Submitted:
16 August 2023
Posted:
18 August 2023
You are already at the latest version
Abstract
Keywords:
Highlights
- Ethyl [5-(4-Chlorophenyl)-2-methyl-1H-imidazol-4-yl}-acetate was found to have a higher docking score, glide score, and glide energy when compared to other compounds sirtuin family proteins.
- Ethyl [5-(4-Chlorophenyl)-2-methyl-1H-imidazol-4-yl}-acetate has higher stability during the itnteraction with sirtuins.
- Imidazole derivative effectively reduces the viability of cancer cell lines A549 and NIC-H460 at lower concentrations compared to imidazole.
- Ethyl [5-(4-Chlorophenyl)-2-methyl-1H-imidazol-4-yl}-acetate greatly affected the gene expression and protein expression of sirtuins family members.
1. Introduction
2. Materials and Methods
2.1. Ligand Preparation
2.2. Protein Preparation and Receptor Grid Generation
2.3. Glide Docking
2.4. Molecular Dynamic Simulation- Desmond
2.5. DFT Analysis
2.6. ADME analysis
2.7. Cell culture maintenance:
2.8. Cell viability assay by MTT:
2.9. Quantitative Real-time PCR (qRT-PCR):
2.10. Western blot analysis:
2.11. Statistical analysis
3. Results
3.1. Structure description
3.2. Molecular Docking
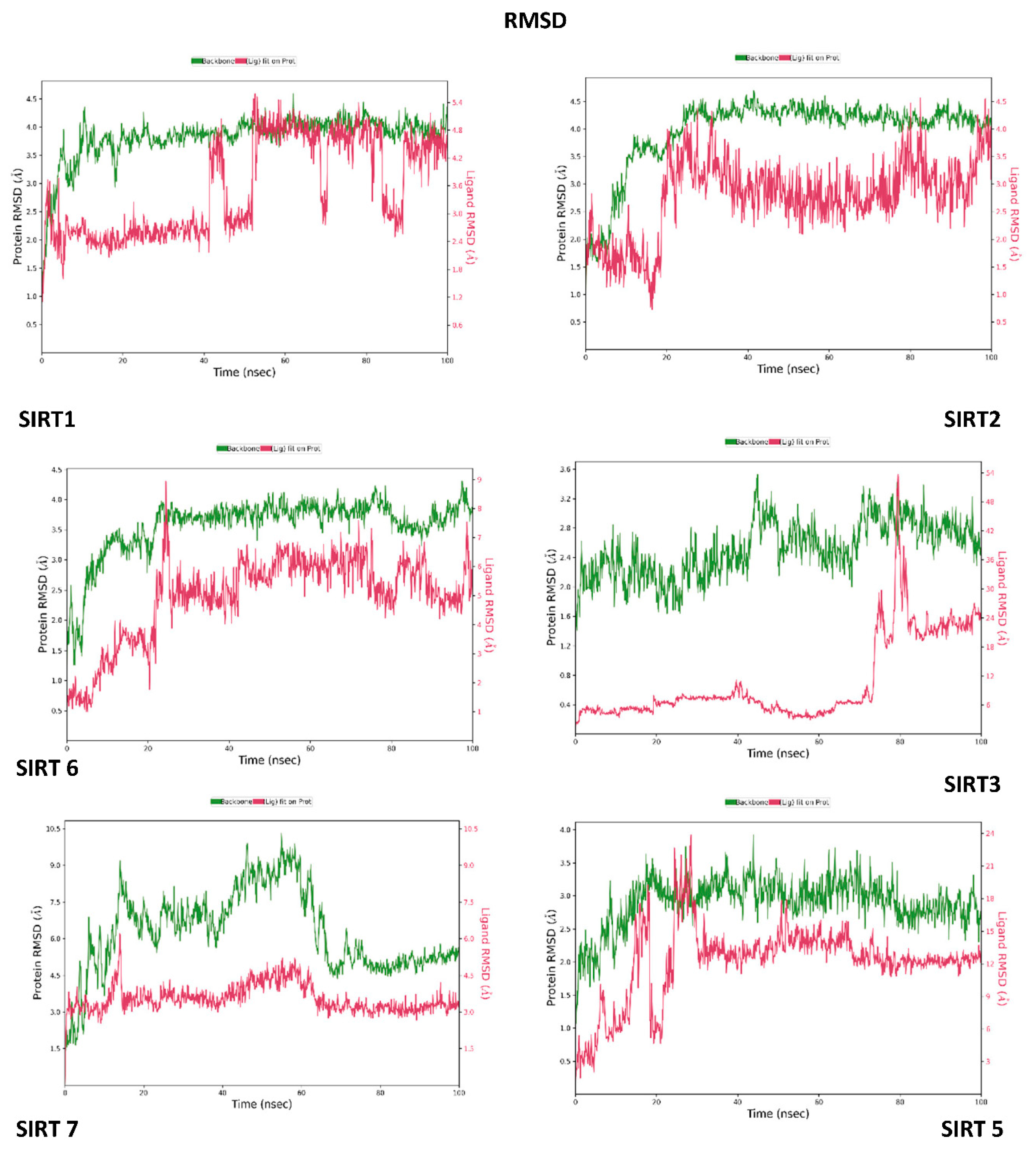
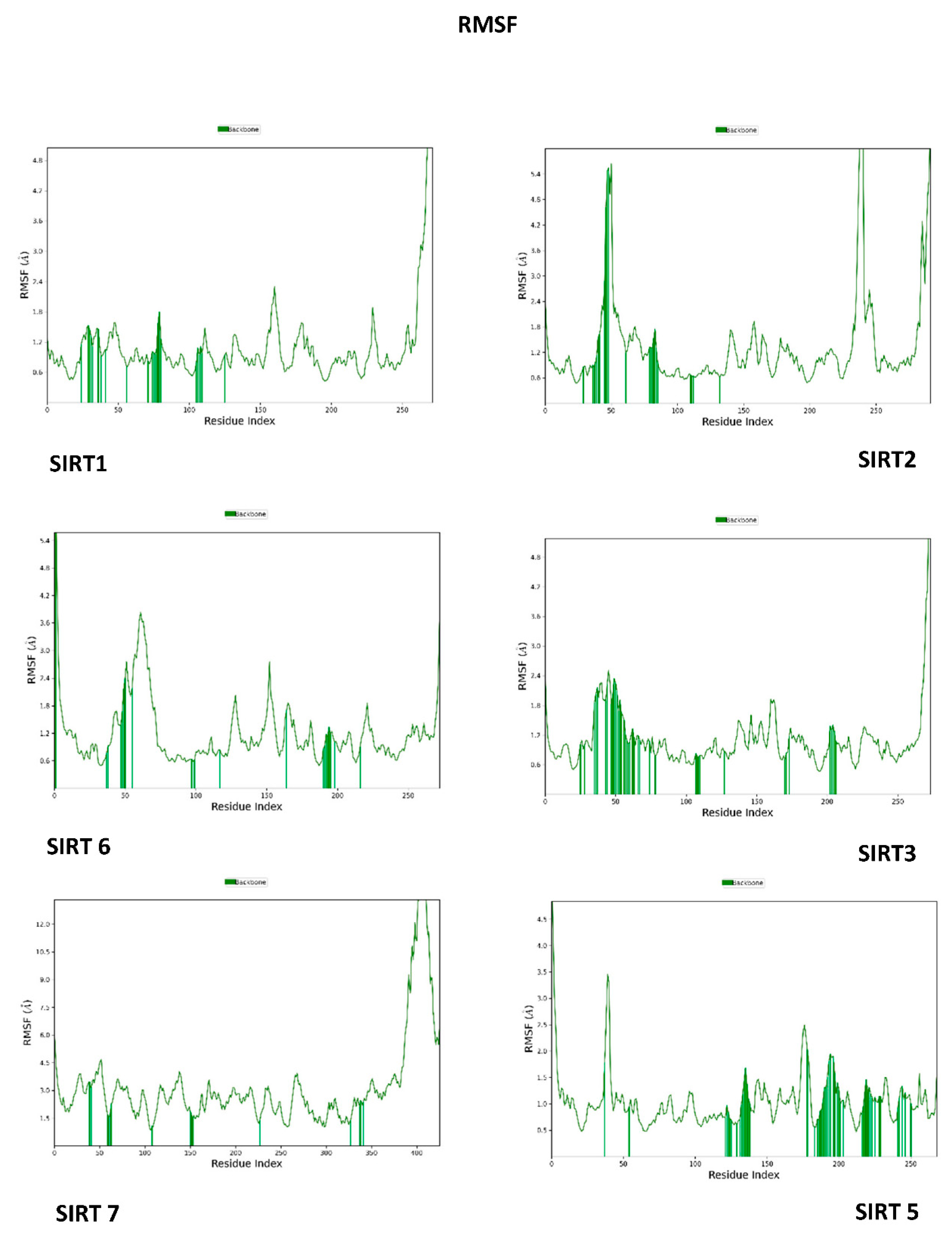
3.3. Molecular Dynamics
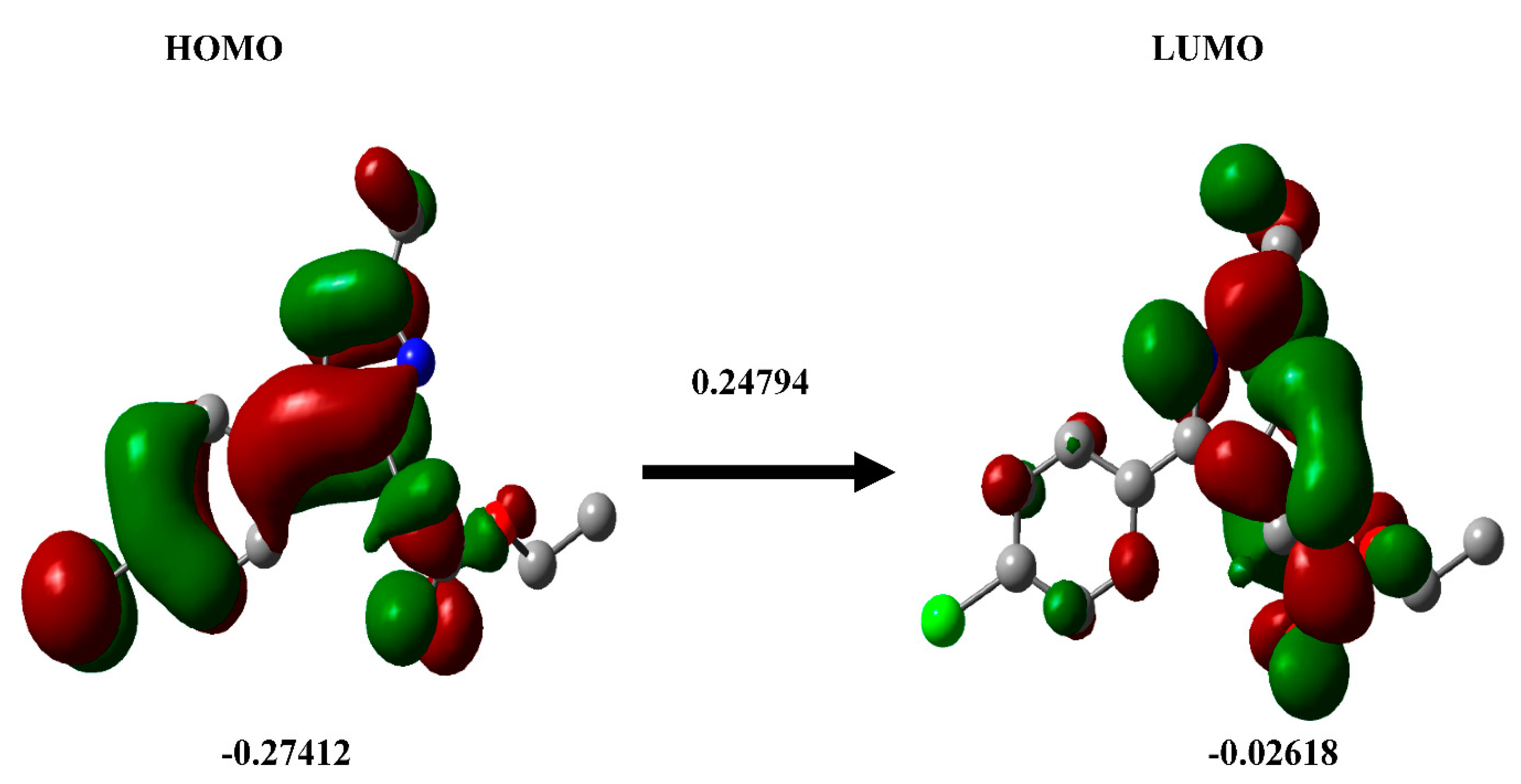
3.4. DFT calculation
3.5. ADME Analysis
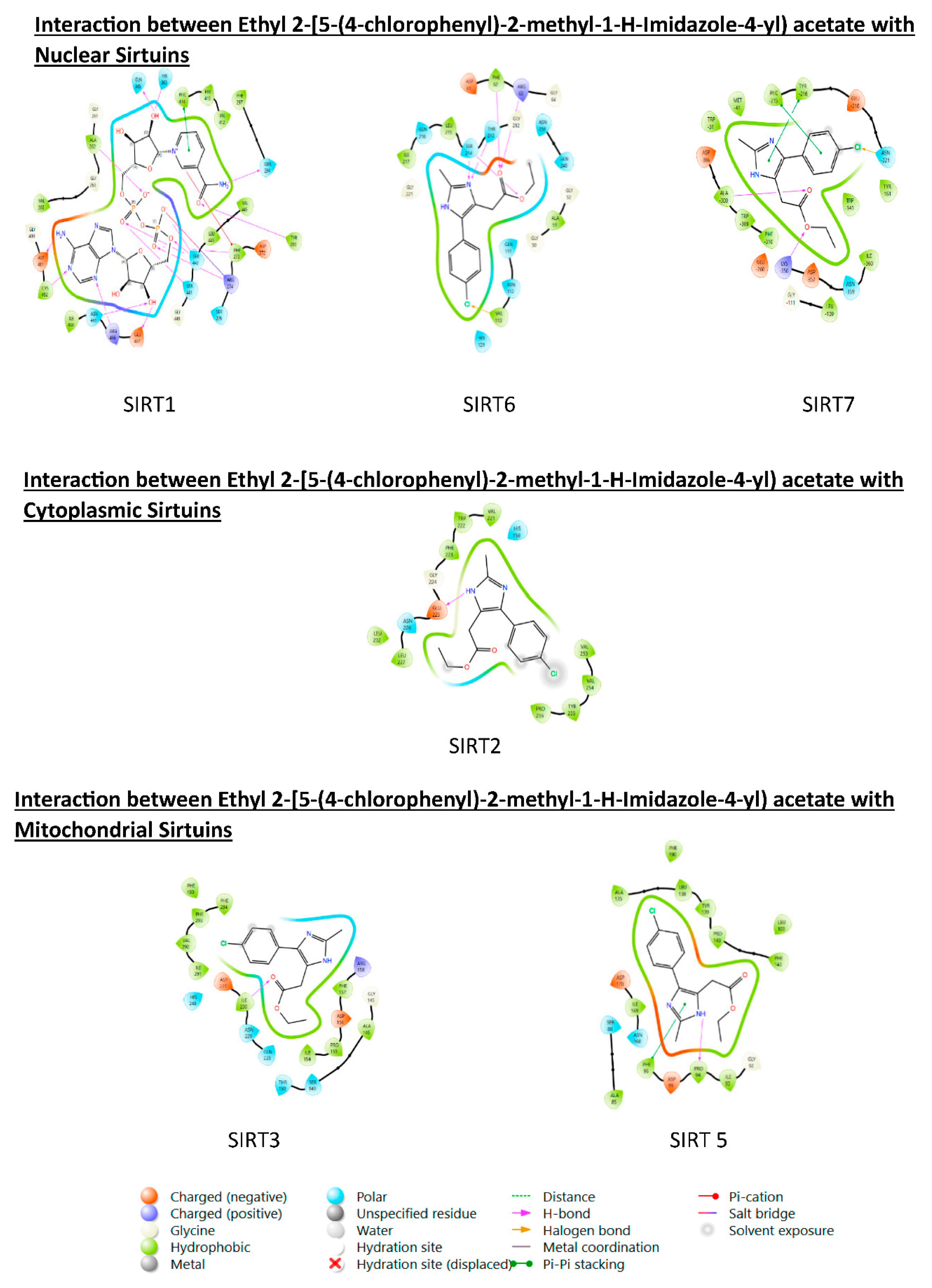
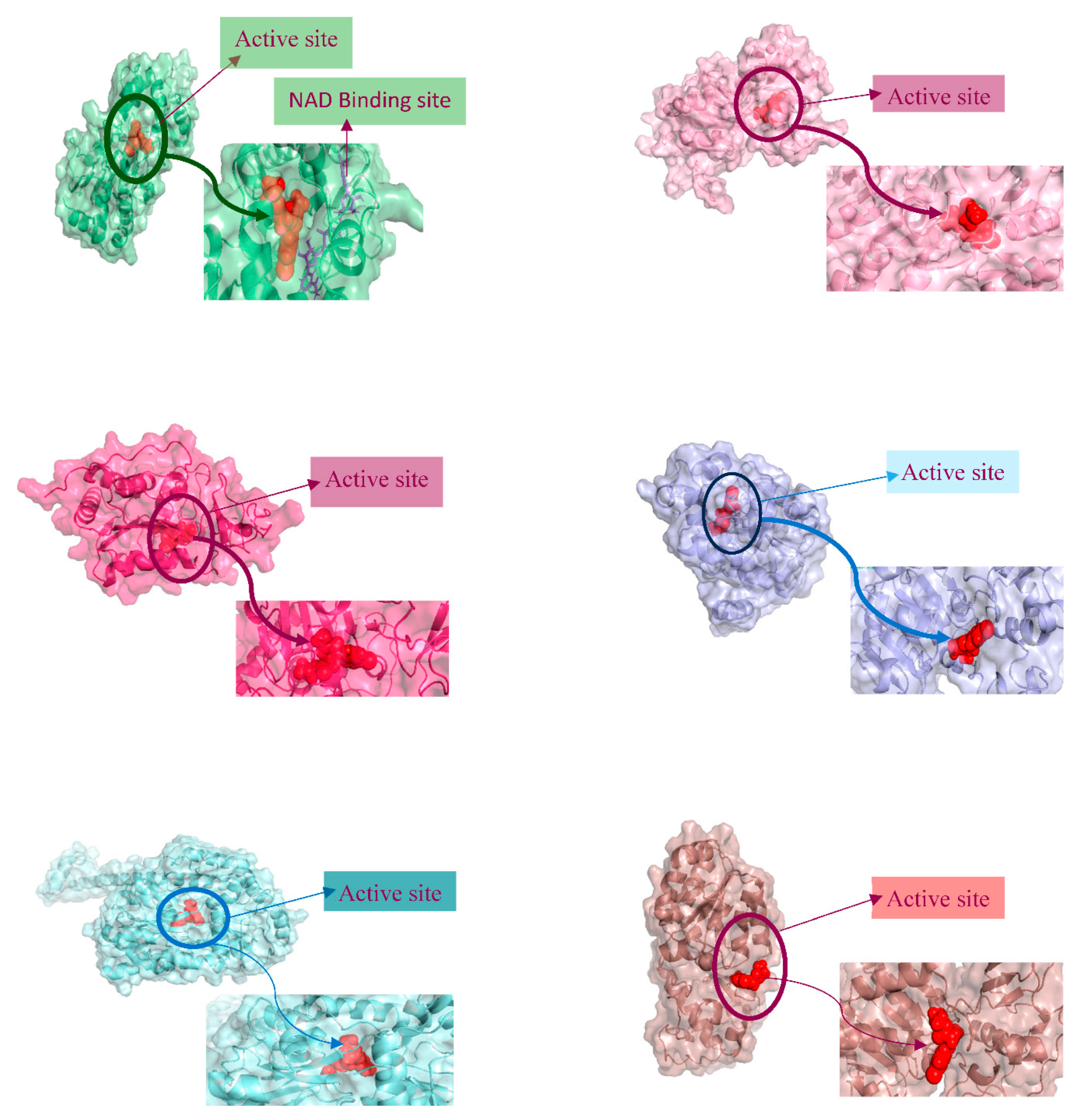
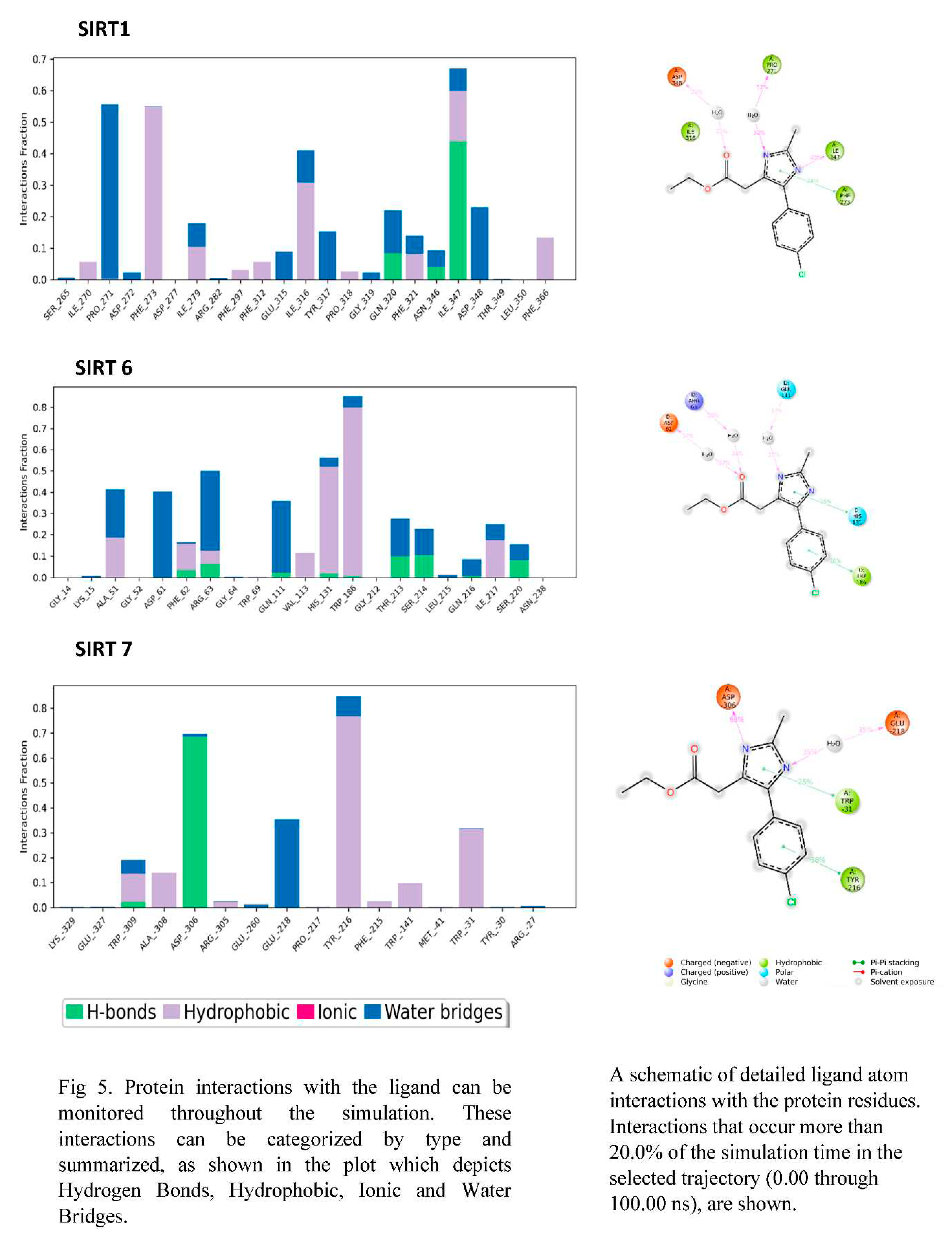
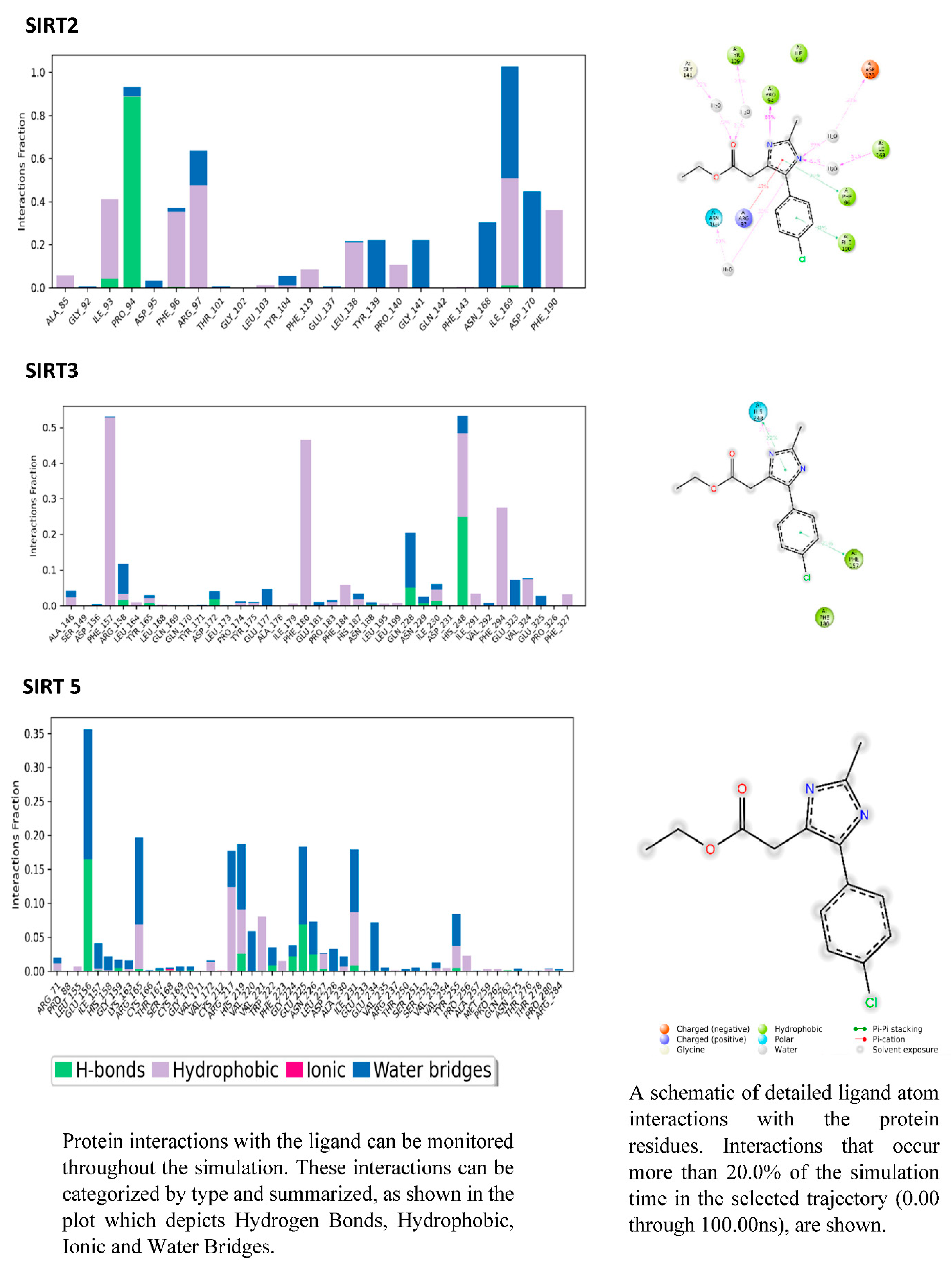
3.6. PROTEIN-LIGAND interaction-3D view
3.7. DFT calculation
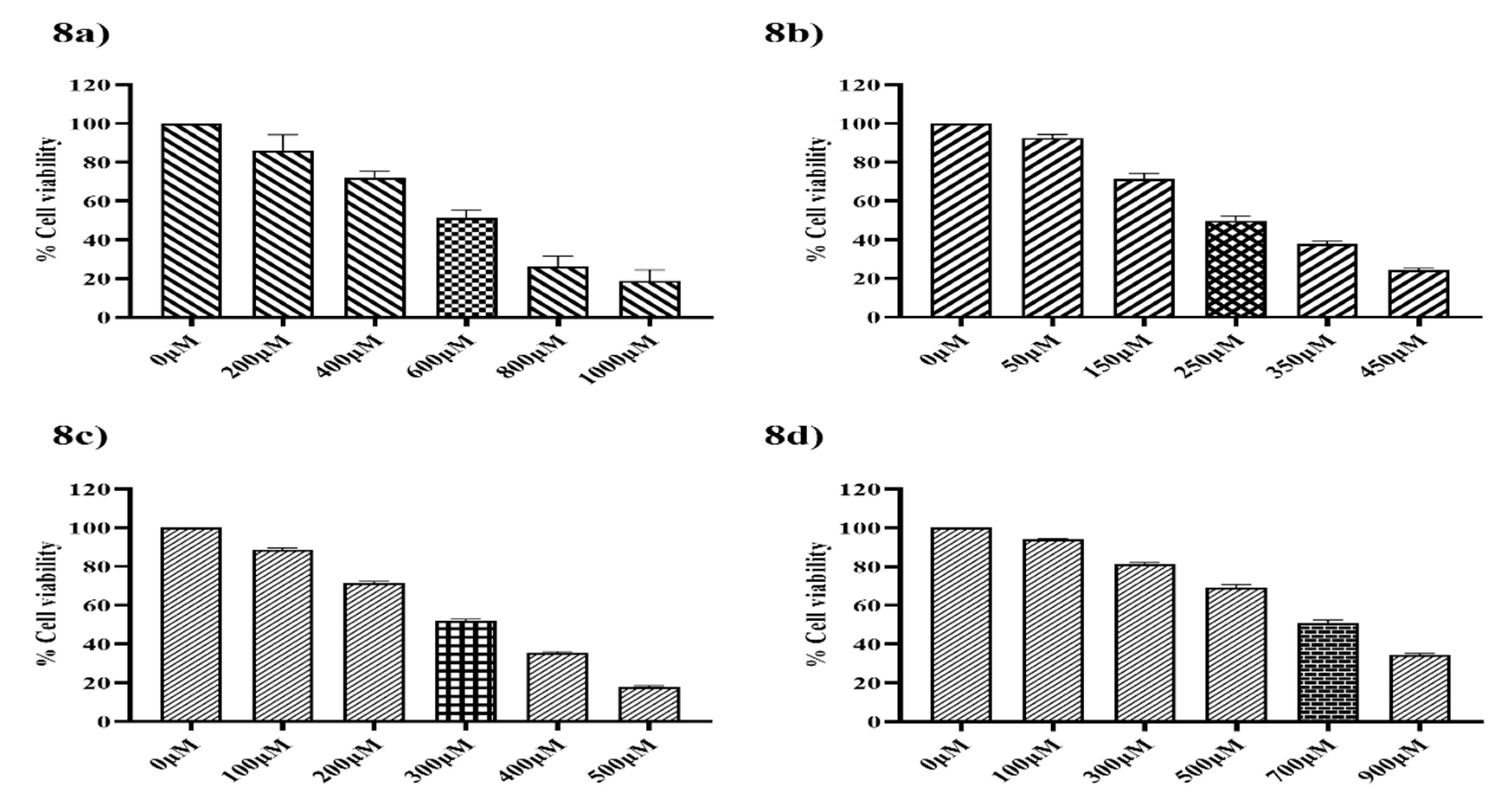
3.8. Cytotoxicity Effect of Ethyl [5-(4- Chlorophenyl)-2-methyl-1H-imidazol-4-yl}-acetate
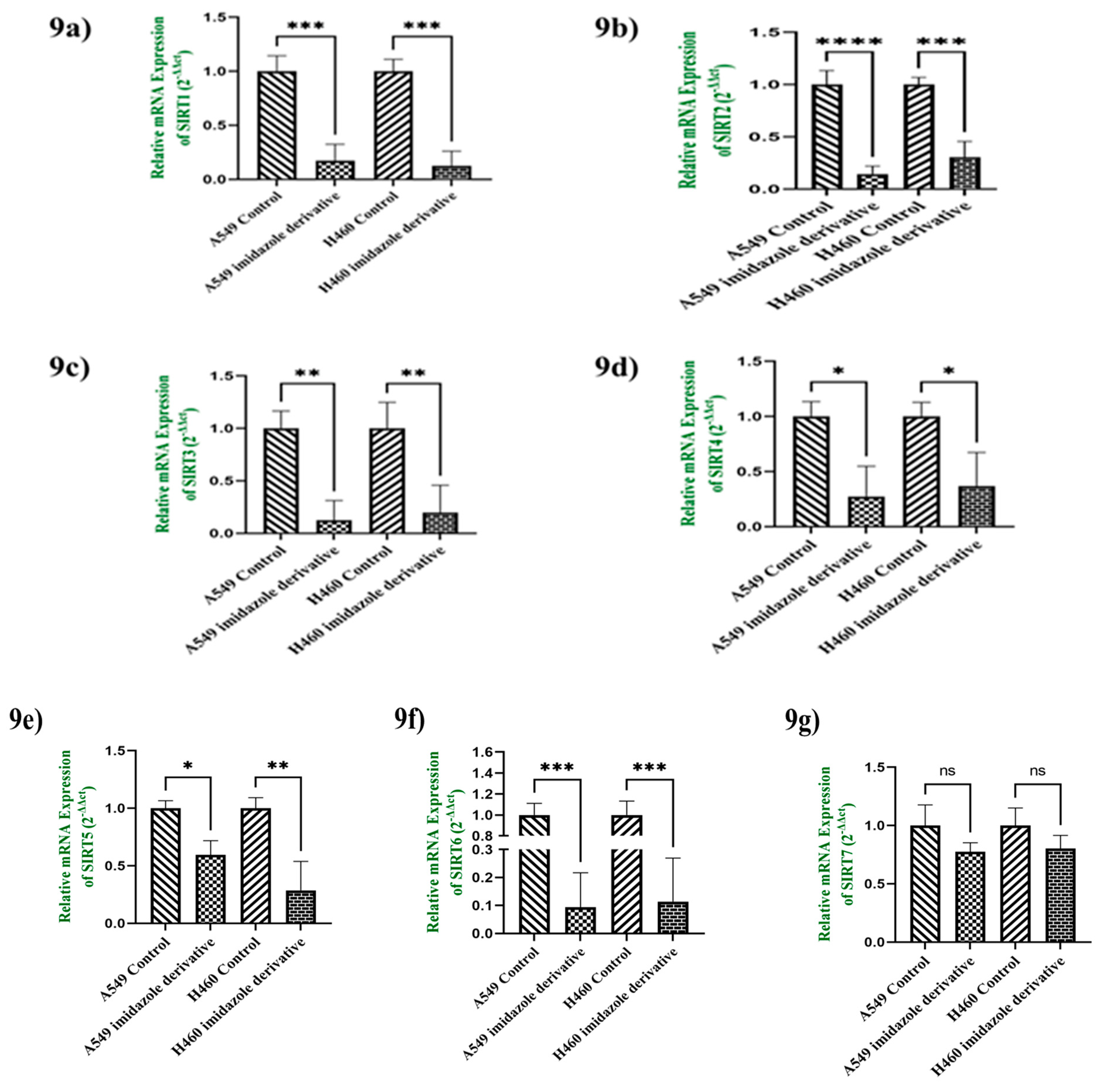
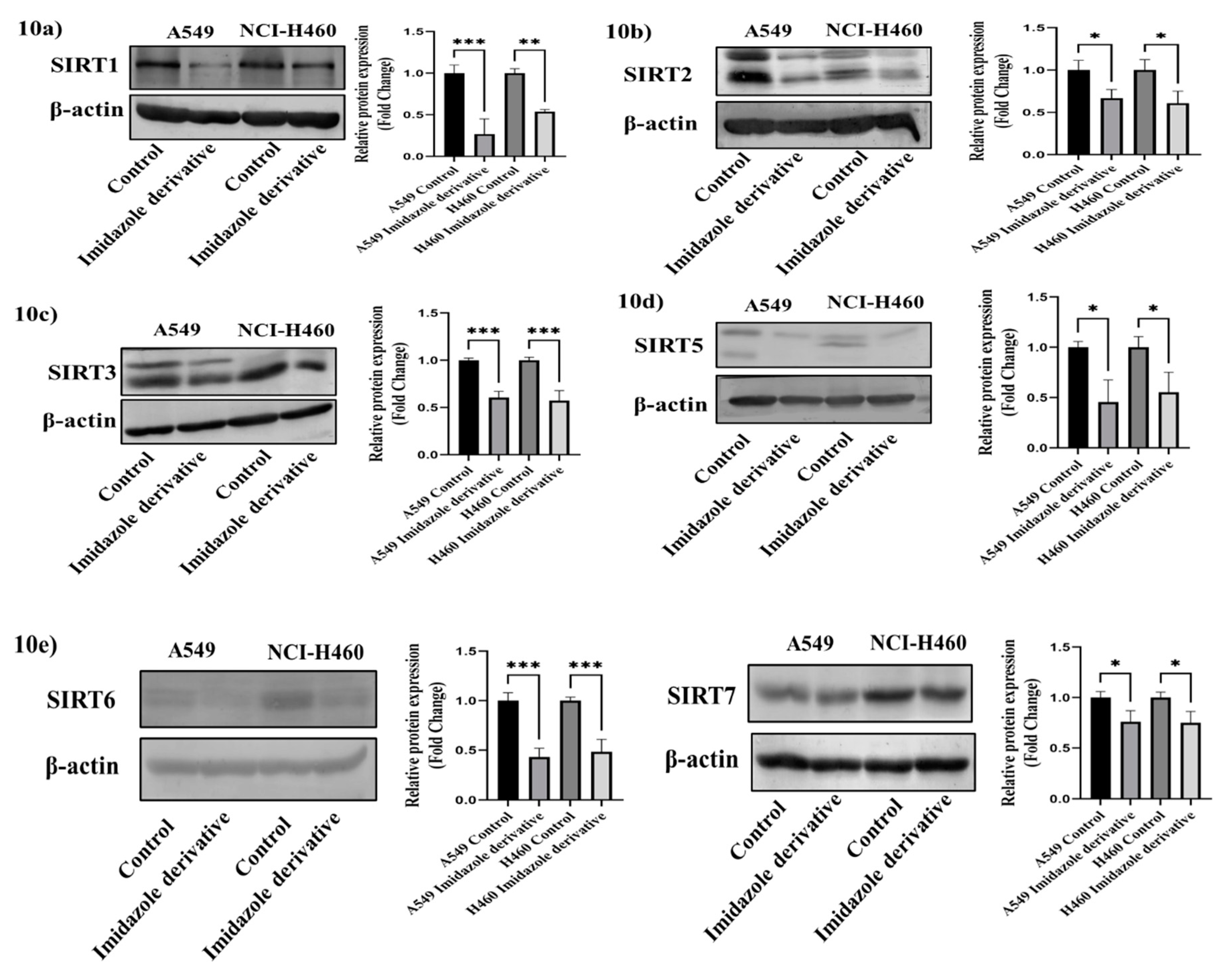
3.9. In-Vitro analysis of Imidazole derivative Ethyl [5-(4-Chlorophenyl)-2-methyl-1H-imidazol-4-yl]-acetate effect on Class III Histone deacetylase family (SIRT)
4. Discussion
5. Conclusion:
Author Contributions
Funding
Ethics approval and consent to participate
Consent for publication
Availability of data and materials
Acknowledgments
Competing interests
Compliance with Ethical Standards
Disclosure of potential conflicts of interest
Research involving human participants and/or animals
Informed consent
Abbreviations
References
- Ali, I.; Lone, M.N.; Aboul-Enein, H.Y. Imidazoles as potential anticancer agents. MedChemComm 2017, 8, 1742–1773. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; LaRosa, C.; Antwi, J.; Govindarajan, R.; Werbovetz, K.A. Imidazoles as Potential Anticancer Agents: An Update on Recent Studies. Molecules 2021, 26, 4213. [Google Scholar] [CrossRef] [PubMed]
- Bryaskova, R.; Georgiev, N.; Philipova, N.; Bakov, V.; Anichina, K.; Argirova, M.; Apostolova, S.; Georgieva, I.; Tzoneva, R. Novel Fluorescent Benzimidazole-Hydrazone-Loaded Micellar Carriers for Controlled Release: Impact on Cell Toxicity, Nuclear and Microtubule Alterations in Breast Cancer Cells. Pharmaceutics 2023, 15, 1753. [Google Scholar] [CrossRef] [PubMed]
- Rajabi, M.; Mousa, S.A. The Role of Angiogenesis in Cancer Treatment. Biomedicines 2017, 5, 34. [Google Scholar] [CrossRef]
- Azad, A.; Kong, A. The Therapeutic Potential of Imidazole or Quinone-Based Compounds as Radiosensitisers in Combination with Radiotherapy for the Treatment of Head and Neck Squamous Cell Carcinoma. Cancers 2022, 14, 4694. [Google Scholar] [CrossRef]
- Tolomeu, H.V.; Fraga, C.A.M. Imidazole: Synthesis, Functionalization and Physicochemical Properties of a Privileged Structure in Medicinal Chemistry. Molecules 2023, 28, 838. [Google Scholar] [CrossRef]
- Sharma, A.; Kumar, V.; Kharb, R.; Kumar, S.; Sharma, P.C.; Pathak, D.P. Imidazole Derivatives as Potential Therapeutic Agents. Curr. Pharm. Des. 2016, 22, 3265–3301. [Google Scholar] [CrossRef]
- Kandasamy, S.; Subramani, P.; Srinivasan, K.; Jayaraj, J.M.; Prasanth, G.; Muthusamy, K.; Rajakannan, V.; Vilwanathan, R. Design and synthesis of imidazole based zinc binding groups as novel small molecule inhibitors targeting Histone deacetylase enzymes in lung cancer. J. Mol. Struct. 2020, 1214, 128177. [Google Scholar] [CrossRef]
- Dai, H.; Sinclair, D.A.; Ellis, J.L.; Steegborn, C. Sirtuin activators and inhibitors: Promises, achievements, and challenges. Pharmacol. Ther. 2018, 188, 140–154. [Google Scholar] [CrossRef]
- Zhu, S.; Dong, Z.; Ke, X.; Hou, J.; Zhao, E.; Zhang, K.; Wang, F.; Yang, L.; Xiang, Z.; Cui, H. The roles of sirtuins family in cell metabolism during tumor development. Semin. Cancer Biol. 2018, 57, 59–71. [Google Scholar] [CrossRef]
- Vu, C.B.; Bemis, J.E.; Disch, J.S.; Ng, P.Y.; Nunes, J.J.; Milne, J.C.; Carney, D.P.; Lynch, A.V.; Smith, J.J.; Lavu, S.; et al. Discovery of Imidazo[1,2-b]thiazole Derivatives as Novel SIRT1 Activators. J. Med. Chem. 2009, 52, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Morris, B.J.; Willcox, B.J.; Donlon, T.A. Genetic And epigenetic regulation of human aging and longevity. Biochim. Biophys. Acta. Mol. Basis Dis. 2019, 1865, 1718–1744. [Google Scholar] [CrossRef] [PubMed]
- Chalkiadaki, A.; Guarente, L. The multifaceted functions of sirtuins in cancer. Nat. Rev. Cancer 2015, 15, 608–624. [Google Scholar] [CrossRef] [PubMed]
- Bosch-Presegue, L.; Vaquero, A. The Dual Role of Sirtuins in Cancer. Genes Cancer 2011, 2, 648–662. [Google Scholar] [CrossRef] [PubMed]
- Alcaín, F.J.; Villalba, J.M. Sirtuin inhibitors. Expert opinion on therapeutic patents 2009, 19, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Liu, J.; Chen, D.; Yan, L.; Zheng, W. Sirtuin inhibition: strategies, inhibitors, and therapeutic potential. Trends in pharmacological sciences 2017, 38, 459–472. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Jing, H.; Lin, H.; Kejík, Z.; Jakubek, M.; Kaplánek, R.; Králová, J.; Mikula, I.; Martásek, P.; Král, V.; et al. Sirtuin inhibitors as anticancer agents. Futur. Med. Chem. 2014, 6, 945–966. [Google Scholar] [CrossRef]
- Wu, Q.-J.; Zhang, T.-N.; Chen, H.-H.; Yu, X.-F.; Lv, J.-L.; Liu, Y.-Y.; Liu, Y.-S.; Zheng, G.; Zhao, J.-Q.; Wei, Y.-F.; et al. The sirtuin family in health and disease. Signal Transduct. Target. Ther. 2022, 7, 1–74. [Google Scholar] [CrossRef]
- Krishnamoorthy, V.; Vilwanathan, R. Silencing Sirtuin 6 induces cell cycle arrest and apoptosis in non-small cell lung cancer cell lines. Genomics 2020, 112, 3703–3712. [Google Scholar] [CrossRef]
- Kirchmair, J.; Markt, P.; Distinto, S.; Wolber, G.; Langer, T. Evaluation of the performance of 3D virtual screening protocols: RMSD comparisons, enrichment assessments, and decoy selection—What can we learn from earlier mistakes? J. Comput. Aided Mol. Des. 2008, 22, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Margreitter, C.; Oostenbrink, C. MDplot: Visualise Molecular Dynamics. R J. 2017, 9, 164–186. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Qian, M.; Peng, B.; Peng, L.; Wang, X.; Zheng, K.; Liu, Z.; Tang, X.; Zhang, S.; Sun, S.; Cao, X. Synergy between SIRT1 and SIRT6 helps recognize DNA breaks and potentiates the DNA damage response and repair in humans and mice. elife 2020, 9, e55828. [Google Scholar] [CrossRef] [PubMed]
- Santos-Barriopedro, I.; Bosch-Presegué, L.; Marazuela-Duque, A.; de la Torre, C.; Colomer, C.; Vazquez, B.N.; Fuhrmann, T.; Martínez-Pastor, B.; Lu, W.; Braun, T.; et al. SIRT6-dependent cysteine monoubiquitination in the PRE-SET domain of Suv39h1 regulates the NF-κB pathway. Nature communications 2018, 9, 101. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Ahmad, F.; Philp, A.; Baar, K.; Williams, T.; Luo, H.; Ke, H.; Rehmann, H.; Taussig, R.; Brown, A.L.; et al. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell 2012, 148, 421–433. [Google Scholar] [CrossRef]
- Pillai, V.B.; Sundaresan, N.R.; Gupta, M.P.; H, L.; M, H.; J, R.; L, T.; J, R.; A, R.; J, G.; et al. Regulation of Akt Signaling by Sirtuins. Circ. Res. 2014, 114, 368–378. [Google Scholar] [CrossRef]
- Qiang, L.; Wang, L.; Kon, N.; Zhao, W.; Lee, S.; Zhang, Y.; Rosenbaum, M.; Zhao, Y.; Gu, W.; Farmer, S.R.; et al. Brown Remodeling of White Adipose Tissue by SirT1-Dependent Deacetylation of Ppary. Cell 2012, 150, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Prabhaharan, M.; Prabakaran, A.R.; Gunasekaran, S.; Srinivasan, S. DFT studies on vibrational spectra, HOMO–LUMO, NBO and thermodynamic function analysis of cyanuric fluoride. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy 2015, 136, 494–503. [Google Scholar] [CrossRef]
- Mathammal, R.; Jayamani, N.; Geetha, N. Molecular Structure, NMR, HOMO, LUMO, and Vibrational Analysis of O-Anisic Acid and Anisic Acid Based on DFT Calculations. Spectrosc. 2013, 2013, 1–18. [Google Scholar] [CrossRef]
- Fatima, S.; Gupta, P.; Sharma, S.; Sharma, A.; Agarwal, S.M. ADMET profiling of geographically diverse phytochemical using chemoinformatic tools. Futur. Med. Chem. 2020, 12, 69–87. [Google Scholar] [CrossRef]
- Telvekar, V.N.; Bairwa, V.K.; Satardekar, K.; Bellubi, A. Novel 2-(2-(4-aryloxybenzylidene) hydrazinyl)benzothiazole derivatives as anti-tubercular agents. Bioorganic Med. Chem. Lett. 2012, 22, 649–652. [Google Scholar] [CrossRef]
- Jhala, D.D.; Chettiar, S.S.; Singh, J.K. Optimization and Validation of an In Vitro Blood Brain Barrier Permeability Assay Using Artificial Lipid Membrane. J. Bioequivalence Bioavailab. 2012, s14. [Google Scholar] [CrossRef]
| S. No | Gene | Primer sequence 5′-3′ | Annealing.Temp | Product Size |
|---|---|---|---|---|
| 1 | GAPDH | F’ P: ATGGGGAAGGTGAAGGTCG R’ P: GGGTCATTGATGGCAACAATATC |
60 | 107 |
| 2 | SIRT1 | F’ P: ACCCAGCTCACCTTCTTT R’ P: CCCAGACTTTCCCACTCT |
57 | 176 |
| 3 | SIRT2 | F’ P: CTCCCCTTCCAGCTTAAC R’ P: TGACACTCACCCCAAGAC |
57 | 175 |
| 4 | SIRT3 | F’ P: GGGAGGGGTACAGTGAGG R’ P: GGGTGACAGAGCGAGATG |
58.1 | 177 |
| 5 | SIRT4 | F’ P: TGGTCATTGCTGGTTTCC R’ P: AGGCAGAGGTTGTGGTGA |
57 | 170 |
| 6 | SIRT5 | F’ P: GGCTTTGCTTTCCCTTAC R’ P: AACCTTGGCGATTAGACC |
58.1 | 171 |
| 7 | SIRT6 | F’ P: TCCCATTGTCTAGCCTCA R’ P: GATGTCGGTGAATTACGC |
58.1 | 181 |
| 8 | SIRT7 | F’ P: TCGGCTCCTCCCTTCTAC R’ P: GGGGGCACTTTAGGAACA |
57 | 180 |
| S. No | Antibody | Molecular weight (kDa) | Brand | Cat. Log |
|---|---|---|---|---|
| 1 | β-Actin | 45 | Novus Biologicals | MAB8929-SP |
| 2 | Rabbit Monoclonal primary SIRT 1 | 120 | Cell Signaling Technology | 9475 |
| 3 | Rabbit Monoclonal primary SIRT 2 | 39,43 | Cell Signaling Technology | 12650 |
| 4 | Rabbit Monoclonal primary SIRT 3 | 28 | Cell Signaling Technology | 5490 |
| 5 | Rabbit Monoclonal primary SIRT 5 | 30 | Santa Cruz Biotechnology | 8782 |
| 6 | Rabbit Monoclonal primary SIRT 6 | 36-42 | Santa Cruz Biotechnology | 12486 |
| 7 | Rabbit Monoclonal primary SIRT 7 | 45 | Santa Cruz Biotechnology | 5360 |
| PDB | SIRT1 (4I5I) | SIRT2(4RMH) | SIRT3 (4JSR) | SIRT5(6LJK) | SIRT6(3K35) | SIRT7(5IQZ) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ligand Name | Glide Score | Glide Energy | Glide Score | Glide Energy | Glide Score | Glide Energy | Glide Score | Glide Energy | Glide Score | Glide Energy | Glide Score | Glide Energy |
| Ethyl 2-(2,5-diphenyl-1H-imidazole-4-yl) acetate | -5.727 | -18.208 | -4.32 | -14.778 | -4.495 | -17.511 | -7.053 | -15.008 | -5.401 | -19.483 | -6.229 | -41.320 |
| Ethyl 2-[2-phenyl-5-(3,4,5-trimethoxyphenyl)-1H-imidazol-4-yl] acetate | -6.655 | -28.841 | -7.289 | -39.668 | -8.186 | -54.470 | -4.912 | -35.961 | -5.773 | -47.432 | -5.000 | -41.870 |
| Ethyl 2-[5-(4-chlorophenyl)-2-phenyl-1H-imidazol-4-yl] acetate | -7.556 | -31.593 | -9.056 | -46.782 | -7.019 | -44.698 | -5.735 | -32.778 | -5.225 | -45.219 | -5.498 | -42.018 |
| Ethyl 2-[5-(4-bromophenyl)-2-phenyl-1H-imidazol-4-yl] acetate | -7.742 | -35.492 | -8.816 | -46.547 | -7.442 | -49.158 | -4.686 | -32.981 | -4.664 | -44.797 | -5.404 | -42.510 |
| Ethyl 2-{2-phenyl-5-[4-(trifluoromethyl) phenyl]-1H-imidazol-4-yl} acetate | -9.656 | -26.563 | -7.711 | -39.699 | -7.714 | -49.695 | -5.584 | -32.798 | -4.825 | -45.74 | -5.558 | -44.785 |
| Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl) acetate | -7.807 | -37.735 | -8.835 | -41.366 | -7.053 | -43.362 | -5.053 | -31.857 | -6.544 | -45.913 | -5.616 | -39.446 |
| Imidazole | -5.723 | -18.112 | -4.318 | -14.734 | -4.495 | -17.511 | -5.277 | -15.008 | -5.385 | -19.497 | -4.092 | -16.893 |
| Ligand Name | Molecular weight | SASA | FOSA | FISA | Donor HB | Acceptor HB | VOLUME | QPlogPw | QPlogP/w | QPlogBB | QPlogS | QPPCaCO | Human Oral Absorption | %Human Oral Absorption |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acceptable Range | (<500 Da) | (300.0 – 1000.0 ) | (0.0 – 750.0) | (7.0 – 330.0) | (<5), g) | (<10), h) | (500.0 – 2000.0) | (4.0 – 45.0) | (−2.0-6.5) | (–3.0 – 1.2) | (−6.5-0.5) | (<25 poor,>500 great) | -- | (<25% is poor and >80% is high) |
| Ethyl 2-(2,5-diphenyl-1H-imidazole-4-yl) acetate | 306.363 | 585.302 | 143.504 | 73.186 | 1 | 3.5 | 1029.252 | 8.373 | 4.212 | -5.184 | -0.284 | 2102.639 | 3 | 100 |
| Ethyl 2-[2-phenyl-5-(3,4,5-trimethoxyphenyl)-1H-imidazol-4-yl] acetate | 396.442 | 683.134 | 374.559 | 72.031 | 0 | 4.75 | 1255.511 | 6.817 | 4.951 | -5.699 | -0.506 | 2055.082 | 3 | 100 |
| Ethyl 2-[5-(4-chlorophenyl)-2-phenyl-1H-imidazol-4-yl] acetate | 340.808 | 646.409 | 176.469 | 57.976 | 1 | 3.5 | 1111.022 | 8.259 | 5.05 | -6.598 | -0.041 | 2793.266 | 1 | 100 |
| Ethyl 2-[5-(4-bromophenyl)-2-phenyl-1H-imidazol-4-yl] acetate | 385.259 | 654.421 | 177.218 | 57.77 | 1 | 3.5 | 1121.383 | 8.291 | 5.14 | -6.768 | -0.033 | 2805.877 | 1 | 100 |
| Ethyl 2-{2-phenyl-5-[4-(trifluoromethyl) phenyl]-1H-imidazol-4-yl} acetate | 374.362 | 666.352 | 171.63 | 59.5 | 1 | 3.5 | 1160.555 | 8.18 | 5.504 | -7.159 | 0.054 | 2701.827 | 1 | 100 |
| Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl) acetate | 278.738 | 558.092 | 272.468 | 72.352 | 1 | 3.5 | 933.926 | 6.793 | 3.6 | -5.01 | -0.145 | 2040.747 | 3 | 100 |
| Imidazole | 68.078 | 228.199 | 0 | 65.325 | 1 | 2 | 309.474 | 5.255 | -0.08 | -0.485 | 0.1 | 2379.132 | 3 | 86.91 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





