Submitted:
05 August 2025
Posted:
07 August 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Challenge to Treatment
1.2. Chemoresistance Mechanisms
2. Protein Methylation
2.1. Types and Function
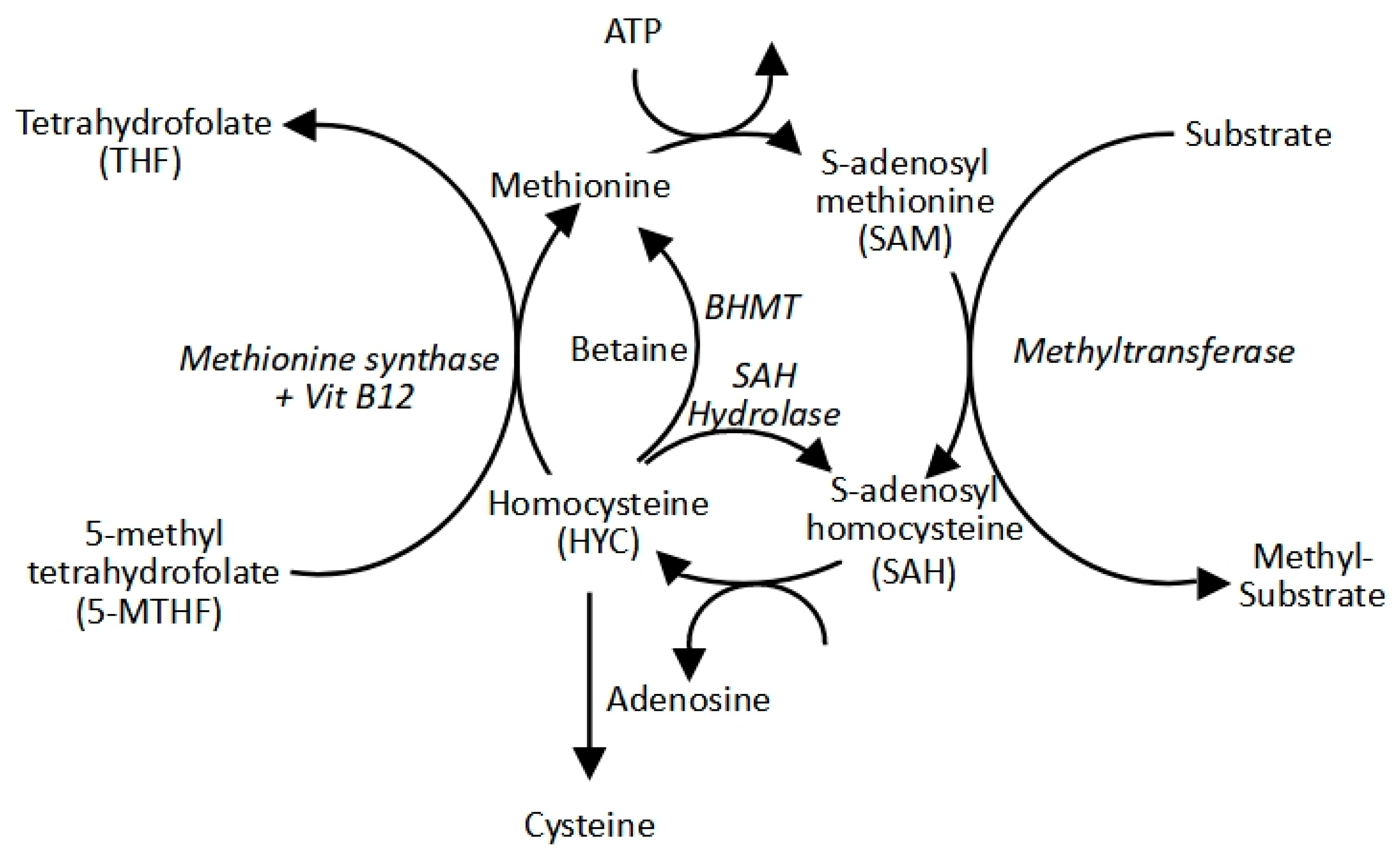
2.2. The Methyl Cycle: S-Adenosylmethionine Production
2.3. Protein Methyltransferases
2.3.1. Protein Lysine Methyltransferases (PKMTs)
2.3.2. Protein Arginine Methyltransferases (PRMTs)
2.3.3. Cancer Dysregulation
2.4. Classes of Methyltransferase Inhibitors
2.4.1. SAM Competitive Inhibitors
2.4.2. Substrate Competitive Inhibitors
2.4.3. Bisubstrate Inhibitors
2.4.4. Allosteric Inhibitors
2.4.5. Complex Disrupting Inhibitors
2.4.6. Covalent Inhibitors
2.4.7. PROTAC Inhibitors
2.4.8. Inhibitor Limitations
3. Failure to Clinic
4. Considerations for Designing Novel PMT Inhibitors
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Abad, E.; Graifer, D.; Lyakhovich, A. DNA damage response and resistance of cancer stem cells. Cancer Lett. 2020, 474, 106–117. [Google Scholar] [CrossRef]
- Ábrányi-Balogh, P.; Petri, L.; Imre, T.; Szijj, P.; Scarpino, A.; Hrast, M.; Mitrović, A.; Fonovič, U.P.; Németh, K.; Barreteau, H.; et al. A road map for prioritizing warheads for cysteine targeting covalent inhibitors. Eur. J. Med. Chem. 2018, 160, 94–107. [Google Scholar] [CrossRef]
- Aebersold, R.; Agar, J.N.; Amster, I.J.; Baker, M.S.; Bertozzi, C.R.; Boja, E.S.; E Costello, C.; Cravatt, B.F.; Fenselau, C.; A Garcia, B.; et al. How many human proteoforms are there? Nat. Chem. Biol. 2018, 14, 206–214. [Google Scholar] [CrossRef]
- Alicea-Velázquez, N.L.; Shinsky, S.A.; Loh, D.M.; Lee, J.H.; Skalnik, D.G.; Cosgrove, M.S. Targeted disruption of the interaction between WD-40 repeat protein 5 (WDR5) and mixed lineage leukemia (MLL)/SET1 family proteins specifically inhibits MLL1 and SETd1A methyltransferase complexes. Journal of Biological Chemistry 2016, 291, 22357–22372. [Google Scholar] [CrossRef]
- Alimbetov, D.; Askarova, S.; Umbayev, B.; Davis, T.; Kipling, D. Pharmacological Targeting of Cell Cycle, Apoptotic and Cell Adhesion Signaling Pathways Implicated in Chemoresistance of Cancer Cells. Int. J. Mol. Sci. 2018, 19, 1690. [Google Scholar] [CrossRef]
- Alinari, L.; Mahasenan, K.V.; Yan, F.; Karkhanis, V.; Chung, J.-H.; Smith, E.M.; Quinion, C.; Smith, P.L.; Kim, L.; Patton, J.T.; et al. Selective inhibition of protein arginine methyltransferase 5 blocks initiation and maintenance of B-cell transformation. Blood 2015, 125, 2530–2543. [Google Scholar] [CrossRef] [PubMed]
- Antonysamy, S., 2017. The structure and function of the PRMT5: MEP50 complex. Macromolecular Protein Complexes: Structure and Function, pp.185-194.
- Arrigoni, E.; Galimberti, S.; Petrini, M.; Danesi, R.; Di Paolo, A. ATP-binding cassette transmembrane transporters and their epigenetic control in cancer: an overview. Expert Opin. Drug Metab. Toxicol. 2016, 12, 1419–1432. [Google Scholar] [CrossRef] [PubMed]
- Asberry, A.M.; Cai, X.; Deng, X.; Santiago, U.; Liu, S.; Sims, H.S.; Liang, W.; Xu, X.; Wan, J.; Jiang, W.; et al. Discovery and Biological Characterization of PRMT5:MEP50 Protein–Protein Interaction Inhibitors. J. Med. Chem. 2022, 65, 13793–13812. [Google Scholar] [CrossRef] [PubMed]
- Baker, T.; Nerle, S.; Pritchard, J.; Zhao, B.; Rivera, V.M.; Garner, A.; Gonzalvez, F. Acquisition of a single EZH2 D1 domain mutation confers acquired resistance to EZH2-targeted inhibitors. Oncotarget 2015, 6, 32646–32655. [Google Scholar] [CrossRef]
- Barsyte-Lovejoy, D.; Li, F.; Oudhoff, M.J.; Tatlock, J.H.; Dong, A.; Zeng, H.; Wu, H.; Freeman, S.A.; Schapira, M.; Senisterra, G.A.; et al. ( R )-PFI-2 is a potent and selective inhibitor of SETD7 methyltransferase activity in cells. Proc. Natl. Acad. Sci. 2014, 111, 12853–12858. [Google Scholar] [CrossRef]
- Bashore, F.M.; Foley, C.A.; Ong, H.W.; Rectenwald, J.M.; Hanley, R.P.; Norris-Drouin, J.L.; Cholensky, S.H.; Mills, C.A.; Pearce, K.H.; Herring, L.E.; et al. PROTAC Linkerology Leads to an Optimized Bivalent Chemical Degrader of Polycomb Repressive Complex 2 (PRC2) Components. ACS Chem. Biol. 2023, 18, 494–507. [Google Scholar] [CrossRef]
- Békés, M.; Langley, D.R.; Crews, C.M. PROTAC targeted protein degraders: the past is prologue. Nat. Rev. Drug Discov. 2022, 21, 181–200. [Google Scholar] [CrossRef]
- Bisserier, M.; Wajapeyee, N. Mechanisms of resistance to EZH2 inhibitors in diffuse large B-cell lymphomas. Blood 2018, 131, 2125–2137. [Google Scholar] [CrossRef]
- Biswas, R.; Bugde, P.; He, J.; Merien, F.; Lu, J.; Liu, D.-X.; Myint, K.; Liu, J.; McKeage, M.; Li, Y. Transport-Mediated Oxaliplatin Resistance Associated with Endogenous Overexpression of MRP2 in Caco-2 and PANC-1 Cells. Cancers 2019, 11, 1330. [Google Scholar] [CrossRef]
- Borkin, D.; He, S.; Miao, H.; Kempinska, K.; Pollock, J.; Chase, J.; Purohit, T.; Malik, B.; Zhao, T.; Wang, J.; et al. Pharmacologic Inhibition of the Menin-MLL Interaction Blocks Progression of MLL Leukemia In Vivo. Cancer Cell 2015, 27, 589–602. [Google Scholar] [CrossRef] [PubMed]
- Brekker, M.A.; Sartawi, T.; Sawatzky, T.M.; Causey, C.P.; Rehman, F.K.; Knuckley, B. A peptoid-based inhibitor of protein arginine methyltransferase 1 (PRMT1) induces apoptosis and autophagy in cancer cells. J. Biol. Chem. 2022, 298, 102205. [Google Scholar] [CrossRef]
- Butler, K.V.; Ma, A.; Yu, W.; Li, F.; Tempel, W.; Babault, N.; Pittella-Silva, F.; Shao, J.; Wang, J.; Luo, M.; et al. Structure-Based Design of a Covalent Inhibitor of the SET Domain-Containing Protein 8 (SETD8) Lysine Methyltransferase. J. Med. Chem. 2016, 59, 9881–9889. [Google Scholar] [CrossRef] [PubMed]
- Campagna-Slater, V.; Mok, M.W.; Nguyen, K.T.; Feher, M.; Najmanovich, R.; Schapira, M. Structural Chemistry of the Histone Methyltransferases Cofactor Binding Site. J. Chem. Inf. Model. 2011, 51, 612–623. [Google Scholar] [CrossRef] [PubMed]
- Chai, H.; Pan, C.; Zhang, M.; Huo, H.; Shan, H.; Wu, J. Histone methyltransferase SETD1A interacts with notch and promotes notch transactivation to augment ovarian cancer development. BMC Cancer 2023, 23, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chan-Penebre, E.; Kuplast, K.G.; Majer, C.R.; Boriack-Sjodin, P.A.; Wigle, T.J.; Johnston, L.D.; Rioux, N.; Munchhof, M.J.; Jin, L.; Jacques, S.L.; et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat. Chem. Biol. 2015, 11, 432–437. [Google Scholar] [CrossRef]
- Chen, L., Zeng, Y. and Zhou, S.F., 2018. Role of apoptosis in cancer resistance to chemotherapy. Current understanding of apoptosis-programmed cell death.
- Chen, W.; Chen, X.; Li, D.; Wang, X.; Long, G.; Jiang, Z.; You, Q.; Guo, X. Discovery of a potent MLL1 and WDR5 protein-protein interaction inhibitor with in vivo antitumor activity. Eur. J. Med. Chem. 2021, 223, 113677. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, H.; Yang, S.; Su, D. Increased ABCC2 expression predicts cisplatin resistance in non-small cell lung cancer. Cell Biochem. Funct. 2020, 39, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Copeland, R.A. Protein methyltransferase inhibitors as precision cancer therapeutics: a decade of discovery. Philos. Trans. R. Soc. B: Biol. Sci. 2018, 373, 20170080. [Google Scholar] [CrossRef]
- Dale, B.; Anderson, C.; Park, K.-S.; Kaniskan, H.Ü.; Ma, A.; Shen, Y.; Zhang, C.; Xie, L.; Chen, X.; Yu, X.; et al. Targeting Triple-Negative Breast Cancer by a Novel Proteolysis Targeting Chimera Degrader of Enhancer of Zeste Homolog 2. ACS Pharmacol. Transl. Sci. 2022, 5, 491–507. [Google Scholar] [CrossRef]
- Damaraju, V.L.; Damaraju, S.; Young, J.D.; A Baldwin, S.; Mackey, J.; Sawyer, M.B.; E Cass, C. Nucleoside anticancer drugs: the role of nucleoside transporters in resistance to cancer chemotherapy. Oncogene 2003, 22, 7524–7536. [Google Scholar] [CrossRef]
- Dempke, W.C.M.; Desole, M.; Chiusolo, P.; Sica, S.; Schmidt-Hieber, M. Targeting the undruggable: menin inhibitors ante portas. J. Cancer Res. Clin. Oncol. 2023, 149, 9451–9459. [Google Scholar] [CrossRef]
- Diehl, C.J.; Ciulli, A. Discovery of small molecule ligands for the von Hippel-Lindau (VHL) E3 ligase and their use as inhibitors and PROTAC degraders. Chem. Soc. Rev. 2022, 51, 8216–8257. [Google Scholar] [CrossRef] [PubMed]
- Dölle, A.; Adhikari, B.; Krämer, A.; Weckesser, J.; Berner, N.; Berger, L.-M.; Diebold, M.; Szewczyk, M.M.; Barsyte-Lovejoy, D.; Arrowsmith, C.H.; et al. Design, Synthesis, and Evaluation of WD-Repeat-Containing Protein 5 (WDR5) Degraders. J. Med. Chem. 2021, 64, 10682–10710. [Google Scholar] [CrossRef]
- Dong, H.; Liu, S.; Zhang, X.; Chen, S.; Kang, L.; Chen, Y.; Ma, S.; Fu, X.; Liu, Y.; Zhang, H.; et al. An Allosteric PRC2 Inhibitor Targeting EED Suppresses Tumor Progression by Modulating the Immune Response. Cancer Res. 2019, 79, 5587–5596. [Google Scholar] [CrossRef]
- Drosos, Y.; Myers, J.A.; Xu, B.; Mathias, K.M.; Beane, E.C.; Radko-Juettner, S.; Mobley, R.J.; Larsen, M.E.; Piccioni, F.; Ma, X.; et al. NSD1 mediates antagonism between SWI/SNF and polycomb complexes and is required for transcriptional activation upon EZH2 inhibition. Mol. Cell 2022, 82, 2472–2489.e8. [Google Scholar] [CrossRef] [PubMed]
- Duncan, K.W.; Rioux, N.; Boriack-Sjodin, P.A.; Munchhof, M.J.; Reiter, L.A.; Majer, C.R.; Jin, L.; Johnston, L.D.; Chan-Penebre, E.; Kuplast, K.G.; et al. Structure and Property Guided Design in the Identification of PRMT5 Tool Compound EPZ015666. ACS Med. Chem. Lett. 2015, 7, 162–166. [Google Scholar] [CrossRef]
- Eggert, E.; Hillig, R.C.; Koehr, S.; Stöckigt, D.; Weiske, J.; Barak, N.; Mowat, J.; Brumby, T.; Christ, C.D.; ter Laak, A.; et al. Discovery and Characterization of a Highly Potent and Selective Aminopyrazoline-Based in Vivo Probe (BAY-598) for the Protein Lysine Methyltransferase SMYD2. J. Med. Chem. 2016, 59, 4578–4600. [Google Scholar] [CrossRef]
- Elton, T.S.; Ozer, H.G.; Yalowich, J.C. Effects of DNA topoisomerase IIα splice variants on acquired drug resistance. Cancer Drug Resist. 2020, 3, 161–170. [Google Scholar] [CrossRef]
- Falnes, P.Ø.; Małecki, J.M.; Herrera, M.C.; Bengtsen, M.; Davydova, E. Human seven-β-strand (METTL) methyltransferases - conquering the universe of protein lysine methylation. J. Biol. Chem. 2023, 299, 104661. [Google Scholar] [CrossRef]
- Fernández-Ramos, D.; Lopitz-Otsoa, F.; Lu, S.C.; Mato, J.M. S-Adenosylmethionine: A Multifaceted Regulator in Cancer Pathogenesis and Therapy. Cancers 2025, 17, 535. [Google Scholar] [CrossRef]
- Froese, D. S., Fowler, B. & Baumgartner, M. R. Vitamin B12, folate, and the methionine remethylation cycle-biochemistry, pathways, and regulation. J. Inherit. Metab. Dis. 42, 673–685 (2019).
- G. Blum, G. Ibanez, X. Rao, D. Shum, C. Radu, H. Djaballah, J. C. Rice and M. Luo, ACS Chem. Biol., 2014, 9, 2471–2478.
- Galmarini, C.M.; Mackey, J.R.; Dumontet, C. Nucleoside analogues: mechanisms of drug resistance and reversal strategies. Leukemia 2001, 15, 875–890. [Google Scholar] [CrossRef] [PubMed]
- Gambini, L., Baggio, C., Udompholkul, P., Jossart, J., Salem, A.F., Perry, J.J.P. and Pellecchia, M., 2019. Covalent inhibitors of protein–protein interactions targeting lysine, tyrosine, or histidine residues. Journal of medicinal chemistry, 62(11), pp.5616-5627.
- Gao, J.; Wang, C.; Wei, W. The effects of drug transporters on the efficacy of methotrexate in the treatment of rheumatoid arthritis. Life Sci. 2021, 268, 118907. [Google Scholar] [CrossRef]
- Gehringer, M.; Laufer, S.A. Emerging and Re-Emerging Warheads for Targeted Covalent Inhibitors: Applications in Medicinal Chemistry and Chemical Biology. J. Med. Chem. 2018, 62, 5673–5724. [Google Scholar] [CrossRef] [PubMed]
- Gerhart, S.V.; Kellner, W.A.; Thompson, C.; Pappalardi, M.B.; Zhang, X.-P.; de Oca, R.M.; Penebre, E.; Duncan, K.; Boriack-Sjodin, A.; Le, B.; et al. Activation of the p53-MDM4 regulatory axis defines the anti-tumour response to PRMT5 inhibition through its role in regulating cellular splicing. Sci. Rep. 2018, 8, 1–15. [Google Scholar] [CrossRef]
- Gibaja, V.; Shen, F.; Harari, J.; Korn, J.; Ruddy, D.; Saenz-Vash, V.; Zhai, H.; Rejtar, T.; Paris, C.G.; Yu, Z.; et al. Development of secondary mutations in wild-type and mutant EZH2 alleles cooperates to confer resistance to EZH2 inhibitors. Oncogene 2015, 35, 558–566. [Google Scholar] [CrossRef]
- Gradl, S.; Steuber, H.; Weiske, J.; Schmees, N.; Siegel, S.; Stoeckigt, D.; Christ, C.D.; Li, F.; Organ, S.; Barsyte-Lovejoy, D.; et al. Abstract 1646: Discovery and characterization of BAY-6035, a novel benzodiazepine-based SMYD3 inhibitor. Cancer Res. 2018, 78, 1646–1646. [Google Scholar] [CrossRef]
- Grebien, F.; Vedadi, M.; Getlik, M.; Giambruno, R.; Grover, A.; Avellino, R.; Skucha, A.; Vittori, S.; Kuznetsova, E.; Smil, D.; et al. Pharmacological targeting of the Wdr5-MLL interaction in C/EBPα N-terminal leukemia. Nat. Chem. Biol. 2015, 11, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Li, Y.; Zhou, Z.; Hou, L.; Liu, W.; Ren, W.; Mi, D.; Sun, J.; Dai, X.; Wu, Y.; et al. Targeting PRMT5 through PROTAC for the treatment of triple-negative breast cancer. J. Exp. Clin. Cancer Res. 2024, 43, 1–15. [Google Scholar] [CrossRef]
- H. Nguyen, A. Allali-Hassani, S. Antonysamy, S. Chang, L. H. Chen, C. Curtis, S. Emtage, L. Fan, T. Gheyi, F. Li, S. Liu, J. R. Martin, D. Mendel, J. B. Olsen, L. Pelletier, T. Shatseva, S. Wu, F. F. Zhang, C. H. Arrowsmith, P. J. Brown, R. M. Campbell, B. A. Garcia, D. Barsyte-Lovejoy, M. Mader and M. Vedadi, J. Biol. Chem., 2015, 290, 13641–13653.
- Halby, L.; Marechal, N.; Pechalrieu, D.; Cura, V.; Franchini, D.-M.; Faux, C.; Alby, F.; Troffer-Charlier, N.; Kudithipudi, S.; Jeltsch, A.; et al. Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors. Philos. Trans. R. Soc. B: Biol. Sci. 2018, 373, 20170072. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Huang, Y.; Qiu, M.; Yin, C.; Ren, H.; Gan, H.; Li, H.; Zhou, Y.; Xia, J.; Li, W.; et al. Immunoassay of S-adenosylmethionine and S-adenosylhomocysteine: the methylation index as a biomarker for disease and health status. BMC Res. Notes 2016, 9, 498. [Google Scholar] [CrossRef]
- He Y et al. 2017 The EED protein–protein interaction inhibitor A-395 inactivates the PRC2 complex. Nat. Chem. Biol. 13, 389–395. [CrossRef]
- He, H.; Li, X.; Su, F.; Jin, H.; Zhang, J.; Wang, Y. Current and Emerging Approaches Targeting G9a for the Treatment of Various Diseases. J. Med. Chem. 2024, 68, 1068–1089. [Google Scholar] [CrossRef]
- He, Y.; Selvaraju, S.; Curtin, M.L.; Jakob, C.G.; Zhu, H.; Comess, K.M.; Shaw, B.; The, J.; Lima-Fernandes, E.; Szewczyk, M.M.; et al. The EED protein–protein interaction inhibitor A-395 inactivates the PRC2 complex. Nat. Chem. Biol. 2017, 13, 389–395. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Selvaraju, S.; Curtin, M.L.; Jakob, C.G.; Zhu, H.; Comess, K.M.; Shaw, B.; The, J.; Lima-Fernandes, E.; Szewczyk, M.M.; et al. The EED protein–protein interaction inhibitor A-395 inactivates the PRC2 complex. Nat. Chem. Biol. 2017, 13, 389–395. [Google Scholar] [CrossRef]
- Hintzen, J.C.J.; Moesgaard, L.; Kwiatkowski, S.; Drozak, J.; Kongsted, J.; Mecinović, J. β-Actin Peptide-Based Inhibitors of Histidine Methyltransferase SETD3. Chem. Med. Chem. 2021, 16, 2695–2702. [Google Scholar] [CrossRef]
- Ho, M.-C.; Wilczek, C.; Bonanno, J.B.; Xing, L.; Seznec, J.; Matsui, T.; Carter, L.G.; Onikubo, T.; Kumar, P.R.; Chan, M.K.; et al. Structure of the Arginine Methyltransferase PRMT5-MEP50 Reveals a Mechanism for Substrate Specificity. PLoS ONE 2013, 8, e57008. [Google Scholar] [CrossRef]
- Hsu, H.; Chen, M.; Baskaran, R.; Lin, Y.; Day, C.H.; Lin, Y.; Tu, C.; Padma, V.V.; Kuo, W.; Huang, C. Oxaliplatin resistance in colorectal cancer cells is mediated via activation of ABCG2 to alleviate ER stress induced apoptosis. J. Cell. Physiol. 2017, 233, 5458–5467. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.H.-R.; Rasmusson, T.; Robinson, J.; Pachl, F.; Read, J.; Kawatkar, S.; O'Donovan, D.H.; Bagal, S.; Code, E.; Rawlins, P.; et al. EED-Targeted PROTACs Degrade EED, EZH2, and SUZ12 in the PRC2 Complex. Cell Chem. Biol. 2020, 27, 41–46.e17. [Google Scholar] [CrossRef]
- Huang, X.; Yan, J.; Zhang, M.; Wang, Y.; Chen, Y.; Fu, X.; Wei, R.; Zheng, X.-L.; Liu, Z.; Zhang, X.; et al. Targeting Epigenetic Crosstalk as a Therapeutic Strategy for EZH2-Aberrant Solid Tumors. Cell 2018, 175, 186–199.e19. [Google Scholar] [CrossRef] [PubMed]
- Hui, R.C.-Y.; Francis, R.E.; Guest, S.K.; Costa, J.R.; Gomes, A.R.; Myatt, S.S.; Brosens, J.J.; Lam, E.W.-F. Doxorubicin activates FOXO3a to induce the expression of multidrug resistance gene ABCB1 (MDR1) in K562 leukemic cells. Mol. Cancer Ther. 2008, 7, 670–678. [Google Scholar] [CrossRef]
- Ju, Y.; Song, H.; He, Y.; Lo, Y.; Fan, Z.; Lu, J. Development of a Selective and Potent PRMT4 PROTAC Degrader with Efficacy against Multiple Myeloma in Vitro and in Vivo. J. Med. Chem. 2025, 68, 13973–13989. [Google Scholar] [CrossRef]
- Ju, Y.; Song, H.; He, Y.; Lo, Y.; Fan, Z.; Lu, J. Development of a Selective and Potent PRMT4 PROTAC Degrader with Efficacy against Multiple Myeloma in Vitro and in Vivo. J. Med. Chem. 2025, 68, 13973–13989. [Google Scholar] [CrossRef]
- K. V. Butler, A. Ma, W. Yu, F. Li, W. Tempel, N. Babault, F. Pittella-Silva, J. Shao, J. Wang, M. Luo, M. Vedadi, P. J. Brown, C. H. Arrowsmith and J. Jin, J. Med. Chem., 2016, 59, 9881–9889.
- Kaniskan, H.Ü.; Martini, M.L.; Jin, J. Inhibitors of Protein Methyltransferases and Demethylases. Chem. Rev. 2017, 118, 989–1068. [Google Scholar] [CrossRef]
- Kaniskan, H.Ü.; Szewczyk, M.M.; Yu, Z.; Eram, M.S.; Yang, X.; Schmidt, K.; Luo, X.; Dai, M.; He, F.; Zang, I.; et al. A potent, selective and cell-active allosteric inhibitor of protein arginine methyltransferase 3 (PRMT3). Angew. Chem. Int. Ed. 2015, 54, 5166–70. [Google Scholar] [CrossRef]
- Kannampuzha, S.; Gopalakrishnan, A.V. Cancer chemoresistance and its mechanisms: Associated molecular factors and its regulatory role. Med Oncol. 2023, 40, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kazansky, Y.; Cameron, D.; Mueller, H.S.; Demarest, P.; Zaffaroni, N.; Arrighetti, N.; Zuco, V.; Kuwahara, Y.; Somwar, R.; Ladanyi, M.; et al. Overcoming Clinical Resistance to EZH2 Inhibition Using Rational Epigenetic Combination Therapy. Cancer Discov. 2024, 14, 965–981. [Google Scholar] [CrossRef]
- Khan, S.U.; Fatima, K.; Aisha, S.; Malik, F. Unveiling the mechanisms and challenges of cancer drug resistance. Cell Commun. Signal. 2024, 22, 1–26. [Google Scholar] [CrossRef]
- Kim, W.; Bird, G.H.; Neff, T.; Guo, G.; A Kerenyi, M.; Walensky, L.D.; Orkin, S.H. Targeted disruption of the EZH2–EED complex inhibits EZH2-dependent cancer. Nat. Chem. Biol. 2013, 9, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Kim, Y.J.; Lee, H.I.; Jeong, S.-H.; Nam, H.J.; Cho, J.H. NRF2 Knockdown Resensitizes 5-Fluorouracil-Resistant Pancreatic Cancer Cells by Suppressing HO-1 and ABCG2 Expression. Int. J. Mol. Sci. 2020, 21, 4646. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Bird, G.H.; Neff, T.; Guo, G.; A Kerenyi, M.; Walensky, L.D.; Orkin, S.H. Targeted disruption of the EZH2–EED complex inhibits EZH2-dependent cancer. Nat. Chem. Biol. 2013, 9, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Knuhtsen, A.; Legrand, B.; Van der Poorten, O.; Amblard, M.; Martinez, J.; Ballet, S.; Kristensen, J.L.; Pedersen, D.S. Conformationally Constrained Peptidomimetics as Inhibitors of the Protein Arginine Methyl Transferases. Chem. – A Eur. J. 2016, 22, 14022–14028. [Google Scholar] [CrossRef]
- Kobayashi, S., Boggon, T.J., Dayaram, T., Jänne, P.A., Kocher, O., Meyerson, M., Johnson, B.E., Eck, M.J., Tenen, D.G. and Halmos, B., 2005. EGFR mutation and resistance of non–small-cell lung cancer to gefitinib. New England Journal of Medicine, 352(8), pp.786-792.
- Konze, K.D.; Ma, A.; Li, F.; Barsyte-Lovejoy, D.; Parton, T.; MacNevin, C.J.; Liu, F.; Gao, C.; Huang, X.-P.; Kuznetsova, E.; et al. An Orally Bioavailable Chemical Probe of the Lysine Methyltransferases EZH2 and EZH1. ACS Chem. Biol. 2013, 8, 1324–1334. [Google Scholar] [CrossRef]
- Konze, K.D.; Pattenden, S.G.; Liu, F.; Barsyte-Lovejoy, D.; Li, F.; Simon, J.M.; Davis, I.J.; Vedadi, M.; Jin, J. A Chemical Tool for In Vitro and In Vivo Precipitation of Lysine Methyltransferase G9a. ChemMedChem 2014, 9, 549–553. [Google Scholar] [CrossRef]
- Kozbial, P.Z.; Mushegian, A.R. Natural history of S-adenosylmethionine-binding proteins. BMC Struct. Biol. 2005, 5, 19–19. [Google Scholar] [CrossRef]
- Kozbial, P.Z.; Mushegian, A.R. Natural history of S-adenosylmethionine-binding proteins. BMC Struct. Biol. 2005, 5, 19–19. [Google Scholar] [CrossRef]
- Kryukov, G.V.; Wilson, F.H.; Ruth, J.R.; Paulk, J.; Tsherniak, A.; Marlow, S.E.; Vazquez, F.; Weir, B.A.; Fitzgerald, M.E.; Tanaka, M.; et al. MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science 2016, 351, 1214–1218. [Google Scholar] [CrossRef]
- Krzyzanowski, A., Esser, L.M., Willaume, A., Prudent, R., Peter, C., ‘t Hart, P. and Waldmann, H., 2022. Development of macrocyclic PRMT5–adaptor protein interaction inhibitors. Journal of Medicinal Chemistry, 65(22), pp.15300-15311.
- Kung, P.P., Bingham, P., Brooun, A., Collins, M., Deng, Y.L., Dinh, D., Fan, C., Gajiwala, K.S., Grantner, R., Gukasyan, H.J. and Hu, W., 2018. Optimization of orally bioavailable enhancer of zeste homolog 2 (EZH2) inhibitors using ligand and property-based design strategies: identification of development candidate (R)-5, 8-Dichloro-7-(methoxy (oxetan-3-yl) methyl)-2-((4-methoxy-6-methyl-2-oxo-1, 2-dihydropyridin-3-yl) methyl)-3, 4-dihydroisoquinolin-1 (2 H)-one (PF-06821497).
- LegaardAndersson, J.; Christensen, J.; Kleine-Kohlbrecher, D.; Comet, I.V.; Støier, J.F.; Antoku, Y.; Poljak, V.; Moretti, L.; Dolberg, J.; Jacso, T.; et al. Discovery of NSD2-Degraders from Novel and Selective DEL Hits. ChemBioChem 2023, 24, e202300515. [Google Scholar] [CrossRef] [PubMed]
- Levy, D. Lysine methylation signaling of non-histone proteins in the nucleus. Cell. Mol. Life Sci. 2019, 76, 2873–2883. [Google Scholar] [CrossRef] [PubMed]
- Li, A., Song, J., Lai, Q., Liu, B., Wang, H., Xu, Y., Feng, X., Sun, X. and Du, Z., 2016. Hypermethylation of ATP-binding cassette B1 (ABCB 1) multidrug resistance 1 (MDR 1) is associated with cisplatin resistance in the A549 lung adenocarcinoma cell line. International journal of experimental pathology, 97(6), pp.412-421.
- Li, B.; Rong, D.; Wang, Y. Targeting Protein-Protein Interaction with Covalent Small-Molecule Inhibitors. Curr. Top. Med. Chem. 2019, 19, 1872–1876. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Yu, X.; Kottur, J.; Gong, W.; Zhang, Z.; Storey, A.J.; Tsai, Y.-H.; Uryu, H.; Shen, Y.; Byrum, S.D.; et al. Discovery of a dual WDR5 and Ikaros PROTAC degrader as an anti-cancer therapeutic. Oncogene 2022, 41, 3328–3340. [Google Scholar] [CrossRef]
- Li, L.-Y.; Guan, Y.-D.; Chen, X.-S.; Yang, J.-M.; Cheng, Y. DNA Repair Pathways in Cancer Therapy and Resistance. Front. Pharmacol. 2021, 11. [Google Scholar] [CrossRef]
- Lin, H.; Luengo, J.I. Nucleoside protein arginine methyltransferase 5 (PRMT5) inhibitors. Bioorganic Med. Chem. Lett. 2019, 29, 1264–1269. [Google Scholar] [CrossRef]
- Lin, H.; Wang, B.; Yu, J.; Wang, J.; Li, Q.; Cao, B. Protein arginine methyltransferase 8 gene enhances the colon cancer stem cell (CSC) function by upregulating the pluripotency transcription factor. J. Cancer 2018, 9, 1394–1402. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Wang, M.; Zhang, Y.W.; Tong, S.; Leal, R.A.; Shetty, R.; Vaddi, K.; Luengo, J.I. Discovery of Potent and Selective Covalent Protein Arginine Methyltransferase 5 (PRMT5) Inhibitors. ACS Med. Chem. Lett. 2019, 10, 1033–1038. [Google Scholar] [CrossRef]
- Liu, F.; Barsyte-Lovejoy, D.; Li, F.; Xiong, Y.; Korboukh, V.; Huang, X.-P.; Allali-Hassani, A.; Janzen, W.P.; Roth, B.L.; Frye, S.V.; et al. Discovery of an in Vivo Chemical Probe of the Lysine Methyltransferases G9a and GLP. J. Med. Chem. 2013, 56, 8931–8942. [Google Scholar] [CrossRef]
- Liu, K.-L.; Zhu, K.; Zhang, H. An overview of the development of EED inhibitors to disable the PRC2 function. RSC Med. Chem. 2021, 13, 39–53. [Google Scholar] [CrossRef]
- Liu, L.; Parolia, A.; Liu, Y.; Hou, C.; He, T.; Qiao, Y.; Eyunni, S.; Luo, J.; Li, C.; Wang, Y.; et al. Discovery of LLC0424 as a Potent and Selective in Vivo NSD2 PROTAC Degrader. J. Med. Chem. 2024, 67, 6938–6951. [Google Scholar] [CrossRef]
- Liu, Z.; Hu, X.; Wang, Q.; Wu, X.; Zhang, Q.; Wei, W.; Su, X.; He, H.; Zhou, S.; Hu, R.; et al. Design and Synthesis of EZH2-Based PROTACs to Degrade the PRC2 Complex for Targeting the Noncatalytic Activity of EZH2. J. Med. Chem. 2021, 64, 2829–2848. [Google Scholar] [CrossRef]
- Loenen, W. S-Adenosylmethionine: jack of all trades and master of everything? Biochem. Soc. Trans. 2006, 34, 330–333. [Google Scholar] [CrossRef]
- Lu, B.; Shen, X.; Zhang, L.; Liu, D.; Zhang, C.; Cao, J.; Shen, R.; Zhang, J.; Wang, D.; Wan, H.; et al. Discovery of EBI-2511: A Highly Potent and Orally Active EZH2 Inhibitor for the Treatment of Non-Hodgkin’s Lymphoma. ACS Med. Chem. Lett. 2018, 9, 98–102. [Google Scholar] [CrossRef]
- Luo, M. Current Chemical Biology Approaches to Interrogate Protein Methyltransferases. ACS Chem. Biol. 2012, 7, 443–463. [Google Scholar] [CrossRef]
- M. J. Thomenius, J. Totman, D. Harvey, L. H. Mitchell, T. V. Riera, K. Cosmopoulos, A. R. Grassian, C. Klaus, M. Foley, E. A. Admirand, H. Jahic, C. Majer, T. Wigle, S. L. Jacques, J. Gureasko, D. Brach, T. Lingaraj, K. West, S. Smith, N. Rioux, N. J. Waters, C. Tang, A. Raimondi, M. Munchhof, J. E. Mills, S. Ribich, M. Porter Scott, K. W. Kuntz, W. P. Janzen, M. Moyer, J. J. Smith, R. Chesworth, R. A. Copeland and P. A. Boriack-Sjodin, PLoS One, 2018, 13, e0197372.
- Ma, A.; Stratikopoulos, E.; Park, K.-S.; Wei, J.; Martin, T.C.; Yang, X.; Schwarz, M.; Leshchenko, V.; Rialdi, A.; Dale, B.; et al. Discovery of a first-in-class EZH2 selective degrader. Nat. Chem. Biol. 2019, 16, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Ma, A.; Yu, W.; Li, F.; Bleich, R.M.; Herold, J.M.; Butler, K.V.; Norris, J.L.; Korboukh, V.; Tripathy, A.; Janzen, W.P.; et al. Discovery of a Selective, Substrate-Competitive Inhibitor of the Lysine Methyltransferase SETD8. J. Med. Chem. 2014, 57, 6822–6833. [Google Scholar] [CrossRef] [PubMed]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef]
- Martin, P.L.; Pérez-Areales, F.J.; Rao, S.V.; Walsh, S.J.; Carroll, J.S.; Spring, D.R. Towards the Targeted Protein Degradation of PRMT1. ChemMedChem 2024, 19, e202400269. [Google Scholar] [CrossRef] [PubMed]
- Marx, V. Inside the chase after those elusive proteoforms. Nat. Methods 2024, 21, 158–163. [Google Scholar] [CrossRef]
- Mato, J.M.; Corrales, F.J.; Lu, S.C.; Avila, M.A. S-Adenosylmethionine: a control switch that regulates liver function. FASEB J. 2002, 16, 15–26. [Google Scholar] [CrossRef]
- Meng, F.; Xu, C.; Park, K.-S.; Kaniskan, H.Ü.; Wang, G.G.; Jin, J. Discovery of a First-in-Class Degrader for Nuclear Receptor Binding SET Domain Protein 2 (NSD2) and Ikaros/Aiolos. J. Med. Chem. 2022, 65, 10611–10625. [Google Scholar] [CrossRef]
- Micallef, I. and Baron, B., 2023. Proteomic strategies for methylation analysis in colorectal cancer chemoresistance. Journal of Proteome Data and Methods, 5, p.16.
- Micallef, I.; Fenech, K.; Baron, B. Therapeutic targeting potential of the protein lysine and arginine methyltransferases to reverse cancer chemoresistance. Front. Mol. Biosci. 2024, 11, 1455415. [Google Scholar] [CrossRef]
- Mirzaei, S.; Gholami, M.H.; Hashemi, F.; Zabolian, A.; Farahani, M.V.; Hushmandi, K.; Zarrabi, A.; Goldman, A.; Ashrafizadeh, M.; Orive, G. Advances in understanding the role of P-gp in doxorubicin resistance: Molecular pathways, therapeutic strategies, and prospects. Drug Discov. Today 2022, 27, 436–455. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, L.H.; Boriack-Sjodin, P.A.; Smith, S.; Thomenius, M.; Rioux, N.; Munchhof, M.; Mills, J.E.; Klaus, C.; Totman, J.; Riera, T.V.; et al. Novel Oxindole Sulfonamides and Sulfamides: EPZ031686, the First Orally Bioavailable Small Molecule SMYD3 Inhibitor. ACS Med. Chem. Lett. 2015, 7, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Mondal, P.; Meeran, S.M. microRNAs in cancer chemoresistance: The sword and the shield. Non-coding RNA Res. 2021, 6, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Mori, S., Iwase, K., Iwanami, N., Tanaka, Y., Kagechika, H. and Hirano, T., 2010. Development of novel bisubstrate-type inhibitors of histone methyltransferase SET7/9. Bioorganic & medicinal chemistry, 18(23), pp.8158-8166.
- Mosca, L.; Minopoli, M.; Pagano, M.; Vitiello, F.; Carriero, M.V.; Cacciapuoti, G.; Porcelli, M. Effects of S-adenosyl-L-methionine on the invasion and migration of head and neck squamous cancer cells and analysis of the underlying mechanisms. Int. J. Oncol. 2020, 56, 1212–1224. [Google Scholar] [CrossRef]
- Mosca, L.; Pagano, M.; Borzacchiello, L.; Mele, L.; Russo, A.; Russo, G.; Cacciapuoti, G.; Porcelli, M. S-Adenosylmethionine Increases the Sensitivity of Human Colorectal Cancer Cells to 5-Fluorouracil by Inhibiting P-Glycoprotein Expression and NF-κB Activation. Int. J. Mol. Sci. 2021, 22, 9286. [Google Scholar] [CrossRef]
- Mukherjee, A., 2025. Design, Synthesis, and Biological Evaluation of (Doctoral dissertation).
- Mulvaney, K.M.; Blomquist, C.; Acharya, N.; Li, R.; Ranaghan, M.J.; O’kEefe, M.; Rodriguez, D.J.; Young, M.J.; Kesar, D.; Pal, D.; et al. Molecular basis for substrate recruitment to the PRMT5 methylosome. Mol. Cell 2021, 81, 3481–3495.e7. [Google Scholar] [CrossRef]
- Naryzhny, S., 2024. Puzzle of proteoform variety—where is a key?. Proteomes, 12(2), p.15.
- Nazki, F.H.; Sameer, A.S.; Ganaie, B.A. Folate: Metabolism, genes, polymorphisms and the associated diseases. Gene 2014, 533, 11–20. [Google Scholar] [CrossRef]
- Ning, J.; Chen, L.; Xiao, G.; Zeng, Y.; Shi, W.; Tanzhu, G.; Zhou, R. The protein arginine methyltransferase family (PRMTs) regulates metastases in various tumors: From experimental study to clinical application. Biomed. Pharmacother. 2023, 167, 115456. [Google Scholar] [CrossRef]
- Pan, S.; Li, Z.; He, Z.; Qiu, J.; Zhou, S. Molecular mechanisms for tumour resistance to chemotherapy. Clin. Exp. Pharmacol. Physiol. 2016, 43, 723–737. [Google Scholar] [CrossRef]
- Pang, F.; Zhang, L.; Li, M.; Yi, X.; Wang, Y.; Yang, P.; Wen, B.; Jiang, J.; Teng, Y.; Yang, X.; et al. Ribosomal S6 protein kinase 4 promotes resistance to EZH2 inhibitors in glioblastoma. Cancer Gene Ther. 2023, 30, 1636–1648. [Google Scholar] [CrossRef]
- Park, K.-S.; Xiong, Y.; Yim, H.; Velez, J.; Babault, N.; Kumar, P.; Liu, J.; Jin, J. Discovery of the First-in-Class G9a/GLP Covalent Inhibitors. J. Med. Chem. 2022, 65, 10506–10522. [Google Scholar] [CrossRef]
- Pascale, R.M.; Simile, M.M.; Calvisi, D.F.; Feo, C.F.; Feo, F. S-Adenosylmethionine: From the Discovery of Its Inhibition of Tumorigenesis to Its Use as a Therapeutic Agent. Cells 2022, 11, 409. [Google Scholar] [CrossRef]
- Peserico, A.; Germani, A.; Sanese, P.; Barbosa, A.J.; Di Virgilio, V.; Fittipaldi, R.; Fabini, E.; Bertucci, C.; Varchi, G.; Moyer, M.P.; et al. A SMYD3 Small-Molecule Inhibitor Impairing Cancer Cell Growth. J. Cell. Physiol. 2015, 230, 2447–2460. [Google Scholar] [CrossRef]
- Pljesa-Ercegovac, M.; Savic-Radojevic, A.; Matic, M.; Coric, V.; Djukic, T.; Radic, T.; Simic, T. Glutathione Transferases: Potential Targets to Overcome Chemoresistance in Solid Tumors. Int. J. Mol. Sci. 2018, 19, 3785. [Google Scholar] [CrossRef] [PubMed]
- Potjewyd, F.; Turner, A.-M.W.; Beri, J.; Rectenwald, J.M.; Norris-Drouin, J.L.; Cholensky, S.H.; Margolis, D.M.; Pearce, K.H.; Herring, L.E.; James, L.I. Degradation of Polycomb Repressive Complex 2 with an EED-Targeted Bivalent Chemical Degrader. Cell Chem. Biol. 2020, 27, 47–56.e15. [Google Scholar] [CrossRef] [PubMed]
- Powers, J.C., Asgian, J.L., Ekici, Ö.D. and James, K.E., 2002. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chemical reviews, 102(12), pp.4639-4750.
- Qi, W.; Zhao, K.; Gu, J.; Huang, Y.; Wang, Y.; Zhang, H.; Zhang, M.; Zhang, J.; Yu, Z.; Li, L.; et al. An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of EED. Nat. Chem. Biol. 2017, 13, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Zhao, K.; Gu, J.; Huang, Y.; Wang, Y.; Zhang, H.; Zhang, M.; Zhang, J.; Yu, Z.; Li, L.; et al. An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of EED. Nat. Chem. Biol. 2017, 13, 381–388. [Google Scholar] [CrossRef]
- Rej, R.K.; Wang, C.; Lu, J.; Wang, M.; Petrunak, E.; Zawacki, K.P.; McEachern, D.; Fernandez-Salas, E.; Yang, C.-Y.; Wang, L.; et al. EEDi-5285: An Exceptionally Potent, Efficacious, and Orally Active Small-Molecule Inhibitor of Embryonic Ectoderm Development. J. Med. Chem. 2020, 63, 7252–7267. [Google Scholar] [CrossRef]
- Roh, Y.-G.; Mun, M.-H.; Jeong, M.-S.; Kim, W.-T.; Lee, S.-R.; Chung, J.-W.; Kim, S.I.; Kim, T.N.; Kil Nam, J.; Leem, S.-H. Drug resistance of bladder cancer cells through activation of ABCG2 by FOXM1. BMB Rep. 2018, 51, 98–103. [Google Scholar] [CrossRef]
- Rossi, A.; Zacchi, F.; Reni, A.; Rota, M.; Palmerio, S.; Menis, J.; Zivi, A.; Milleri, S.; Milella, M. Progresses and Pitfalls of Epigenetics in Solid Tumors Clinical Trials. Int. J. Mol. Sci. 2024, 25, 11740. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.M.; Kim, K.B.; Kumagai, A.; Mercurio, F.; Crews, C.M.; Deshaies, R.J. Protacs: Chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. USA 2001, 98, 8554–8559. [Google Scholar] [CrossRef] [PubMed]
- Sakthivel, K.M.; Hariharan, S. Regulatory players of DNA damage repair mechanisms: Role in Cancer Chemoresistance. Biomed. Pharmacother. 2017, 93, 1238–1245. [Google Scholar] [CrossRef] [PubMed]
- Schapira, M. and de Freitas, R.F., 2014. Structural biology and chemistry of protein arginine methyltransferases. Medchemcomm, 5(12), pp.1779-1788.
- Shailesh, H.; Siveen, K.S.; Sif, S. Protein arginine methyltransferase 5 (PRMT5) activates WNT/β-catenin signalling in breast cancer cells via epigenetic silencing of DKK1 and DKK3. J. Cell. Mol. Med. 2021, 25, 1583–1600. [Google Scholar] [CrossRef]
- Shen, Y.; Gao, G.; Yu, X.; Kim, H.S.; Wang, L.; Xie, L.; Schwarz, M.; Chen, X.; Guccione, E.; Liu, J.; et al. Discovery of First-in-Class Protein Arginine Methyltransferase 5 (PRMT5) Degraders. J. Med. Chem. 2020, 63, 9977–9989. [Google Scholar] [CrossRef]
- Shen, Y.; Li, F.; Szewczyk, M.M.; Halabelian, L.; Park, K.-S.; Chau, I.; Dong, A.; Zeng, H.; Chen, H.; Meng, F.; et al. Discovery of a First-in-Class Protein Arginine Methyltransferase 6 (PRMT6) Covalent Inhibitor. J. Med. Chem. 2020, 63, 5477–5487. [Google Scholar] [CrossRef]
- Shi, Y.; Shen, Q.; Long, R.; Mao, Y.; Tong, S.; Yang, Y.; Gao, J.; Zhou, H.; Chen, Y.; Zhou, B. Discovery of Potent and Selective G9a Degraders for the Treatment of Pancreatic Cancer. J. Med. Chem. 2024, 67, 13271–13285. [Google Scholar] [CrossRef]
- Shui, X., Tian, L., Zhou, Y. and Zhao, B., 2024. Targeting ACSS2 activity suspends the formation of chemoresistance through suppressed histone H3 acetylation in human breast cancer.
- Smith, L.M.; Kelleher, N.L. Proteoforms as the next proteomics currency. Science 2018, 359, 1106–1107. [Google Scholar] [CrossRef] [PubMed]
- Spitzwieser, M.; Pirker, C.; Koblmüller, B.; Pfeiler, G.; Hacker, S.; Berger, W.; Heffeter, P.; Cichna-Markl, M. Promoter methylation patterns of ABCB1, ABCC1 and ABCG2 in human cancer cell lines, multidrug-resistant cell models and tumor, tumor-adjacent and tumor-distant tissues from breast cancer patients. Oncotarget 2016, 7, 73347–73369. [Google Scholar] [CrossRef] [PubMed]
- Sumarpo, A.; Ito, K.; Saiki, Y.; Ishizawa, K.; Wang, R.; Chen, N.; Sunamura, M.; Horii, A. Genetic and epigenetic aberrations of ABCB1 synergistically boost the acquisition of taxane resistance in esophageal squamous cancer cells. Biochem. Biophys. Res. Commun. 2020, 526, 586–591. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, Y.; Chen, X.; Yu, A.; Du, W.; Huang, Y.; Wu, F.; Yu, L.; Li, J.; Wen, C.; et al. Discovery of a potent and selective proteolysis targeting chimera (PROTAC) degrader of NSD3 histone methyltransferase. Eur. J. Med. Chem. 2022, 239, 114528. [Google Scholar] [CrossRef]
- Sweis, R.F.; Pliushchev, M.; Brown, P.J.; Guo, J.; Li, F.; Maag, D.; Petros, A.M.; Soni, N.B.; Tse, C.; Vedadi, M.; et al. Discovery and Development of Potent and Selective Inhibitors of Histone Methyltransferase G9a. ACS Med. Chem. Lett. 2014, 5, 205–209. [Google Scholar] [CrossRef]
- Sweis, R.F.; Wang, Z.; Algire, M.; Arrowsmith, C.H.; Brown, P.J.; Chiang, G.G.; Guo, J.; Jakob, C.G.; Kennedy, S.; Li, F.; et al. Discovery of A-893, A New Cell-Active Benzoxazinone Inhibitor of Lysine Methyltransferase SMYD2. ACS Med. Chem. Lett. 2015, 6, 695–700. [Google Scholar] [CrossRef]
- Takemoto, Y.; Ito, A.; Niwa, H.; Okamura, M.; Fujiwara, T.; Hirano, T.; Handa, N.; Umehara, T.; Sonoda, T.; Ogawa, K.; et al. Identification of Cyproheptadine as an Inhibitor of SET Domain Containing Lysine Methyltransferase 7/9 (Set7/9) That Regulates Estrogen-Dependent Transcription. J. Med. Chem. 2016, 59, 3650–3660. [Google Scholar] [CrossRef]
- Talibov, V.O.; Fabini, E.; FitzGerald, E.A.; Tedesco, D.; Cederfelt, D.; Talu, M.J.; Rachman, M.M.; Mihalic, F.; Manoni, E.; Naldi, M.; et al. Discovery of an Allosteric Ligand Binding Site in SMYD3 Lysine Methyltransferase. ChemBioChem 2021, 22, 1597–1608. [Google Scholar] [CrossRef] [PubMed]
- Talukdar, A.; Mukherjee, A.; Bhattacharya, D. Fascinating Transformation of SAM-Competitive Protein Methyltransferase Inhibitors from Nucleoside Analogues to Non-Nucleoside Analogues. J. Med. Chem. 2022, 65, 1662–1684. [Google Scholar] [CrossRef] [PubMed]
- Talukdar, A.; Mukherjee, A.; Bhattacharya, D. Fascinating Transformation of SAM-Competitive Protein Methyltransferase Inhibitors from Nucleoside Analogues to Non-Nucleoside Analogues. J. Med. Chem. 2022, 65, 1662–1684. [Google Scholar] [CrossRef]
- Taylor, A.P., Swewczyk, M., Kennedy, S., Trush, V.V., Wu, H., Zeng, H., Dong, A., Ferreira de Freitas, R., Tatlock, J., Kumpf, R.A. and Wythes, M., 2019. Selective, small-molecule co-factor binding site inhibition of a Su (var) 3–9, enhancer of zeste, trithorax domain containing lysine methyltransferase. Journal of medicinal chemistry, 62(17), pp.7669-7683.
- Tong, C.; Chang, X.; Qu, F.; Bian, J.; Wang, J.; Li, Z.; Xu, X. Overview of the development of protein arginine methyltransferase modulators: Achievements and future directions. Eur. J. Med. Chem. 2024, 267, 116212. [Google Scholar] [CrossRef]
- Tu, Y.; Sun, Y.; Qiao, S.; Luo, Y.; Liu, P.; Jiang, Z.-X.; Hu, Y.; Wang, Z.; Huang, P.; Wen, S. Design, Synthesis, and Evaluation of VHL-Based EZH2 Degraders to Enhance Therapeutic Activity against Lymphoma. J. Med. Chem. 2021, 64, 10167–10184. [Google Scholar] [CrossRef] [PubMed]
- Vaidyanathan, A.; Sawers, L.; Gannon, A.-L.; Chakravarty, P.; Scott, A.L.; E Bray, S.; Ferguson, M.J.; Smith, G. ABCB1 (MDR1) induction defines a common resistance mechanism in paclitaxel- and olaparib-resistant ovarian cancer cells. Br. J. Cancer 2016, 115, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, Rishi G., Victor S. Gehling, Les A. Dakin, Andrew S. Cook, Christopher G. Nasveschuk, Martin Duplessis, Priyadarshini Iyer et al. "Identification of (R)-N-((4-Methoxy-6-methyl-2-oxo-1, 2-dihydropyridin-3-yl) methyl)-2-methyl-1-(1-(1-(2, 2, 2-trifluoroethyl) piperidin-4-yl) ethyl)-1 H-indole-3-carboxamide (CPI-1205), a Potent and Selective Inhibitor of Histone Methyltransferase EZH2, Suitable for Phase I Clinical Trials for B-Cell Lymphomas." Journal of medicinal chemistry 59, no. 21 (2016): 9928-9941.
- Velez, J.; Dale, B.; Park, K.-S.; Kaniskan, H.Ü.; Yu, X.; Jin, J. Discovery of a novel, highly potent EZH2 PROTAC degrader for targeting non-canonical oncogenic functions of EZH2. Eur. J. Med. Chem. 2024, 267, 116154–116154. [Google Scholar] [CrossRef]
- Velez, J.; Han, Y.; Yim, H.; Yang, P.; Deng, Z.; Park, K.-S.; Kabir, M.; Kaniskan, H.Ü.; Xiong, Y.; Jin, J. Discovery of the First-in-Class G9a/GLP PROTAC Degrader. J. Med. Chem. 2024, 67, 6397–6409. [Google Scholar] [CrossRef] [PubMed]
- Vougiouklakis, T.; Bernard, B.J.; Nigam, N.; Burkitt, K.; Nakamura, Y.; Saloura, V. Clinicopathologic significance of protein lysine methyltransferases in cancer. Clin. Epigenetics 2020, 12, 1–19. [Google Scholar] [CrossRef]
- Wang, C.; Chen, X.; Liu, X.; Lu, D.; Li, S.; Qu, L.; Yin, F.; Luo, H.; Zhang, Y.; Luo, Z.; et al. Discovery of precision targeting EZH2 degraders for triple-negative breast cancer. Eur. J. Med. Chem. 2022, 238, 114462. [Google Scholar] [CrossRef]
- Wang, J.; Yu, X.; Gong, W.; Liu, X.; Park, K.-S.; Ma, A.; Tsai, Y.-H.; Shen, Y.; Onikubo, T.; Pi, W.-C.; et al. EZH2 noncanonically binds cMyc and p300 through a cryptic transactivation domain to mediate gene activation and promote oncogenesis. Nat. Cell Biol. 2022, 24, 384–399. [Google Scholar] [CrossRef]
- Wang, M.Y.; Liow, P.; Guzman, M.I.T.; Qi, J. Exploring Methods of Targeting Histone Methyltransferases and Their Applications in Cancer Therapeutics. ACS Chem. Biol. 2022, 17, 744–755. [Google Scholar] [CrossRef]
- Wang, S.E.; Xiong, Y.; Jang, M.-A.; Park, K.-S.; Donahue, M.; Velez, J.; Jin, J.; Jiang, Y.-H. Newly developed oral bioavailable EHMT2 inhibitor as a potential epigenetic therapy for Prader-Willi syndrome. Mol. Ther. 2024, 32, 2662–2675. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, Z.; Szyf, M. S-adenosyl-methionine (SAM) alters the transcriptome and methylome and specifically blocks growth and invasiveness of liver cancer cells. Oncotarget 2017, 8, 111866–111881. [Google Scholar] [CrossRef] [PubMed]
- Waters, N.J. Preclinical Pharmacokinetics and Pharmacodynamics of Pinometostat (EPZ-5676), a First-in-Class, Small Molecule S-Adenosyl Methionine Competitive Inhibitor of DOT1L. Eur. J. Drug Metab. Pharmacokinet. 2017, 42, 891–901. [Google Scholar] [CrossRef]
- Wei, L.; Mei, D.; Hu, S.; Du, S. Dual-target EZH2 inhibitor: latest advances in medicinal chemistry. Futur. Med. Chem. 2024, 16, 1561–1582. [Google Scholar] [CrossRef]
- Wu, H.; Zheng, W.; Eram, M.S.; Vhuiyan, M.; Dong, A.; Zeng, H.; He, H.; Brown, P.; Frankel, A.; Vedadi, M.; et al. Structural basis of arginine asymmetrical dimethylation by PRMT6. Biochem. J. 2016, 473, 3049–3063. [Google Scholar] [CrossRef]
- Xu, C.; Meng, F.; Park, K.-S.; Storey, A.J.; Gong, W.; Tsai, Y.-H.; Gibson, E.; Byrum, S.D.; Li, D.; Edmondson, R.D.; et al. A NSD3-targeted PROTAC suppresses NSD3 and cMyc oncogenic nodes in cancer cells. Cell Chem. Biol. 2021, 29, 386–397.e9. [Google Scholar] [CrossRef]
- Xu, S., Aguilar, A., Huang, L., Xu, T., Zheng, K., McEachern, D., Przybranowski, S., Foster, C., Zawacki, K., Liu, Z. and Chinnaswamy, K., 2020. Discovery of M-808 as a highly potent, covalent, small-molecule inhibitor of the Menin–MLL interaction with strong in vivo antitumor activity. Journal of medicinal chemistry, 63(9), pp.4997-5010.
- Yang, Y.; Hadjikyriacou, A.; Xia, Z.; Gayatri, S.; Kim, D.; Zurita-Lopez, C.; Kelly, R.; Guo, A.; Li, W.; Clarke, S.G.; et al. PRMT9 is a Type II methyltransferase that methylates the splicing factor SAP145. Nat. Commun. 2015, 6, 1–12. [Google Scholar] [CrossRef]
- Ye, P.; Xing, H.; Lou, F.; Wang, K.; Pan, Q.; Zhou, X.; Gong, L.; Li, D. Histone deacetylase 2 regulates doxorubicin (Dox) sensitivity of colorectal cancer cells by targeting ABCB1 transcription. Cancer Chemother. Pharmacol. 2016, 77, 613–621. [Google Scholar] [CrossRef]
- Yokoyama, A.; Somervaille, T.C.; Smith, K.S.; Rozenblatt-Rosen, O.; Meyerson, M.; Cleary, M.L. The Menin Tumor Suppressor Protein Is an Essential Oncogenic Cofactor for MLL-Associated Leukemogenesis. Cell 2005, 123, 207–218. [Google Scholar] [CrossRef]
- Yu, X.; Li, D.; Kottur, J.; Kim, H.S.; Herring, L.E.; Yu, Y.; Xie, L.; Hu, X.; Chen, X.; Cai, L.; et al. Discovery of Potent and Selective WDR5 Proteolysis Targeting Chimeras as Potential Therapeutics for Pancreatic Cancer. J. Med. Chem. 2023, 66, 16168–16186. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, D.; Kottur, J.; Shen, Y.; Kim, H.S.; Park, K.-S.; Tsai, Y.-H.; Gong, W.; Wang, J.; Suzuki, K.; et al. A selective WDR5 degrader inhibits acute myeloid leukemia in patient-derived mouse models. Sci. Transl. Med. 2021, 13, eabj1578–eabj1578. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Wang, Q.; Paulk, J.; Kubicek, S.; Kemp, M.M.; Adams, D.J.; Shamji, A.F.; Wagner, B.K.; Schreiber, S.L. A Small-Molecule Probe of the Histone Methyltransferase G9a Induces Cellular Senescence in Pancreatic Adenocarcinoma. ACS Chem. Biol. 2012, 7, 1152–1157. [Google Scholar] [CrossRef]
- Zeng, H.; and Xu, W., 2015. Enzymatic assays of histone methyltransferase enzymes. In Epigenetic technological applications (pp. 333-361). Academic Press.
- Zhang, H.; Sun, Z.; Liu, Z.; Song, C. Overcoming the Emerging Drug Resistance of Smoothened: An Overview of Small-Molecule SMO Antagonists with Antiresistance Activity. Futur. Med. Chem. 2018, 10, 2855–2875. [Google Scholar] [CrossRef]
- Zhang, J.; Qian, K.; Yan, C.; He, M.; Jassim, B.A.; Ivanov, I.; Zheng, Y.G. Discovery of decamidine as a new and potent PRMT1 inhibitor. MedChemComm 2017, 8, 440–444. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Chen, X.; Hu, X.; Duan, X.; Wan, G.; Li, L.; Feng, Q.; Zhang, Y.; Wang, N.; Yu, L. Covalent inhibitors of EZH2: Design, synthesis and evaluation. Biomed. Pharmacother. 2022, 147, 112617. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ma, Y.; Hu, X.; Zheng, Y.; Chen, X. Targeting PRMT5/Akt signalling axis prevents human lung cancer cell growth. J. Cell. Mol. Med. 2018, 23, 1333–1342. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Kim, H.; Qian, C.; Xie, L.; Chen, X.; Xiong, Y.; Hu, J.; Chen, M.; Guccione, E.; Shen, Y.; et al. Discovery of a Potent and Selective Protein Arginine Methyltransferase 5 (PRMT5) PROTAC Degrader. J. Med. Chem. 2025. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Xia, T.; Chen, D.-Q.; Xiong, X.; Shi, L.; Zuo, Y.; Xiao, H.; Liu, L. Promising role of protein arginine methyltransferases in overcoming anti-cancer drug resistance. Drug Resist. Updat. 2023, 72, 101016. [Google Scholar] [CrossRef]
- Zou, W.; Li, M.; Wan, S.; Ma, J.; Lian, L.; Luo, G.; Zhou, Y.; Li, J.; Zhou, B. Discovery of PRMT3 Degrader for the Treatment of Acute Leukemia. Adv. Sci. 2024, 11, e2405963. [Google Scholar] [CrossRef]
| Substrate | Inhibitor | Reference |
|---|---|---|
| EHMT2 | UNC0642 | Liu et al., 2013 |
| UNC0925 | Konze et al., 2014 | |
| A-366 | Sweis et al., 2014 | |
| MS152 | Wang et al., 2024 | |
| EZH2 | UNC1999 | Konze et al., 2013 |
| CPI-1205 | Vaswani et al., 2016 | |
| PF-06821497 | Kung et al., 2018 | |
| EBI-2511 | Lu et al., 2018 | |
| SETD7 | (R)-PFI-2 | Barsyte-Lovejoy et al., 2014 |
| Cyproheptadine | Takemoto et al., 2016 | |
| SMYD2 | BAY-598 | Eggery et al., 2016 |
| EPZ030456 | Mitchell et al., 2016 | |
| A-893 | Sweis et al., 2015 | |
| LLY-507 | Nguyen et al., 2015 | |
| EPZ033294 | Thomenius et al., 2018 | |
| SMYD3 | BCI-121 | Peserico et al., 2015 |
| EPZ028862 | Thomenius et al., 2018 | |
| BAY-6035 | Gradl et al., 2018 | |
| SETD8 | UNC0379 | Ma et al., 2014 |
| SPS8I1 | Blum et al., 2014 | |
| MS2177 | Butler et al., 2016 | |
| PRMT5 | GSK3326595/EPZ015938 (Pemrametostat) | Gerhart et al., 2018 |
| GSK3235025/EPZ015666 | Chan-Penebre et al., 2015 | |
| GSK3203591/EPZ015866 | Duncan et al., 2016 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





