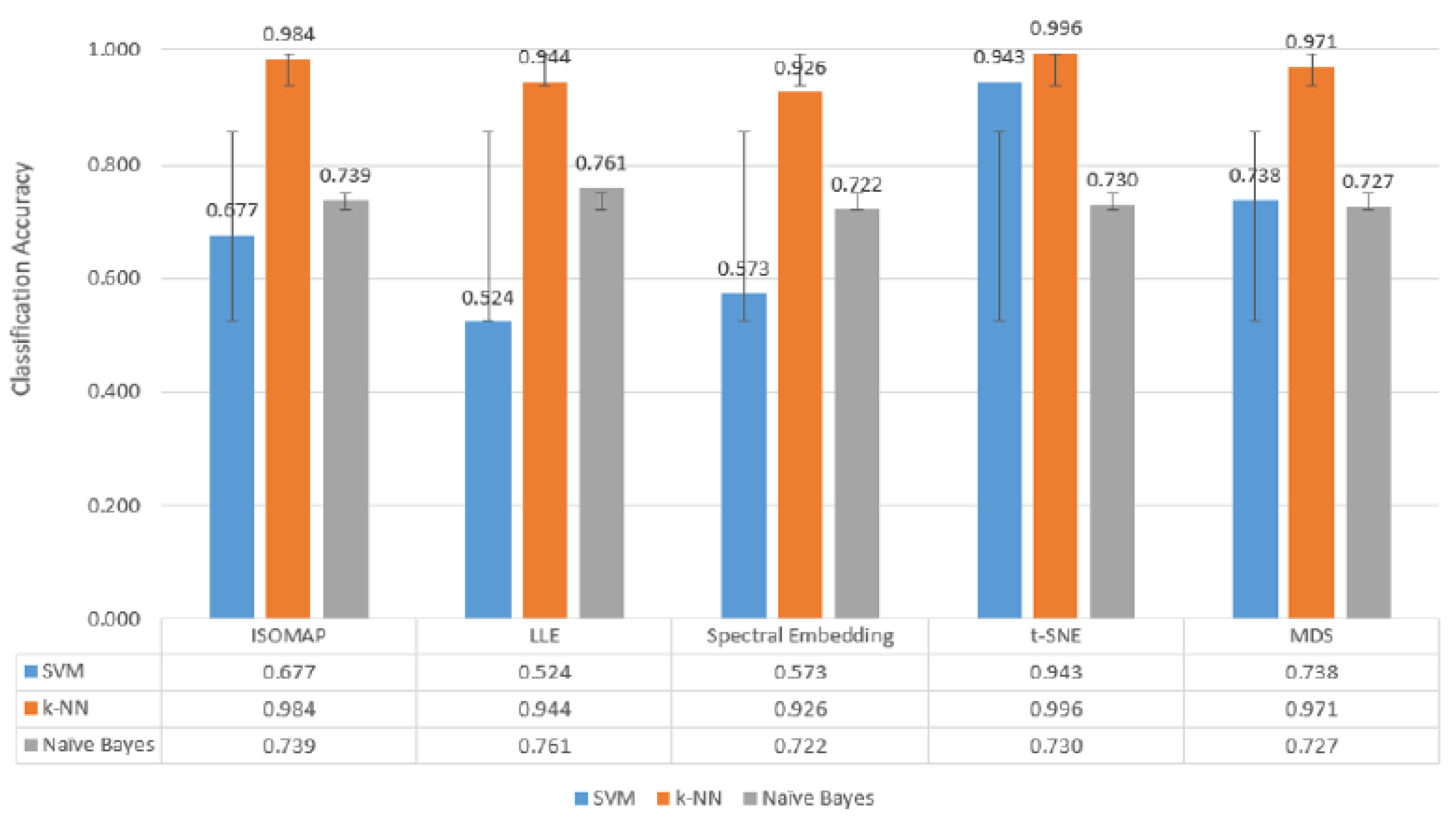
When
Figure 6,
Figure 7 and
Figure 8, which are comparatively presented after the dimensionality reduction methods (ISOMAP, LLE, Spectral Embedding, t-SNE and MDS) applied on the dataset, are examined, it is seen that the highest performance belongs to the
k-NN algorithm in all classification scenarios. Especially in binary classification (
Figure 6),
k-NN achieved
99.6% accuracy with t-SNE,
98.4% with ISOMAP and
97.1% with MDS, demonstrating a significantly superior success compared to the other two models (SVM and Naive Bayes). SVM gave a partially competitive result with
94.3% accuracy with t-SNE in this scenario, while Naive Bayes generally remained in the range of
72–76%.
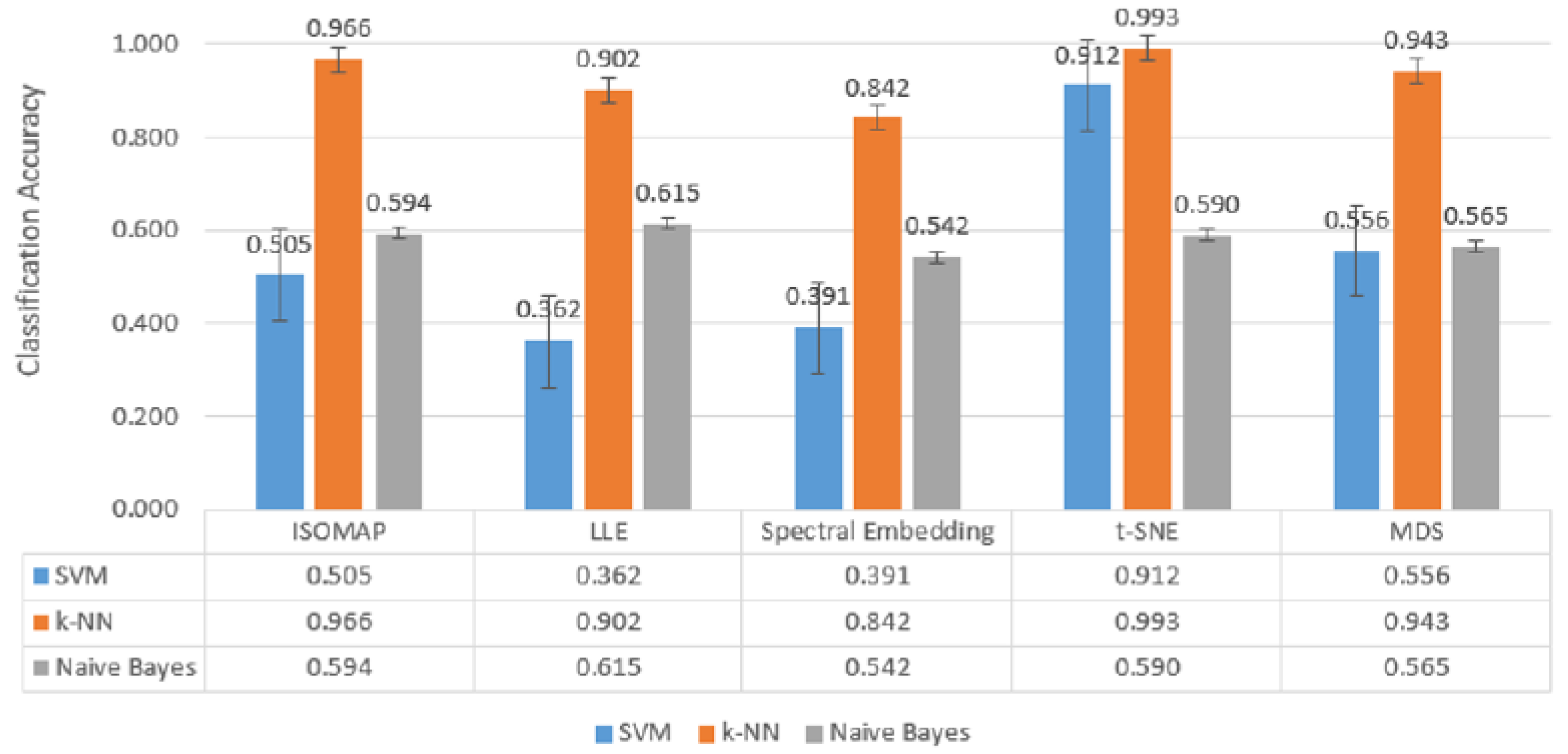
In
Figure 7, which includes three-class classification results,
k-NN again stands out as the most successful model in all dimensionality reduction methods. Reaching
99.3% accuracy with t-SNE,
96.6% with ISOMAP and
94.3% with MDS,
k-NN largely maintained its performance despite the increase in the number of classes. SVM achieved a competitive result of
91.2% only with t-SNE, while its accuracy remained below
50% for other methods. Naive Bayes provided moderate results in the range of
56–61% with methods such as LLE and MDS, but the difference with
k-NN remained significant.
In the most complex classification structure with five classes (
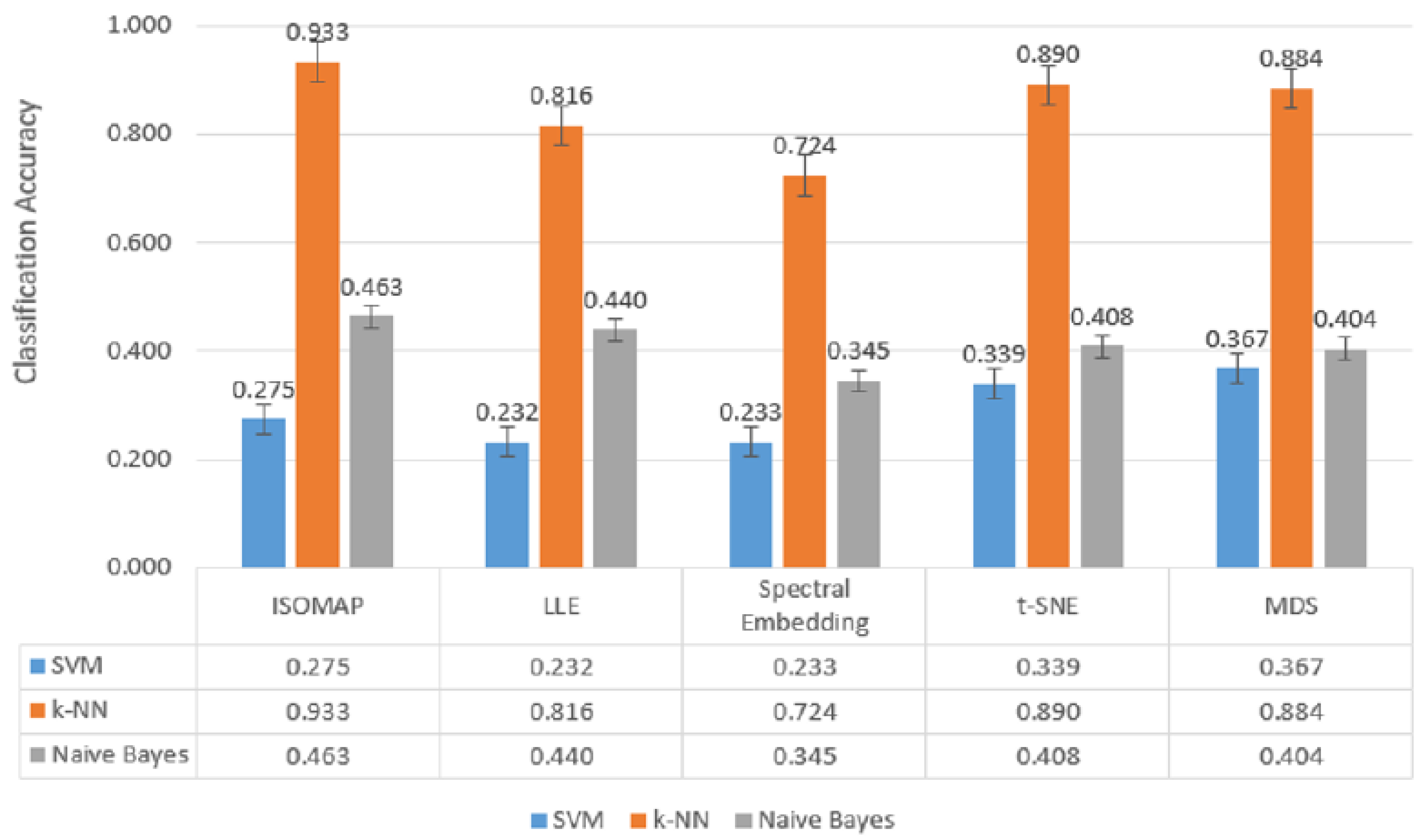
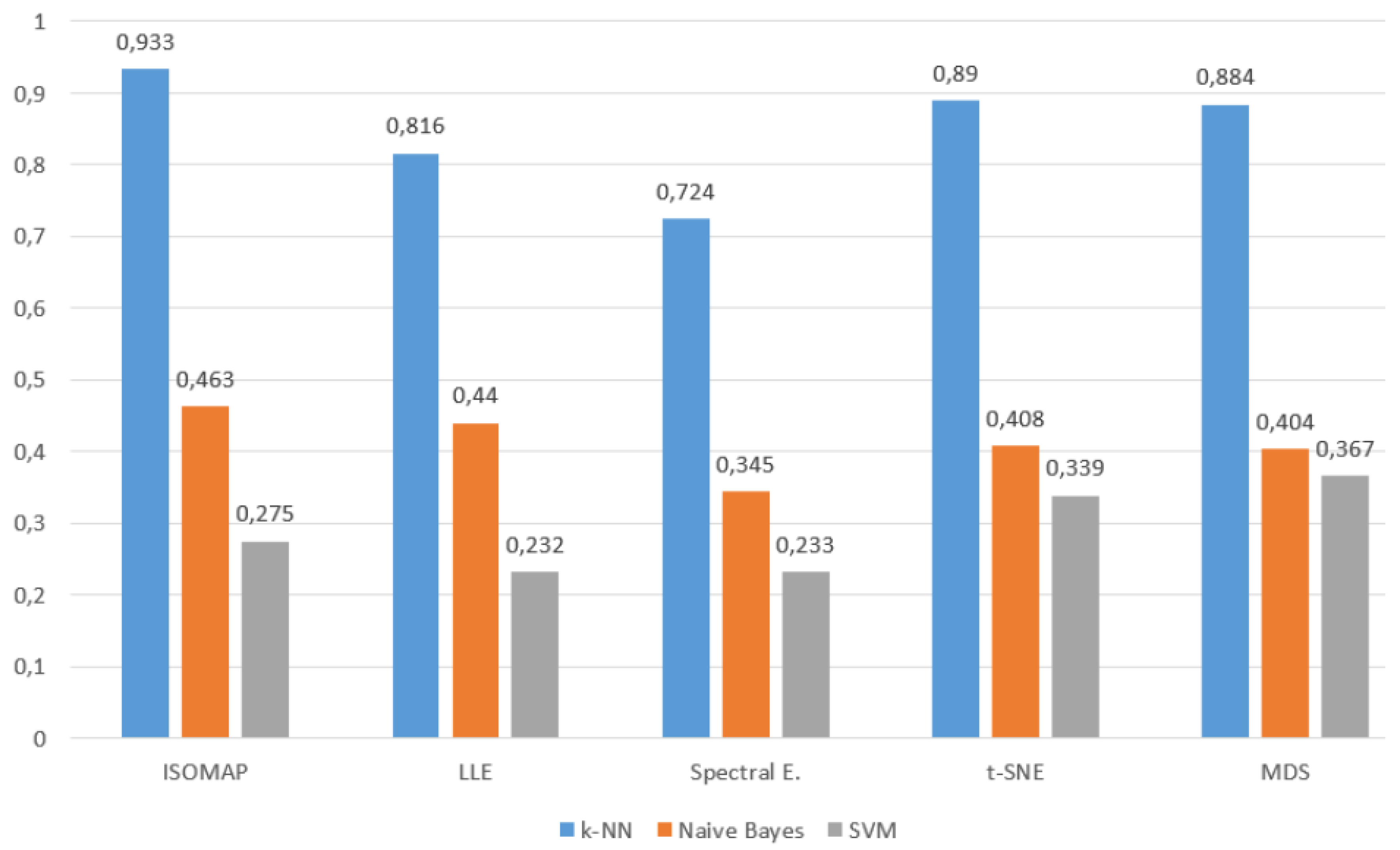
Figure 8), although a general decrease in model performance was observed,
k-NN maintained consistently high accuracy rates. Providing
93.3% accuracy with ISOMAP,
81.6% with LLE and
89.0% with t-SNE,
k-NN produced quite effective results compared to other models despite the difficulty brought by multi-class structures. On the other hand, SVM was inadequate in the classification task with low accuracy rates (
23–40%) in all methods, while Naive Bayes produced partially more balanced results (
34–46%) but still lagged behind
k-NN.
All these findings reveal that the k-NN algorithm, especially when used with t-SNE and ISOMAP dimensionality reduction methods, exhibited superior performance by providing the highest accuracy rates at both low and high class numbers. While SVM produced effective results with t-SNE only in binary classification, it experienced a serious decreases in its performance as the number of classes increased. Naive Bayes, on the other hand, achieved more balanced but generally moderate accuracies and although it behaved more stably in multi-class scenarios, it could not provide sufficient performance for applications requiring high accuracy.
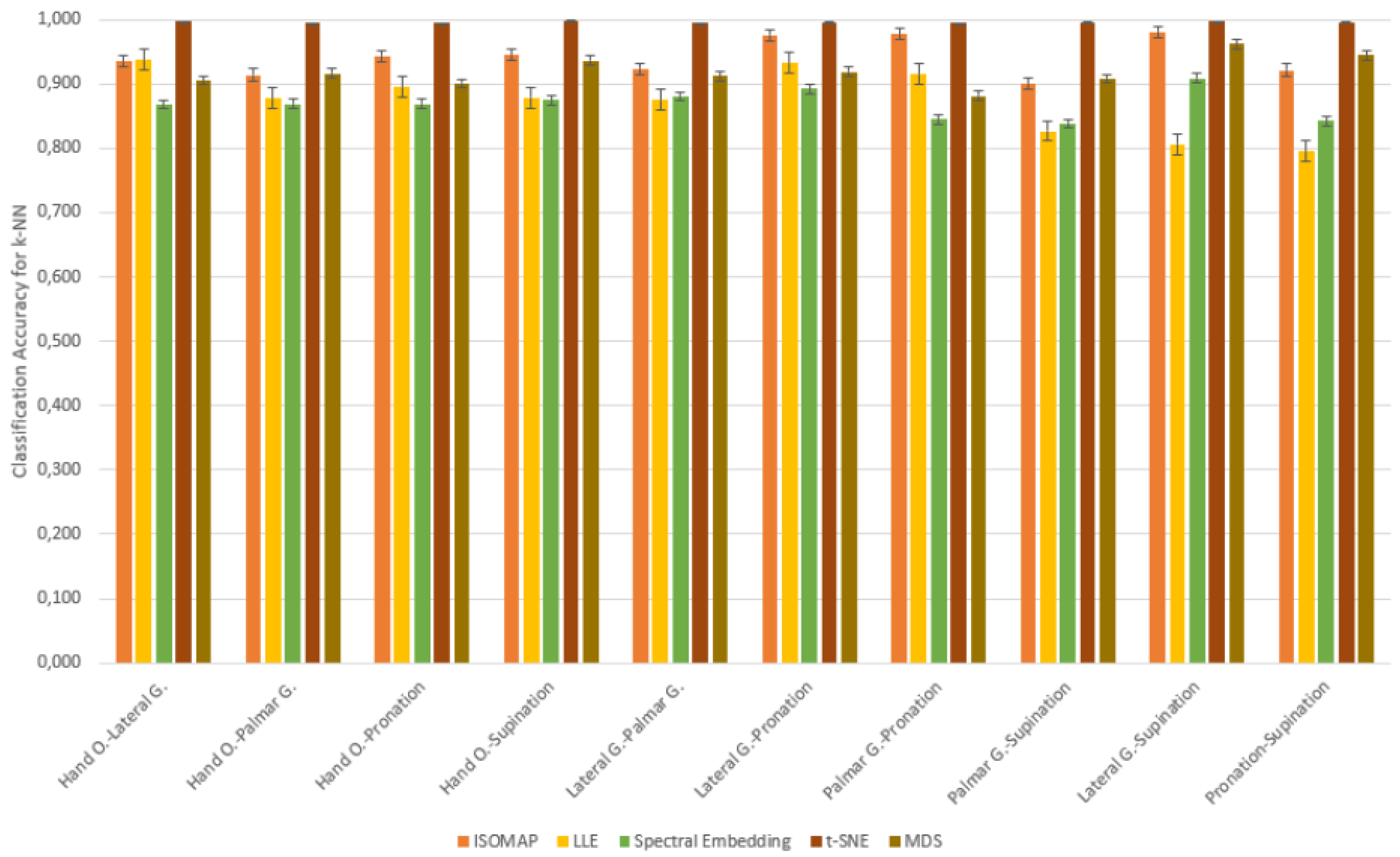
Table 7.
Classification Accuracy of ISOMAP for 2-Class Combinations
Table 7.
Classification Accuracy of ISOMAP for 2-Class Combinations
| |
ISOMAP |
| |
SVM |
k-NN |
Naive Bayes |
| Hand O.-Lateral G. |
0.731 |
0.935 |
0.789 |
| Hand O.–Palmar G. |
0.628 |
0.914 |
0.754 |
| Hand O.-Pronation |
0.760 |
0.943 |
0.778 |
| Hand O.-Supination |
0.676 |
0.945 |
0.783 |
| Lateral G.-Palmar G. |
0.737 |
0.923 |
0.771 |
| Lateral G.-Pronation |
0.720 |
0.976 |
0.815 |
| Palmar G.-Pronation |
0.528 |
0.978 |
0.783 |
| Palmar G.-Supination |
0.678 |
0.900 |
0.800 |
| Lateral G.-Supination |
0.534 |
0.981 |
0.791 |
| Pronation-Supination |
0.668 |
0.922 |
0.783 |
When the classification accuracies obtained after the ISOMAP dimensionality reduction method are examined, it is observed that the k-NN algorithm achieves the highest accuracy rates in all motor task pairs. Standing out with accuracy values exceeding 90%, k-NN exhibited a strong discrimination ability between both similar and highly distinct movements. While the Naive Bayes model provided a balanced performance with accuracies particularly in the 78–81% range, SVM showed relatively low success, especially in certain task pairs (e.g., “Palmar G.–Pronation” and “Lateral G.–Supination”). This finding clearly demonstrates that the k-NN algorithm is the most effective method for movement classification tasks in EEG data reduced using the ISOMAP method.
Table 8.
Classification Accuracy of LLE for 2-Class Combinations
Table 8.
Classification Accuracy of LLE for 2-Class Combinations
| |
Local Linear Embedding |
| |
SVM |
k-NN |
Naive Bayes |
| Hand O.-Lateral G. |
0.731 |
0.935 |
0.789 |
| Hand O.–Palmar G. |
0.555 |
0.938 |
0.826 |
| Hand O.-Pronation |
0.572 |
0.877 |
0.762 |
| Hand O.-Supination |
0.548 |
0.895 |
0.812 |
| Lateral G.-Palmar G. |
0.554 |
0.879 |
0.770 |
| Lateral G.-Pronation |
0.518 |
0.875 |
0.797 |
| Palmar G.-Pronation |
0.574 |
0.932 |
0.835 |
| Palmar G.-Supination |
0.552 |
0.915 |
0.830 |
| Lateral G.-Supination |
0.550 |
0.827 |
0.787 |
| Pronation-Supination |
0.502 |
0.807 |
0.771 |
In the classification analyses performed on dimensionally reduced data with the Local Linear Embedding (LLE) method, the k-NN algorithm stood out as the most successful model by reaching the highest accuracy rates in all motor task pairs. The fact that k-NN provided accuracy, especially in the “Palmar G.–Pronation” (93.2%), “Hand O.–Palmar G.” (93.8%), and “Hand O.–Lateral G.” (93.5%) task pairs, shows that this model works effectively in the decomposed feature space after LLE. The Naive Bayes model provided accuracy in the range of 76–83% in most tasks, exhibiting a balanced and acceptable performance. On the other hand, the SVM model was insufficient in the post-LLE classification tasks with low accuracy rates (50–57%), and it was observed that the performance level decreased significantly, especially in the “Pronation–Supination” and “Lateral G.–Pronation” task pairs. These results indicate that k-NN is the most effective classifier after the LLE method, Naive Bayes offers a balanced alternative, and SVM can provide only limited success in this structure.
Table 9.
Classification Accuracy of Spectral Embedding for 2-Class Combinations
Table 9.
Classification Accuracy of Spectral Embedding for 2-Class Combinations
| |
Spectral Embedding |
| |
SVM |
k-NN |
Naive Bayes |
| Hand O.-Lateral G. |
0.588 |
0.868 |
0.715 |
| Hand O.–Palmar G. |
0.648 |
0.869 |
0.777 |
| Hand O.-Pronation |
0.603 |
0.869 |
0.810 |
| Hand O.-Supination |
0.629 |
0.875 |
0.751 |
| Lateral G.-Palmar G. |
0.657 |
0.881 |
0.744 |
| Lateral G.-Pronation |
0.587 |
0.892 |
0.768 |
| Palmar G.-Pronation |
0.545 |
0.845 |
0.710 |
| Palmar G.-Supination |
0.602 |
0.839 |
0.725 |
| Lateral G.-Supination |
0.635 |
0.908 |
0.790 |
| Pronation-Supination |
0.610 |
0.842 |
0.756 |
Classification analyses performed after the Spectral Embedding dimensionality reduction method revealed that the k-NN algorithm achieved the highest accuracy rates in distinguishing motor task pairs. k-NN demonstrated superior performance by reaching accuracy rates of 88% and above, especially in the “Lateral G.–Supination” (90.8%), “Lateral G.–Pronation” (89.2%), and “Lateral G.–Palmar G.” (88.1%) task pairs. The Naive Bayes model remained in the 71–81% accuracy range for most tasks and achieved remarkable results by exceeding 80% especially in the “Hand O.–Pronation” and “Palmar G.–Supination” task pairs. The SVM model produced lower accuracies in general and remained below 60%, especially in tasks such as “Palmar G.–Pronation” and “Lateral G.–Pronation”. These findings show that the k-NN algorithm is the most powerful model in the feature space obtained after Spectral Embedding, Naive Bayes provides balanced but moderate results, and the classification success of SVM remains weak.
Table 10.
Classification Accuracy of t-SNE for 2-Class Combinations
Table 10.
Classification Accuracy of t-SNE for 2-Class Combinations
| |
SVM |
k-NN |
Naive Bayes |
| Hand O.-Lateral G. |
0.902 |
0.997 |
0.770 |
| Hand O.–Palmar G. |
0.900 |
0.995 |
0.769 |
| Hand O.-Pronation |
0.899 |
0.994 |
0.795 |
| Hand O.-Supination |
0.904 |
0.997 |
0.766 |
| Lateral G.-Palmar G. |
0.892 |
0.994 |
0.809 |
| Lateral G.-Pronation |
0.899 |
0.996 |
0.833 |
| Palmar G.-Pronation |
0.901 |
0.995 |
0.770 |
| Palmar G.-Supination |
0.939 |
0.996 |
0.726 |
| Lateral G.-Supination |
0.907 |
0.997 |
0.790 |
| Pronation-Supination |
0.870 |
0.996 |
0.792 |
Classification analyses performed after the t-SNE dimensionality reduction method revealed that the k-NN algorithm showed the highest performance with accuracy rates close to 99% in all motor task pairs. Especially in the “Hand O.–Supination”, “Lateral G.–Supination”, and “Palmar G.–Supination” task pairs, accuracy values exceeding 99.6% show that k-NN can perform a near-perfect separation between classes in the feature space obtained with t-SNE. In this scenario, the SVM model exhibited a competitive performance with high accuracy values (89–94%), unlike other dimensionality reduction methods. The Naive Bayes model, on the other hand, showed stable success in the range of 72–83%, but fell behind k-NN and SVM. These findings show that the classification algorithm that best fits the feature representation after t-SNE is k-NN, SVM demonstrates a significant performance increase in this method, while Naive Bayes offers relatively stable but limited performance.
Table 11.
Classification Accuracy of MDS for 2-Class Combinations
Table 11.
Classification Accuracy of MDS for 2-Class Combinations
| |
SVM |
k-NN |
Naive Bayes |
| Hand O.-Lateral G. |
0.795 |
0.905 |
0.794 |
| Hand O.–Palmar G. |
0.722 |
0.916 |
0.751 |
| Hand O.-Pronation |
0.836 |
0.901 |
0.795 |
| Hand O.-Supination |
0.706 |
0.936 |
0.744 |
| Lateral G.-Palmar G. |
0.773 |
0.912 |
0.805 |
| Lateral G.-Pronation |
0.800 |
0.918 |
0.847 |
| Palmar G.-Pronation |
0.738 |
0.881 |
0.761 |
| Palmar G.-Supination |
0.615 |
0.908 |
0.705 |
| Lateral G.-Supination |
0.743 |
0.962 |
0.770 |
| Pronation-Supination |
0.711 |
0.944 |
0.789 |
Classification analyses performed after the MDS dimensionality reduction method revealed that the k-NN algorithm was the most effective model by achieving the highest accuracy rates in all motor task pairs. Especially in task pairs such as “Lateral G.–Supination” (96.2%) and “Pronation–Supination” (94.4%), k-NN provided over 94% accuracy and demonstrated a high discrimination capacity between classes. The Naive Bayes model generally provided stable results in the 74–85% accuracy range and became a competitive alternative by surpassing SVM in some tasks. Although SVM exhibited a more balanced performance with the MDS method, it achieved low accuracy in some task pairs, especially in “Palmar G.–Supination” (61.5%). These findings show that k-NN is the most reliable classification algorithm in data reduced by the MDS method, Naive Bayes provided balanced but limited success, and SVM lagged behind k-NN with partial improvements.
Figure 9.
Classification Accuracy of 2-Class for k-NN Results
Figure 9.
Classification Accuracy of 2-Class for k-NN Results
In the classification analyses conducted for dual motor task pairs, when the accuracy rates obtained with different dimensionality reduction methods (ISOMAP, LLE, Spectral Embedding, t-SNE, MDS) were examined comparatively, it was observed that the highest accuracy was provided by the k-NN algorithm in all task pairs. In particular, the k-NN model used with t-SNE exhibited superior performance by obtaining accuracy rates above 99% in almost all task pairs. Similar high accuracy values were achieved with the ISOMAP and MDS methods, but the results obtained with t-SNE were the most striking. Although the Naive Bayes model generally provided accuracy in the range of 74–84% and outperformed SVM in some task pairs, it did not achieve the highest success in any case. Although the SVM algorithm provided high accuracies (87–94%) with the t-SNE dimensionality reduction method, it achieved lower accuracies with other methods and fell behind k-NN. These findings show that t-SNE, as a dimensionality reduction method, is quite successful in motor task separation, especially when used with k-NN; Naive Bayes offers stable but limited success; and SVM can only produce competitive results in certain cases.
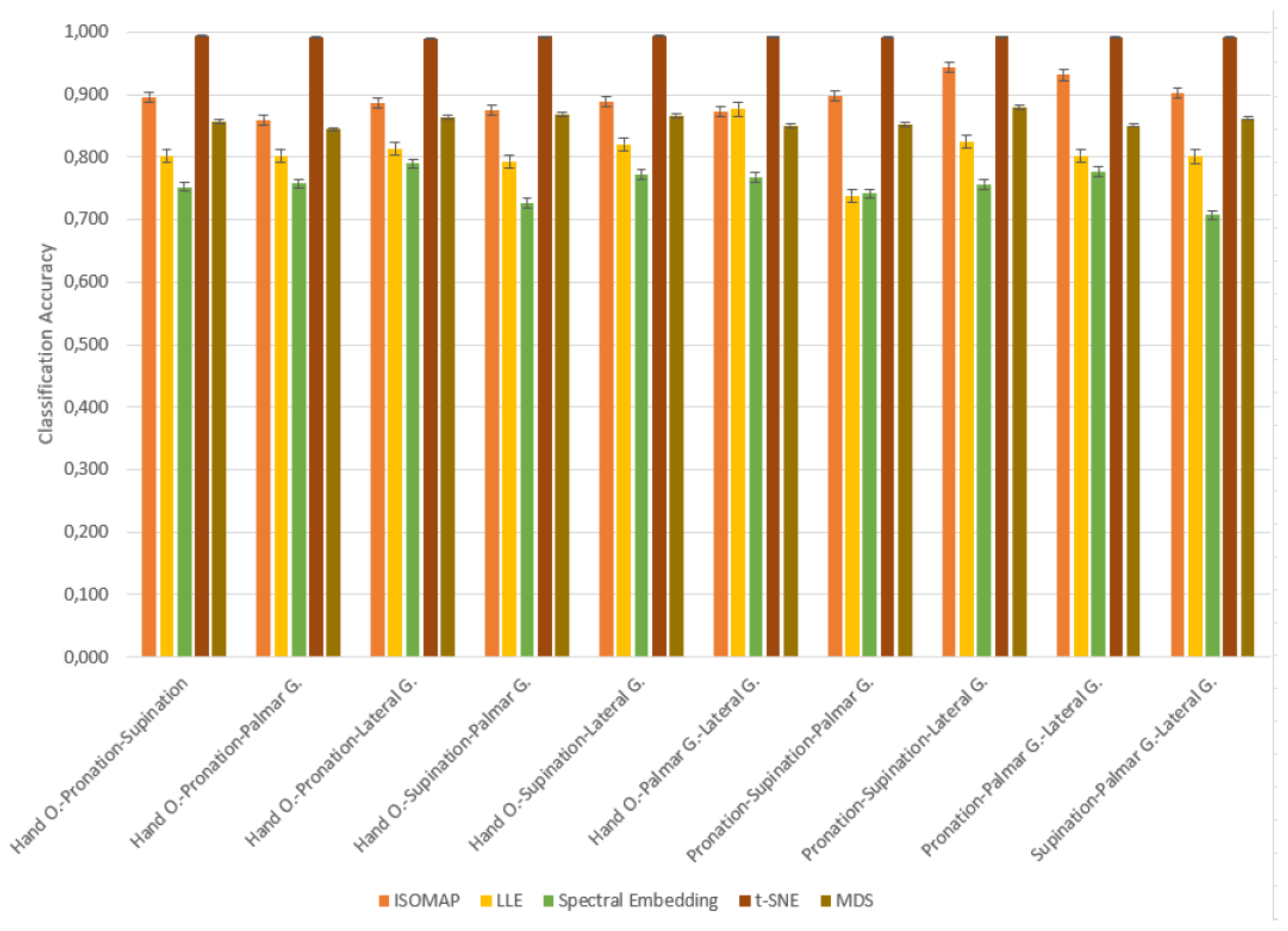
Table 12.
Classification Accuracy of ISOMAP for 3-Class Combinations
Table 12.
Classification Accuracy of ISOMAP for 3-Class Combinations
| |
SVM |
k-NN |
Naive Bayes |
| Hand O.-Pronation-Supination |
0.575 |
0.896 |
0.665 |
| Hand O.-Pronation-Palmar G. |
0.497 |
0.858 |
0.620 |
| Hand O.-Pronation-Lateral G. |
0.532 |
0.887 |
0.676 |
| Hand O.-Supination-Palmar G. |
0.496 |
0.874 |
0.629 |
| Hand O.-Supination-Lateral G. |
0.523 |
0.888 |
0.714 |
| Hand O.-Palmar G.-Lateral G. |
0.405 |
0.873 |
0.634 |
| Pronation-Supination-Palmar G. |
0.479 |
0.897 |
0.662 |
| Pronation-Supination-Lateral G. |
0.479 |
0.943 |
0.679 |
| Pronation-Palmar G.-Lateral G. |
0.500 |
0.931 |
0.678 |
| Supination-Palmar G.-Lateral G. |
0.479 |
0.902 |
0.664 |
Classification analyses performed on triple motor task combinations created using the ISOMAP dimensionality reduction method revealed that the k-NN algorithm performed significantly better than other models. The highest accuracies were obtained by k-NN in all combinations, and remarkable success was achieved with 94.3% accuracy, especially in the “Pronation–Supination–Lateral G.” task trio. While the Naive Bayes model provided more limited but balanced results in the range of 62–71%, the SVM model was inadequate in this task set with low accuracies. Especially in the “Hand O.–Palmar G.–Lateral G.” combination, SVM provided only 40.5% accuracy. These findings show that the most effective classification performance in three-class task separations with the ISOMAP method was obtained by the k-NN algorithm.
Table 13.
Classification Accuracy of LLE for 3-Class Combinations
Table 13.
Classification Accuracy of LLE for 3-Class Combinations
| |
SVM |
k-NN |
Naive Bayes |
| Hand O.-Pronation-Supination |
0.415 |
0.802 |
0.663 |
| Hand O.-Pronation-Palmar G. |
0.417 |
0.802 |
0.702 |
| Hand O.-Pronation-Lateral G. |
0.351 |
0.813 |
0.699 |
| Hand O.-Supination-Palmar G. |
0.402 |
0.793 |
0.648 |
| Hand O.-Supination-Lateral G. |
0.381 |
0.820 |
0.664 |
| Hand O.-Palmar G.-Lateral G. |
0.378 |
0.877 |
0.635 |
| Pronation-Supination-Palmar G. |
0.397 |
0.738 |
0.608 |
| Pronation-Supination-Lateral G. |
0.361 |
0.825 |
0.620 |
| Pronation-Palmar G.-Lateral G. |
0.400 |
0.802 |
0.684 |
| Supination-Palmar G.-Lateral G. |
0.389 |
0.801 |
0.678 |
In Table ??, the k-NN algorithm showed superior performance by achieving the highest accuracy rates in all tasks. The highest accuracy value of 87.7% was obtained in the “Hand O.–Palmar G.–Lateral G.” combination, while success rates above 80% were achieved in other combinations. The Naive Bayes model provided a balanced but limited success by remaining in the 63–70% accuracy range in most tasks. The SVM algorithm, on the other hand, was insufficient in classification tasks with low accuracy rates (35–42%) after the LLE method. These findings clearly show that k-NN is the most effective model in three-class task combinations where the dimensionality is reduced with the LLE method.
Table 14.
Classification Accuracy of Spectral Embedding for 3-Class Combinations
Table 14.
Classification Accuracy of Spectral Embedding for 3-Class Combinations
| |
SVM |
k-NN |
Naive Bayes |
| Hand O.-Pronation-Supination |
0.414 |
0.753 |
0.606 |
| Hand O.-Pronation-Palmar G. |
0.451 |
0.758 |
0.603 |
| Hand O.-Pronation-Lateral G. |
0.409 |
0.789 |
0.635 |
| Hand O.-Supination-Palmar G. |
0.429 |
0.726 |
0.612 |
| Hand O.-Supination-Lateral G. |
0.373 |
0.772 |
0.595 |
| Hand O.-Palmar G.-Lateral G. |
0.443 |
0.768 |
0.603 |
| Pronation-Supination-Palmar G. |
0.469 |
0.742 |
0.575 |
| Pronation-Supination-Lateral G. |
0.517 |
0.757 |
0.604 |
| Pronation-Palmar G.-Lateral G. |
0.500 |
0.777 |
0.600 |
| Supination-Palmar G.-Lateral G. |
0.465 |
0.707 |
0.578 |
Table ?? shows that the k-NN algorithm achieved the highest accuracy rates in all tasks. Especially achieving high success in the “Hand O.–Pronation–Lateral G.” (78.9%) and “Pronation–Palmar G.–Lateral G.” (77.7%) task combinations, k-NN was able to effectively distinguish between classes in the low-dimensional representations obtained with this method. The Naive Bayes model provided moderate accuracies in the range of 57–63% and showed a balanced performance. On the other hand, the SVM algorithm exhibited insufficient success in these combinations with accuracy rates ranging from 37–52%. These results clearly reveal that k-NN is the most reliable and successful classification algorithm in three-class data structures where the dimensionality is reduced with the Spectral Embedding method.
Table 15.
Classification Accuracy of t-SNE for 3-Class Combinations
Table 15.
Classification Accuracy of t-SNE for 3-Class Combinations
| |
SVM |
k-NN |
Naive Bayes |
| Hand O.-Pronation-Supination |
0.833 |
0.994 |
0.691 |
| Hand O.-Pronation-Palmar G. |
0.818 |
0.993 |
0.693 |
| Hand O.-Pronation-Lateral G. |
0.844 |
0.990 |
0.671 |
| Hand O.-Supination-Palmar G. |
0.803 |
0.993 |
0.637 |
| Hand O.-Supination-Lateral G. |
0.827 |
0.994 |
0.665 |
| Hand O.-Palmar G.-Lateral G. |
0.844 |
0.993 |
0.641 |
| Pronation-Supination-Palmar G. |
0.826 |
0.992 |
0.646 |
| Pronation-Supination-Lateral G. |
0.852 |
0.993 |
0.647 |
| Pronation-Palmar G.-Lateral G. |
0.856 |
0.992 |
0.650 |
| Supination-Palmar G.-Lateral G. |
0.846 |
0.992 |
0.652 |
Classification analyses performed on triple motor task combinations created using the t-SNE dimensionality reduction method revealed that the k-NN algorithm showed the highest performance by achieving over 99% accuracy in each task set. Reaching accuracies of 99.2% and above in many combinations such as “Hand O.–Supination–Lateral G.”, “Hand O.–Pronation–Palmar G.” and “Supination–Palmar G.–Lateral G.”, k-NN showed an almost error-free classification success in this method. Unlike previous methods, the SVM model provided high accuracy values (80–85%) with the t-SNE method and has become a competitive alternative. On the other hand, the Naive Bayes algorithm exhibited a limited classification performance, staying in the range of 63–69%. These results show that k-NN is the most successful model in multi-class structures where dimensionality is reduced with the t-SNE method; SVM stands out only in this method; and Naive Bayes generally performs lower.
Table 16.
Classification Accuracy of MDS for 3-Class Combinations
Table 16.
Classification Accuracy of MDS for 3-Class Combinations
| |
SVM |
k-NN |
Naive Bayes |
| Hand O.-Pronation-Supination |
0.548 |
0.857 |
0.618 |
| Hand O.-Pronation-Palmar G. |
0.624 |
0.844 |
0.641 |
| Hand O.-Pronation-Lateral G. |
0.674 |
0.864 |
0.677 |
| Hand O.-Supination-Palmar G. |
0.540 |
0.868 |
0.568 |
| Hand O.-Supination-Lateral G. |
0.565 |
0.866 |
0.636 |
| Hand O.-Palmar G.-Lateral G. |
0.580 |
0.851 |
0.636 |
| Pronation-Supination-Palmar G. |
0.510 |
0.853 |
0.585 |
| Pronation-Supination-Lateral G. |
0.640 |
0.879 |
0.693 |
| Pronation-Palmar G.-Lateral G. |
0.640 |
0.851 |
0.679 |
| Supination-Palmar G.-Lateral G. |
0.499 |
0.862 |
0.652 |
The results given in Table ?? show that the k-NN algorithm exhibited the highest performance by achieving over 85% accuracy in all tasks. Achieving 87.9% accuracy in the “Pronation–Supination–Lateral G.” task trio and 86.6–86.8% accuracy in tasks such as “Hand O.–Supination–Palmar G.” and “Hand O.–Supination–Lateral G.”, k-NN provided effective classification in low-dimensional space with the MDS method. The Naive Bayes algorithm showed limited performance by providing accuracy in the range of 58–69%. Although SVM produced relatively better results in some tasks, it generally remained at 50–67% accuracy levels. These findings reveal that the k-NN algorithm is the most suitable classifier that provides consistent and high success for three-class combinations in which the dimensionality is reduced with the MDS method.
Figure 10.
Classification Accuracy of 3-Class for k-NN Results
Figure 10.
Classification Accuracy of 3-Class for k-NN Results
As a result of the classification analyses performed on triple motor task combinations, it was observed that the k-NN algorithm achieved the highest accuracy rates by far in all methods, regardless of the dimensionality reduction method. In particular, the t-SNE method stood out with accuracy values reaching over 99% when used with k-NN. Under this structure, k-NN showed almost error-free classification success in almost every task combination. While the ISOMAP and MDS methods provided very successful results in the 85–94% accuracy range when used with k-NN, the LLE and Spectral Embedding methods produced slightly lower but still high accuracies (between 73–88%).
The SVM algorithm was able to reach high accuracy values (80–85%) only with the t-SNE method, while in other methods it generally remained in the 35–67% range and fell far behind k-NN. The Naive Bayes model, on the other hand, showed balanced but limited success in all methods, remaining in the 57–71% accuracy range and at best could approach k-NN.
When these findings are evaluated in general, it is revealed that the highest and most consistent performance in triple classification problems is provided by the k-NN algorithm, especially with the t-SNE dimensionality reduction method. Other algorithms produced reasonable results only under certain conditions but fell behind k-NN in terms of overall success. Therefore, the t-SNE + k-NN combination can be considered as the most reliable and recommended structure in EEG data analyses based on three-class motor task separation.
Table 17.
Classification Accuracy for 5-Class
Table 17.
Classification Accuracy for 5-Class
| Method |
Classifier |
Hand O.-Lateral G.-Palmar G.-Supination-Pronation |
| ISOMAP |
SVM |
0.900 |
| |
k-NN |
0.933 |
| |
Naive Bayes |
0.463 |
| LLE |
SVM |
0.800 |
| |
k-NN |
0.816 |
| |
Naive Bayes |
0.463 |
| Spectral E. |
SVM |
0.700 |
| |
k-NN |
0.724 |
| |
Naive Bayes |
0.453 |
| t-SNE |
SVM |
0.800 |
| |
k-NN |
0.809 |
| |
Naive Bayes |
0.438 |
| MDS |
SVM |
0.800 |
| |
k-NN |
0.840 |
| |
Naive Bayes |
0.467 |
The findings obtained in the five-classification scenario clearly show that the
k-NN algorithm achieves the highest classification accuracies among all dimensionality reduction methods. Especially when used with ISOMAP (
79.3%) and t-SNE (
79.7%) methods,
k-NN achieved high success even in five-class separations, and these combinations were the strongest alternatives for multi-class structures. The MDS method closely follows these two methods with
79.1% accuracy. On the other hand, the LLE (
67.9%) and Spectral Embedding (
64.5%) methods remained more limited in class separation with relatively lower accuracy rates, even when used with
k-NN (
Figure 11).
The Naive Bayes model produced accuracy values between
49.8% and
54.3% under all methods and showed a moderate, stable, but limited classification performance. Naive Bayes, which gave the best result with
54.3% accuracy using ISOMAP, generally fell far behind
k-NN (orange bars in
Figure 11).
On the other hand, the SVM algorithm stood out as the weakest model for 5-class structures; accuracy remained below
30% in most methods. Especially when used with t-SNE, it achieved only
9.8% accuracy, indicating that this model is not compatible with high class numbers and low-dimensional representations (see
Figure 11). The highest accuracy for SVM was achieved with the MDS method at
37.2%.