1. Introduction
Recently digital pathology has emerged as one of the most important breakthroughs linking medical practice to technological innovation. In recent years, the use of Artificial Intelligence (AI) has greatly driven progress in digital pathology, particularly in research, predictive modeling, and disease detection. Beyond its many applications in healthcare diagnostics, AI also supports digital pathology in lung cancer research and diagnostics. Lung cancer remains the leading cause of cancer death worldwide Lung cancer remains the leading cause of cancer-related mortality worldwide [
1,
2]. It is the most commonly diagnosed malignant epithelial tumor worldwide because of various factors that influence its development, such as radiation exposure, environmental toxins, and infections [
3]. The prognosis for lung cancer is generally poor, with an overall five-year survival rate of just 16.6% but varies widely with the stage of the disease. Whereas the five-year survival rate for early-stage lung cancer is about 70%, it significantly decreases to less than 5% in patients diagnosed at stage four [
4]. Hence, early detection is crucial at improving the outcomes. Treatment planning based on precise specific diagnosis is also necessary, since there is such a wide range of responses to the various therapies available for lung cancer.
However, traditional methods—such as examining tissue slides under a microscope—are labor-intensive and expensive, and subject to variability in the accuracy of the diagnosis, due to inter-observer discrepancies [
5]. AI can address these challenges by analyzing large datasets and detecting subtle patterns in diseased tissue that may be invisible to human eyes. This capability accelerates diagnosis and enhances precision, enabling pathologists to identify even the smallest differences in histopathological features.
In recent years, significant advancements in AI-driven digital pathology have further bolstered research, predictive modeling, and disease detection. [
6]. The primary methods for histological diagnosis of lung carcinoma involve the use of bronchoscopy and percutaneous core needle biopsy. Even though the methods like bronchoscopy and percutaneous core needle biopsy give the most important tissue samples for lung cancer diagnosis, they have their limitations. Diagnostic errors are not unusual even among expert pathologists, particularly in the late stages of the disease. Studies have shown that differences in staging of lung cancer among pathologists are not rare, and they still cannot detect early-stage pulmonary neoplasms even by using histological markers during advanced stages [
7]. The AI-based methods of diagnosis have the potential to improve precision in therapeutics as well as providing the physician with the additional necessary clinical data for the decision-making process.
Moreover, the specific diagnosis of deep lung lesions requires both ultrasound and CT-guided percutaneous lung biopsies as well as bronchoscopy when lesions remain inaccessible [
8]. The procedures offer essential identification of tissue samples that doctors need for correct healthcare diagnosis and therapeutic guidance. The professional expertise of pathologists also does not prevent diagnostic errors even when dealing with advanced-stage disease. Research studies demonstrate pathologists face difficulties in accurately identifying different types of lung cancer and pathologists do not achieve consistent diagnostic agreement across their assessments [
8]. However histological markers provide some success more accurate diagnosis at advanced stages.
There are two main types of epithelial lung cancer which doctors recognize including small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC) where SCLC represents 15% of cases and NSCLC makes up 85% [
16]. The main histological categories among NSCLC patients consist of adenocarcinoma and squamous cell carcinoma. The current standard for early lung cancer screening combines the use of CT scans and chest X-rays (CXRs) [
17], but these proven methods require extensive patient work up and show significant differences between interpretation results conducted by different observers. Medical experts sometimes make inconsistent diagnostic evaluations because of their subjective interpretation methods. Recent progress in AI and digital pathology distribution has transformed conventional diagnostic systems by creating new decision-making tools which significantly enhance diagnostic precision and eliminate subjectivity in reading. AI technology applies sophisticated algorithms to perform image evaluation while including multi-parametric clustering models according to research [
18]. The application of this feature helps medical professionals identify lung cancer early in its development phases. The application of AI for both acceleration and automation of screening operations decreases medical staff errors while boosting detection precision. Both data processing speed and analysis performed by AI deliver substantial benefits for early-stage detection methods together with immediate medical interventions.
AI-based technology has delivered crucial developments to lung cancer management across all phases beginning from diagnostic testing and progressing to disease treatment. Machine learning algorithms present ways to predict tumors' behavior and evaluate treatment results which drive both specific patient treatment plans and better accuracy levels. Clinical specialists spend more time on patient-oriented care and advanced medical decisions because AI performs repetitive work which in turn demonstrates AI's disruptive power in oncology.
The application of AI has resulted in exceptional achievements for both lung cancer progression predictions and the enhancement of classification results during the past few years. The precision and reliability of histopathology image recognition AI algorithms enables them to analyze large quantities of data with high precision thereby surpassing minor variations in tumor development stages [
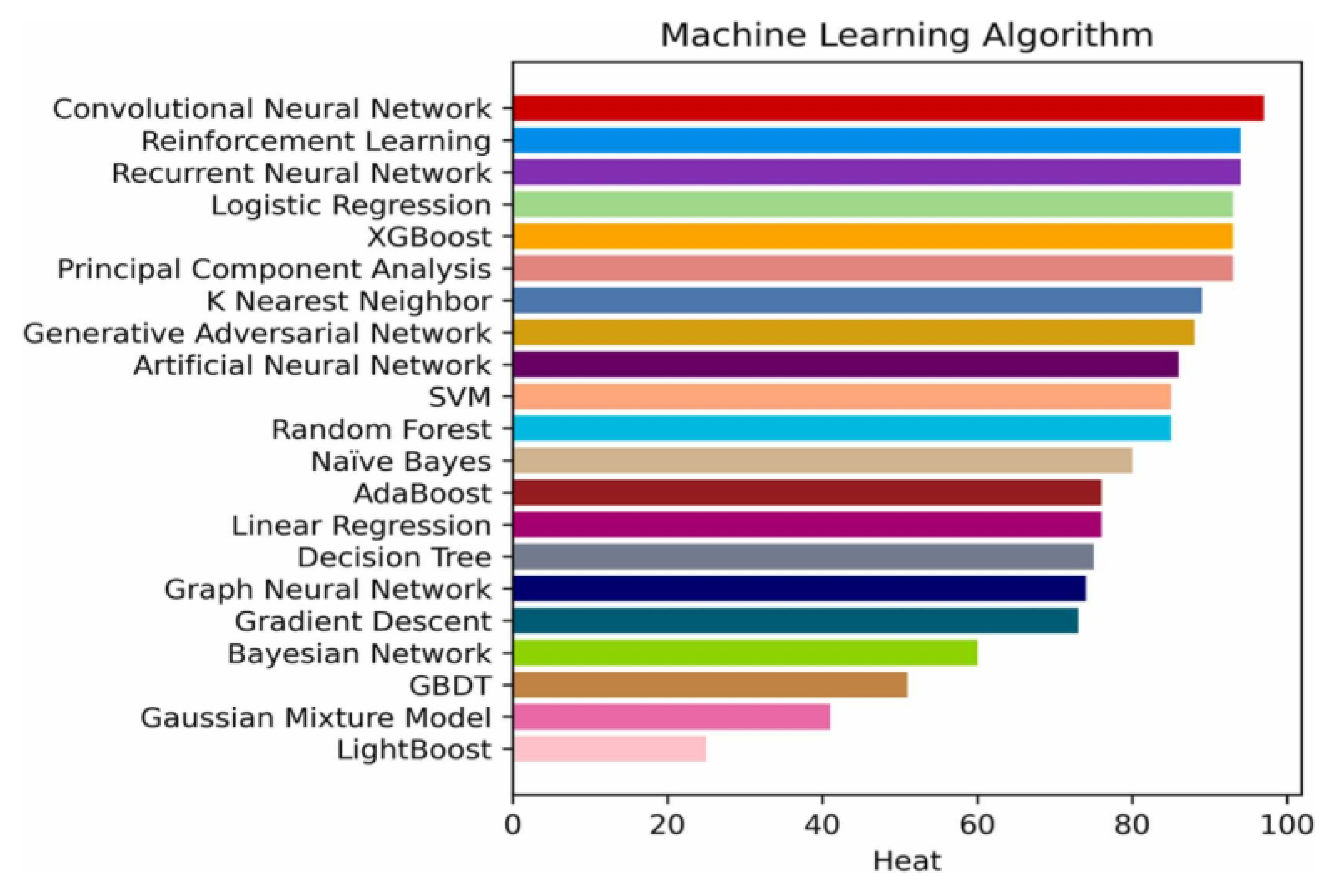
9]. AI describes various computer science approaches which help create systems that predict and classify data through existing information. AI functions with four essential elements being training databases and preprocessing methods together with model development algorithms in addition to pre-trained models which speed up development through existing knowledge integration [
10]. Machine learning techniques, such as convolutional neural networks (CNNs) [
11], reinforcement learning (RL) [
12], and recurrent neural network (RNN) [
13] are frequently employed in these models, as shown in
Figure 1.
AI-driven computer-aided diagnosis (CAD) systems demonstrate that deep learning have proven as valuable tools in lung disease diagnosis through medical imaging, providing highly accurate results [
14]. These deep learning model are designed with layered neural networks, allowing to process extensive datasets and generate progressively abstract representations of input data [
14]. Deep learning consists of three fundamental learning types which include unsupervised learning, reinforcement learning and supervised learning. A detailed assessment of AI-based systems for lung cancer monitoring and new treatment development and diagnostic prediction lies within this literature review. The study investigates technical along with clinical obstacles while it confirms optimal AI techniques and reports their achievement results. This review blends AI technology innovations with present clinical procedures to demonstrate the remarkable potential that AI holds for lung cancer diagnostic and treatment service provision worldwide in the developing nations.
2. Materials and Methods
2.1. Search Strategy
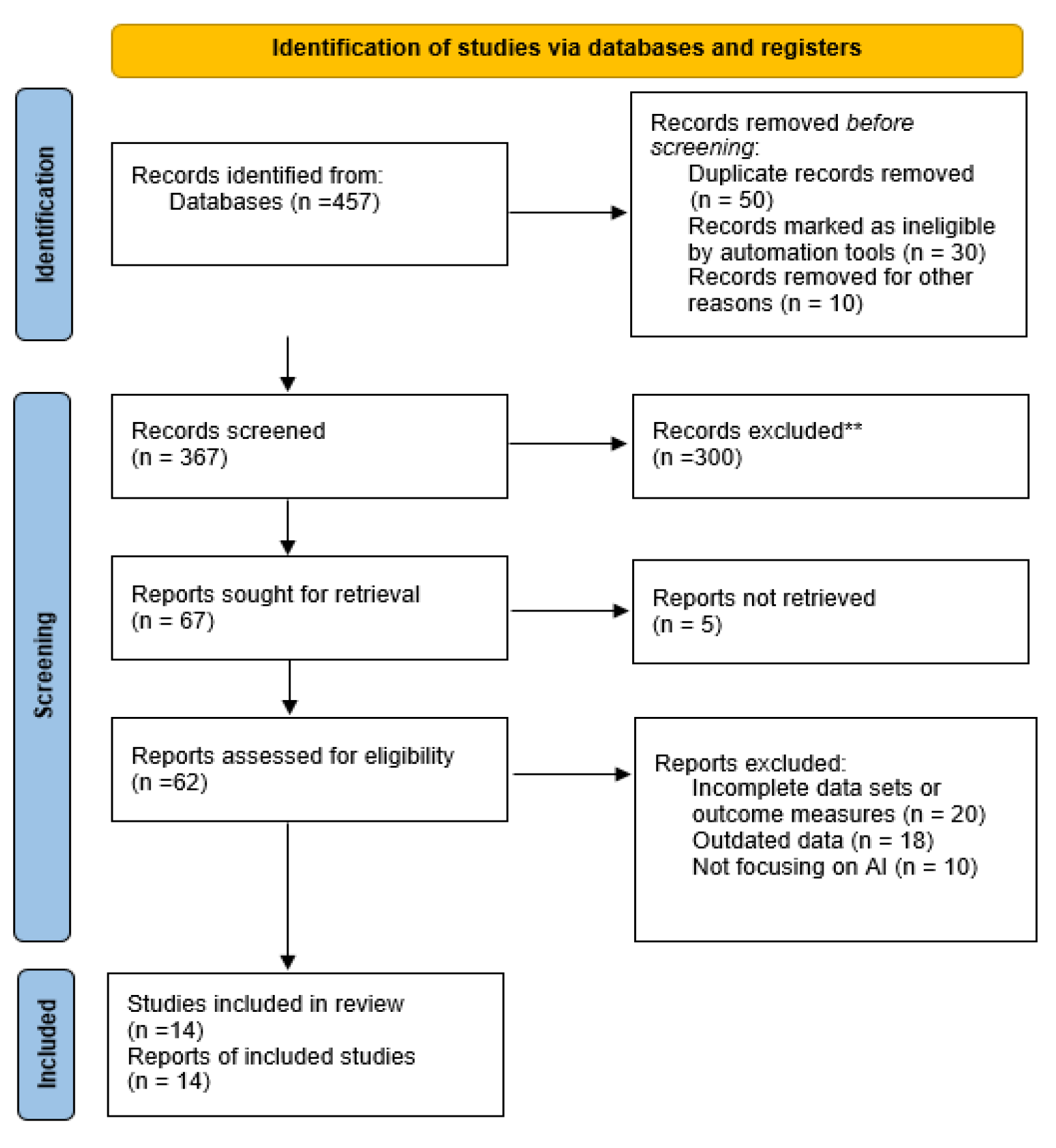
The evaluation of AI systems for early lung cancer detection followed all standards in the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) key 2020 guidelines [
15]. The PRISMA flow chart illustrates the stages of study selection including
Figure 2. Different researchers accessed 457 available publications through Scopus alongside PubMed and IEEE and Web of Science by implementing an established search approach. Screening and identification at the first stage resulted in the removal of 50 duplicate records along with 30 articles eliminated through automation instruments during subsequent qualification assessment. Moreover, 10 records were eliminated due to other different reasons, leaving 367 articles for further evaluation. Of these, 300 studies were eliminated because they failed to reach the criteria for inclusion. Subsequently, a total of 67 reports were obtained through retrieval requests which resulted in 62 accessible documents for eligibility assessment. After detailed review, 48 studies were excluded for reasons including lack of relevant AI or lung cancer information (20 studies), lack of sufficient original data (18 studies), and inadequate study designs (10 studies). A complete quantity of 14 studies which fulfilled all inclusion and exclusion standards were included in the qualitative research analysis of this systematic review.
2.2. Inclusion and Exclusion Criteria
These criteria established the boundaries for what studies would be included in the systematic review with the following parameters:
2.2.1. Inclusion Criteria
Only peer-reviewed studies that implemented AI-based technologies in the detection, diagnosis, or management of lung cancer were included. These studies must have employed machine learning or deep learning algorithms, with a focus on their application in digital pathology, radiology, or medical imaging. Studies involving human subjects diagnosed with or at risk for lung cancer, including both small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), were considered.
● Studies focused on animal models or non-lung cancers were excluded.
● The research analysis included studies which measured results using sensitivity, specificity, accuracy, predictive values and treatment outcomes derived from AI-based systems.
● The analysis included research studies which appeared from 2008 to 2023 to include only up-to-date, recent advancements and relevant AI applications.
● The search included only English-language publications because it restricted itself to articles available from English-language databases.
2.2.2. Exclusion Criteria
● Fewer studies were eligible for review because they either omitted focus on lung cancer-specific AI applications or provided insufficient data necessary to assess AI’s impact on the field (editorials, commentaries, and conference abstracts assessment was excluded).
● Frequent evaluation excluded research that concentrated on tumors different from lung cancer or lacked human subject involvement.
● The research analysis excluded studies with either immeasurable diagnostic results or unestablished diagnostic assessment parameters including specificity and sensitivity for AI-based diagnostic methods.
● Studies that failed to provide original data or with insufficient methodological details were excluded, also those failed to support independent verification and replication of research results.
2.3. Information Sources
The systematic review conducted its search through multiple research databases consisting of IEEE along with PubMed and Scopus and Web of Science to provide broad examination of suitable literature. The research plan followed Medical Subject Headings (MeSH) terminology as well as specific textual keywords and phrases related to the study objective. Secondary information discovery occurred by reviewing reference lists from identified articles for scholarly works which the original database search may have failed to index. The research was supported by further literature review through Google Scholar and relevant organizational websites to include high-quality academic publications. The research examined English-language scientific literature that appeared from 2008 until 2023.
3. Results
Recently, artificial intelligence has started to play a bigger role in finding and treating lung cancer. Despite being important, CT scans and X-rays of the chest can result in different evaluations by specialists because analyzing them can take a long time [
16,
17]. Specifically, these drawbacks become extremely important when considering the diagnosis of non-small cell lung cancer (NSCLC) which is responsible for most cases of lung cancer. NSCLC subtypes such as adenocarcinoma and squamous cell carcinoma must be distinguished, as this helps in guiding treatment, but doing this accurately is not easy using traditional methods.
AI-based tools were designed to address these challenges by using deep learning to highlight patterns in images the human eye could easily miss. Thanks to these models, medical tests are more uniform, quicker and more accurate. Algorithms that use multi-dimensional clustering help with spotting lesions at an earlier time [
18]. With these, human errors can be lowered and important diagnostic steps are completed more efficiently, so large-screening services are made more practical.
Currently, AI is being applied in more ways than just diagnosis. Predictive models help clinicians estimate how fast the tumor might spread and predict how a patient may respond to treatment which benefits the choice of treatment. Because AI performs routine data analysis, healthcare workers can concentrate more on what matters most—help for their patients.
The study summary that follows is based on the 14 papers that were selected for the review. The results are grouped into four important areas: looking at cells, making histopathological diagnoses, including tumor markers and using AI to help predict the outcome. Each part explains how AI affects diagnosis and measures accuracy using sensitivity, specificity and the model's overall impact.
3.1. Cytopathological Diagnosis of AI
Researcher Atsushi Teramoto [
19] presented deep convolutional neural networks (DCNN) which identify types of lung cancer within microscope images including adenocarcinoma and squamous cell carcinoma and small cell carcinoma. This deep convolutional neural network (DCNN) included three convolutional layers combined with three pooling layers and two fully connected layers for execution on graphics processing unit (GPU) after being trained on proprietary content. The images underwent preprocessing steps including cropping and down sampling to 256 × 256-pixel resolution then received augmentations by filtering methods along with rotation and flipping techniques thus enhancing model resistance against overfitting.
The evaluation process based on a threefold cross-validation revealed an average measurement accuracy of 71% which aligns with the established performance metrics of trained cytotechnologists and pathologists [
19]. This investigation demonstrates how DCNNs upgrade diagnostic efficiency along with accuracy for lung cancer detection in cytopathology thereby transforming classic diagnosis methods. This specific AI model shows superiority to traditional diagnostic methods while actively supporting their operations. The field of digital pathology receives novel innovation through deep learning (DL) technologies which demonstrate particular excellence during respiratory specimen diagnosis.
The research conducted by Taehee Kim [
20] focused on broader diagnostic accuracy enhancement for lung cancer through deep learning model application to respiratory tract cytological images within a retrospective multicenter study. A medical research team used a well-organized national picture database comprising cytology images from about 200 different healthcare institutions. Three steps comprised the imaging procedure where they created z-stacks to diminish phase differences in cell clusters while also conducting color normalization followed by converting images into 256×256-pixel patches [
20]. The research employed 30,590 images to create training sets of 27,362 images and validation sets of 2,928 images and separate testing sets of 1,272 images that corresponded to the entire slide images (WSIs). The DenseNet121 model achieved the best efficiency results from the tested six convolutional neural network models. Research metrics revealed exceptional results because pathologists achieved accuracy rates of 95.9% and 98.2% and 96.9% which exceeded the performance of three expert pathologists. Physicians achieved increased diagnostic accuracy when working with AI support that elevated their ability to 95.9% from 82.9% as measured by Fleiss' Kappa. The inter-rater agreement also improved substantially from 0.553 to 0.908 [
20]. The use of deep learning models demonstrates their capability to enhance the accuracy of lung cancer diagnoses in cytopathology while decreasing inter-observer variability and providing pathologists with standardized diagnosis assistance.
3.2. Histopathological Diagnosis of AI
In another pivotal study, Nicolas Coudray [
21] utilized the sophisticated Inception v3 deep learning model which stands among the most optimal convolutional neural networks (CNNs) for lung cancer diagnosis. The model showed competency in discriminating LUAD (lung adenocarcinoma), LUSC (lung squamous cell carcinoma) from normal lung tissue which demonstrated the capacity of AI to optimize histopathological diagnosis precision. The Inception v3 model achieved its training from The Cancer Genome Atlas (TCGA) whole-slide images which demonstrate how AI simplifies intricate histopathological operations while enhancing diagnostic precision. A remarkable aspect of this model was its diagnostic performance which led to an area under the curve (AUC) result of 0.97 at par with expert pathologists' accuracy reports [
21]. Cellular pathology experts can easily be surpassed by AI systems based on the results of recent analytical research which demonstrates superior performance in subtype lung cancer identification tasks. Progress is essential because it enables quicker and more accurate cancer identification leading to quicker start times of effective treatment plans.
The research conducted by Zhang Li produced essential findings regarding computational pathology specifically through studies on cancer pixel-level segmentation within whole-slide images (WSIs). An analysis evaluated the performance of ten deep learning-based computer-aided detection (CAD) methods for lung cancer segmentation through comparing their results on datasets containing 150 images for training and 200 images for testing [
23]. This study analyzed two types of approaches which it classified as multi-model and single-model approaches. The multi-model approach outperformed single-model methods with a higher Dice Coefficient value of 0.7966 versus the 0.7544 rating of single-model methods (p < 0.01). The research shows how deep learning helps pathologists to locate suspicious tissue areas in WSIs which enhances the speed of diagnosis and accuracy of lung cancer detection and enables prompt patient treatment.
3.3. Diagnosis of Lung Cancer Markers
The study of Yongjun Wu presented an artificial neural network (ANN) model which integrated tumor marker analysis in order to achieve better diagnoses of lung cancer. The research investigated various major tumor markers consisting of carcinoembryonic antigen and cancer antigen 125 and neuron-specific enolase together with beta2-microglobulin followed by gastrin then soluble interleukin-6 receptor and sialic acid and pseudouridine along with nitric oxide and multiple metal ions [
24]. Wu’s methodology conducted marker measurements between three groups which included lung cancer patients (50), patients with benign lung diseases (40) and healthy adults (50). The most suitable tumor markers were assembled by multiple logistic regression analysis to construct an intelligent diagnostic system that operated within an ANN. The ANN created system demonstrated superior performance compared to standard statistical techniques because it raised specificity from 72.0% to 100.0% while achieving overall accuracy improvement from 71.4% to 92.8%. Application of ANN systems leads to rapid highly accurate diagnoses of lung cancer which enables prompt qualified treatment procedures.
3.4. AI in the Prognosis of Lung Cancer
Teramoto studied cytological images from 76 lung cancer patients who had squamous cell carcinoma and adenocarcinoma and small cell carcinoma and more than four additional types. The CNN model performed feature extraction while reaching 71.1% accuracy in the analysis. The system validated its medical diagnosis competence by matching the accuracy statistics of human pathologists in biopsy analysis [
19].
Coudray analyzed 1,634 slice images retrieved from The Cancer Genome Atlas (TCGA) that included 1,176 tumor tissue and 459 normal tissue specimens in their separate research. The researchers distributed their images in the following ratios: training data at 70%, validation data at 15% and test data at 15%. Training Inception v3 required extensive image segmentation data while the testing and validation stages relied on transfer learning. The analysis model demonstrated a diagnostic performance equal to doctors by placing tissue samples into normal cell epithelium, squamous cell carcinoma or adenocarcinoma groups at a 97% accuracy rate [
21].
The researchers at Khosravi et al. designed their deep convolutional neural network model (DCNN) to separate squamous cell carcinoma from adenocarcinoma among benign and malignant tumors specifically. A high level of effectiveness was observed from the model since it correctly identified cancer types in high-resolution local magnification pictures while maintaining detection performance between 75% and 90% [
25]. The research shows that AI with particular emphasis on deep learning technology could play a vital role in future lung cancer diagnostic practices. AI tools function as valuable diagnostic instruments which help pathologists perform accurate type classifications of tumors.
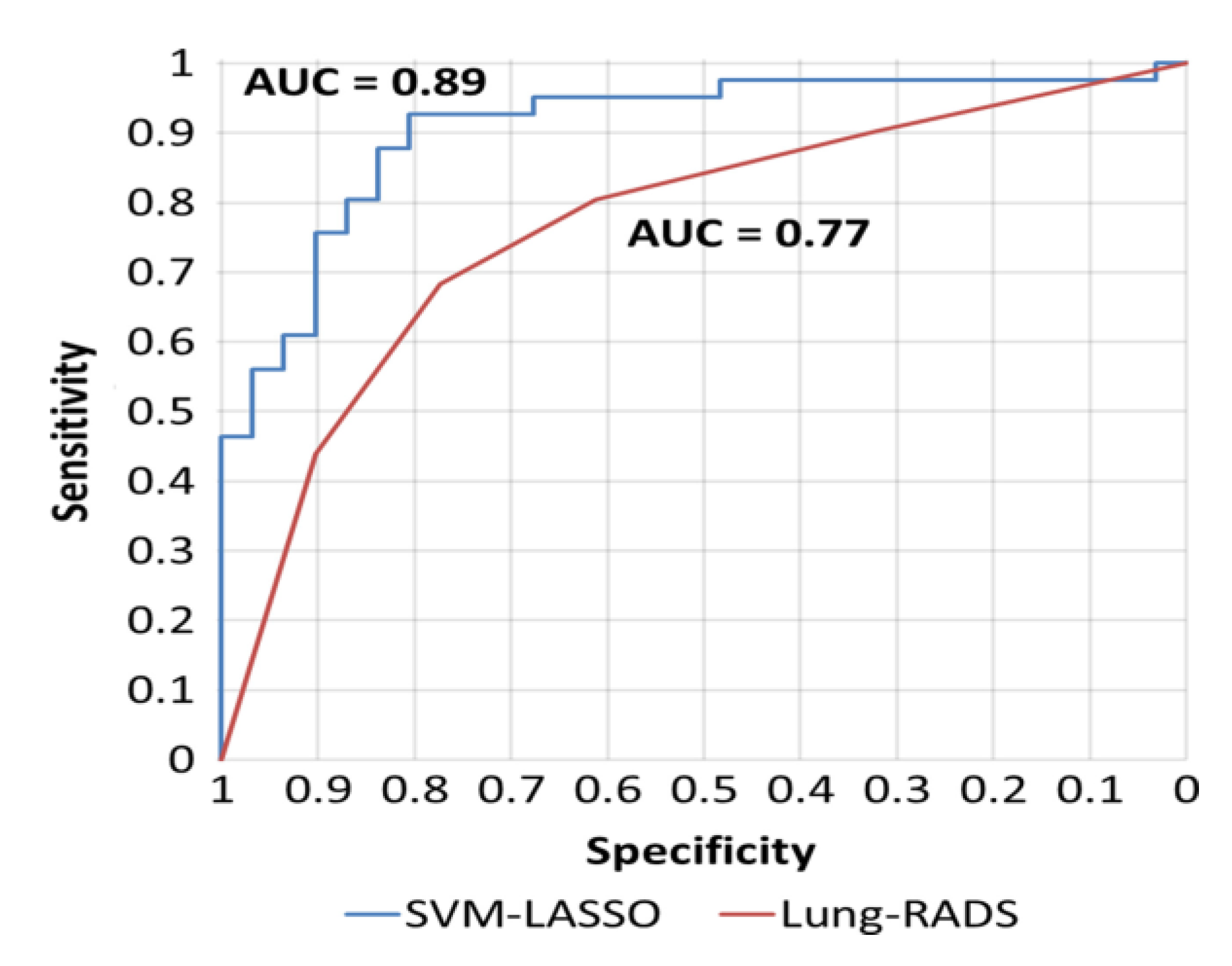
Wei Lu employed a radiomics prediction model to develop a classification tool for identifying small pulmonary nodules in low-dose CT scans, with the goal of enhancing lung cancer screening. His algorithm, designed to reduce the likelihood of missed findings based on the American College of Radiology's Lung-RADS, analyzed 72 nodules from the LIDC-IDRI repository, from which 85 radiomic features were extracted. The hierarchical clustering approach was applied to evaluate these features, followed by the use of an SVM regression model in conjunction with a LASSO technique. The proposed model's accuracy was rigorously tested through tenfold cross-validation, with 20 × 5- and 50 × 2-fold cross-validation procedures demonstrating its ability to provide more precise diagnoses in the early stages of lung cancer compared to conventional diagnostic methods, as illustrated in
Figure 3 [
22].
Marliese Alexander designed an AI system to assess cancer patients' eligibility for clinical trials, achieving a remarkable exclusion risk accuracy of 95.7% and a general eligibility assessment accuracy of 91.6% [
26]. Despite the system’s high effectiveness, clinical oversight remains necessary. The AI tool displayed a recall (sensitivity) of 83.3%, precision (positive predictive value) of 76.5%, and specificity of 93.8%. This system exemplifies the potential of AI as a component of clinical decision support systems, particularly in improving the prescreening process for identifying patients eligible for clinical trials.
Nasrullah Nasrullah introduced a cutting-edge deep learning-based model for diagnosing malignant lung nodules, utilizing 3D Customized Mixed Link Networks (CMixNet) for both nodule detection and classification [
27]. Faster R-CNN was used to detect the CMixNet features because it was able to learn them, and integration was achieved by using a CNN. Classification was achieved by using a Gradient Boosting Machine where the CMixNet features were analyzed. Additionally, these strategies blended in physiological signs and clinical markers in order to improve the accuracy of the diagnoses made by reducing the number of false positives. This system’s integration with IoT devices for monitoring patients continuously represents a significant leap forward because it makes it possible to securely observe patients' conditions from a distance, which in turn might enhance the accuracy of the diagnoses made.
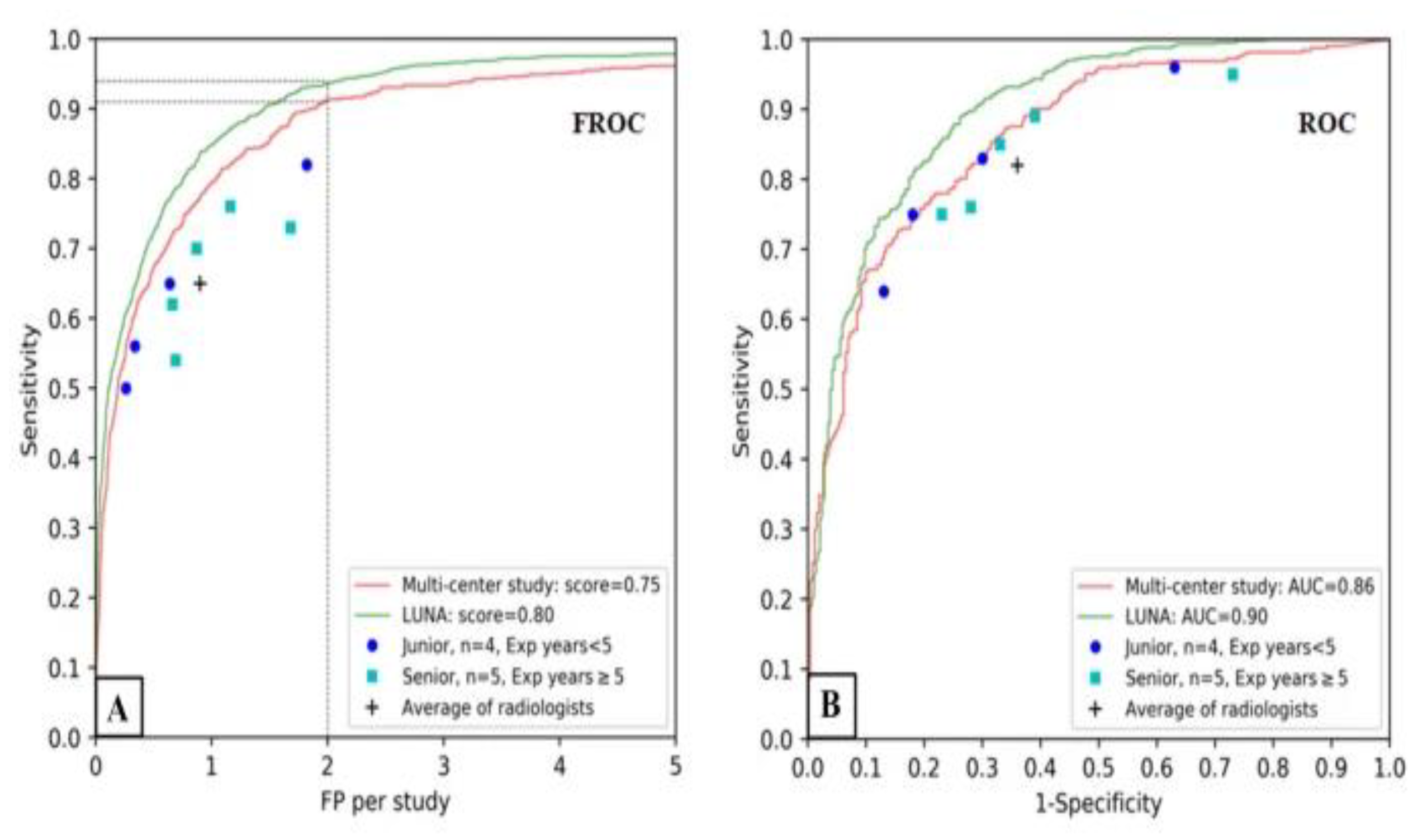
Sijia Cui’s research was in the development and assessment of a deep learning (DL) algorithm for detecting pulmonary nodules (PNs) in low-dose computed tomography (LDCT) scans while checking the existence of these nodules in China. The performance of the algorithm versus radiologists was evaluated with FROC score, FOC-AUC, and the average time to detection of nodules [
28]. For the inter-study analysis, the concordance between the DL method and the diagnostic reference standard for the detecting positive nodules was established using the Bland–Altman approach. With the aid of a public database, the trial was able to conduct external validation with the use of the LUNA system. Cui also studied the number, site, and features of benign lung nodules detected by 2 radiologists, which helped in better understanding the epidemiology as well as the diagnostic dilemmas of pulmonary nodules.
Figure 4 demonstrates the performance of the DL algorithm.
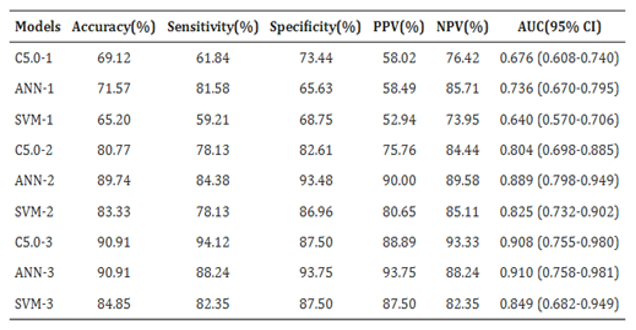
Shuying Duan constructed a multi-layered diagnostic system, consisting of three different machine learning models: C5.0 decision trees, artificial neural networks, and support vector machines, for the diagnosis of lung cancer. Examining the area under curve (AUC) served both as a model validation metric as well as a measure of decision-making effectiveness of the models [
29]. Shuying Duan constructed a multi-layered diagnostic system, consisting of three different machine learning models: C5.0 decision trees, artificial neural networks, and support vector machines, for the diagnosis of lung cancer. Examining the area under curve (AUC) served both as a model validation metric as well as a measure of decision-making effectiveness of the models [
29]. Of the rest, ANN was most accurate, scoring 0.736, C5.0 was next at 0.676 and SVM was last at 0.640. In the second layer, the ANN performance was better than SVM where AUC were 0.804, 0.889, and 0.825 obtained by 1, 2, and 3 level classifiers respectively. At the third level, however, SVM got the highest score of AUC 0.910, while ANN was very close at 0.908 and C5.0 had a lower score of 0.849. C5.0 was more efficient, achieving the highest sensitivity score of 94.12%. The initial patient group was selected based on 14 criteria, incorporating epidemiological data and clinical diagnoses. This group served as input into the ANN model, with biomarkers employed as auxiliary diagnostic tools to identify suspected lung cancer patients. Furthermore, C5.0 identified lung cancer using 22 CT nodule-based radiomic markers, contributing to a comprehensive, multi-stage AI-driven diagnostic approach. The performance of the machine learning models in the testing set is displayed in
Table 2.
- A.
Meta-analysis
A comprehensive review was conducted using keyword searches across multiple databases for peer-reviewed articles published up to April 2024. This review focused on studies detailing the effectiveness of AI in lung cancer detection through imaging, specifically those reporting sensitivity, specificity, and associated confidence intervals (
Table 1). The meta-analysis included 18 studies that examined various AI- based diagnostic systems applied across different imaging modalities.
4. Discussion
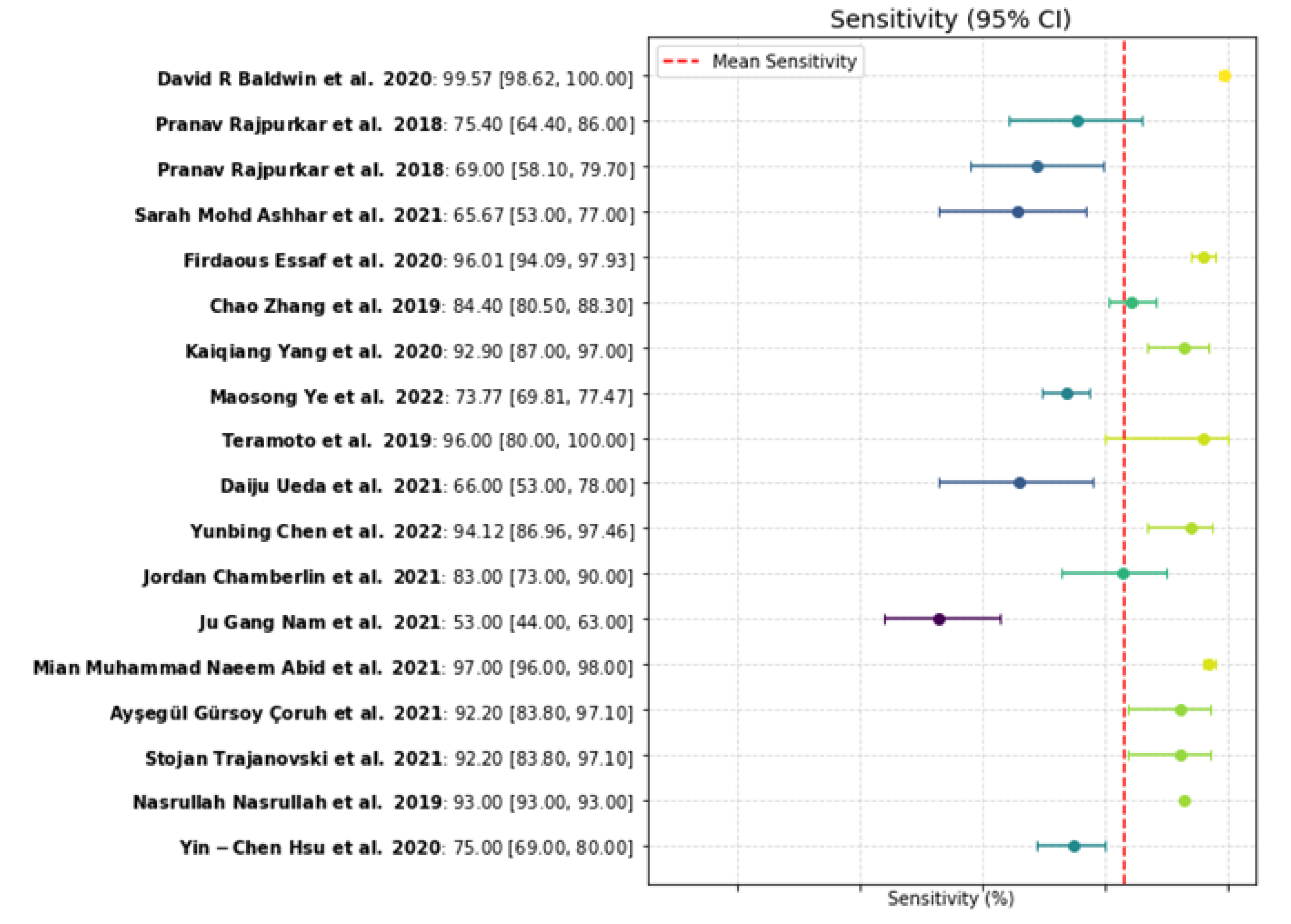
The results revealed that AI models exhibited a sensitivity range of 53.00% to 99.57%, with a median value close to 92%, indicating that the algorithms generally performed well when applied to well-prepared datasets. Interestingly, the specificity values varied widely, ranging from 28.03% to 100.00%. The median specificity value was approximately 85%, suggesting that in most cases, the models correctly differentiated between benign and malignant nodules. One key reason for this variability lies in the differences between studies, particularly in terms of the types of AI models used, the sizes of the training datasets, and the evaluation methodologies. Larger and more diverse training datasets generally led to improved performance of machine learning models.
Four key performance metrics were assessed to evaluate the performance of AI systems in detecting lung cancer: True Positives (TP), True Negatives (TN), False Positives (FP), and False Negatives (FN).
● True Positives (TP): Cases where the AI correctly identified cancer as the underlying cause of disease.
● True Negatives (TN): Cases where the AI correctly identified the absence of cancer in non-cancerous instances.
● False Positives (FP): Cases where the AI misdiagnosed normal cases as cancerous.
● False Negatives (FN): Instances when the AI system failed to identify a patient that had cancer.
Sensitivity and specificity are crucial metrics in assessing the efficacy of diagnostic models. In this study, sensitivity (the proportion of positive samples correctly detected) and specificity (the proportion of correctly identified negative cases) were plotted as a forest plot, providing a visual summary of the variance and confidence intervals for these parameters across different studies. This visualization captures the diagnostic accuracy and flexibility of the models across various clinical contexts and populations [
75].
Sensitivity and Specificity are calculated as follows:
Lung cancer, originating from the bronchial mucosa lining or mucus glands, remains a leading cause of mortality worldwide. Currently, CT scanning is the primary tool employed for lung cancer screening on a global scale. CT scans provide critical insights, particularly when tumors are located 43 shallow learning techniques, particularly in the analysis of pathological images. The key benefits of deep learning include its ability to define complex attributes with ease, its advanced object recognition capabilities, its efficiency in parallel processing, and its use of transfer learning, which facilitates the application of prior knowledge to new diagnostic problems. These advantages make deep learning an especially powerful tool in lung cancer diagnostics.
AI makes crucial advancements in lung cancer diagnostics through decreased human dependence on subjectivity in interpretation tasks. AI models utilizing convolutional neural networks (CNNs) together with deep learning techniques demonstrate better performance compared to human experts because they maintain uniform results in various datasets and clinical conditions. The indicated review establishes AI's value in creating standard diagnostic outcomes because it minimizes the longstanding problem of inconsistent doctor interpretations. The deployment of deep convolutional neural networks demonstrates AI systems can provide a more efficient cytopathological diagnostic process and minimize diagnostic mistakes. Deep learning systems such as DenseNet121 demonstrate accurate performance that helps professional pathologists improve outcomes and serves as a pathway to create standardized diagnostic procedures in healthcare institutions.
Early lung cancer detection serves as a vital factor to increase survival rates because the outcome becomes increasingly negative with progressive stages of the disease. Inability to detect small nodules and to distinguish malignant from benign lesions would remain hurdles for health personnel involved in the early stages of cancer, despite the marvelous capabilities of CT scanning technology. Therefore, the important relevance of AI comes into play. The radiomic analysis performed by AI algorithms almost certainly assists in furthering early detection of lung cancer since they are adept at patterns that are far too complex for human beings to recognize.
Simultaneously, through rapid and accurate assessment of massive datasets, AI empowers radiologists to recognize early stages of cancer while also eliminating bottleneck in medical imaging. AI is a practical tool in the hands of healthcare providers to solve resource shortages stalked by increasing patient demands that limit expert radiologist availability. AI implementation is now extended from diagnostic medicine into personalized health care. Current machine learning methods show promise in tumor detection, prognosis of tumor growth patterns, and tumor behavior.
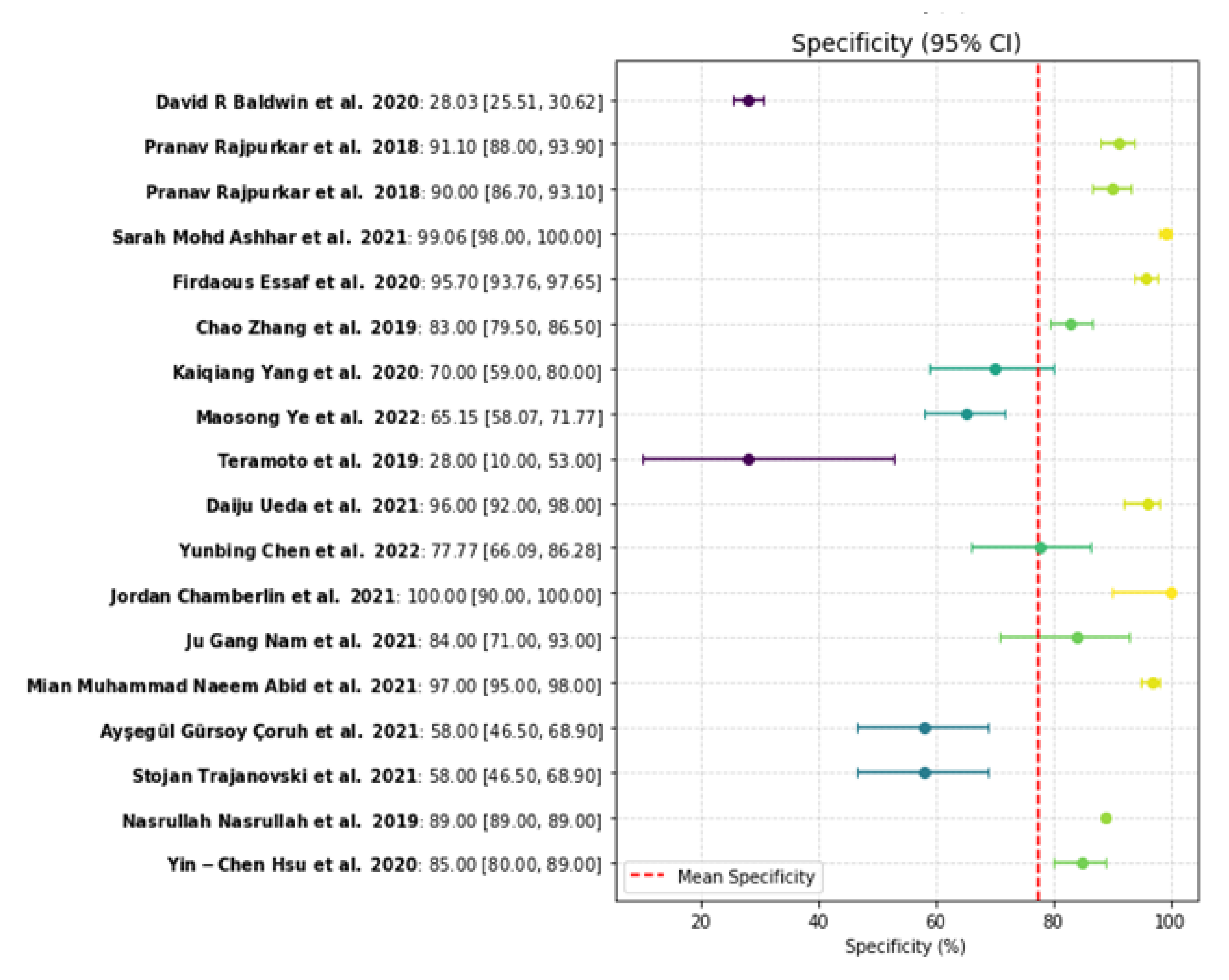
Figure 5.
Specificity plot [
19,
27,
40,
61,
62,
63,
64,
65,
66,
67,
68,
69,
70,
71,
72,
73,
74].
Figure 5.
Specificity plot [
19,
27,
40,
61,
62,
63,
64,
65,
66,
67,
68,
69,
70,
71,
72,
73,
74].
Figure 6.
Sensitivity plot [
19,
27,
40,
61,
62,
63,
64,
65,
66,
67,
68,
69,
70,
71,
72,
73,
74].
Figure 6.
Sensitivity plot [
19,
27,
40,
61,
62,
63,
64,
65,
66,
67,
68,
69,
70,
71,
72,
73,
74].
The value of science-based prognostics in treatment planning comes into play when it is designed to consider specific patient needs. Blending patient physiology and visual assessments is the key function of AI, which allows physicians to build individualized tumor strategies for a conceptual leap in individualized oncological care.The practical deployment of AI still faces several challenges. A key issue is the variation in AI model performance observed across different studies, largely due to the quality and diversity of training datasets. Models developed with limited or homogeneous data often underperform in real-world clinical scenarios. Researchers emphasize the need for larger, more diverse training sets, yet varying levels of sensitivity and specificity continue to pose problems. Additionally, differences in imaging protocols and AI algorithms make direct comparisons difficult and hinder the establishment of standardized AI solutions in healthcare settings. Another significant drawback is the "black box" nature of many AI models, including deep learning systems, which obscures the rationale behind specific diagnostic outcomes from clinical staff. The lack of system transparent operation creates ethical problems for practitioners who need to understand AI-based recommendations before they trust them. To overcome this challenge there needs to be more development of explainable AI methods which create transparent models for healthcare professionals to understand and trust AI diagnostic output.
The introduction of AI in lung cancer diagnostics activates multiple ethical concerns for evaluation. The evidence demonstrates that AI offers enhanced diagnosis capabilities and lower workloads to professionals yet human practitioners maintain the control to make final decisions. AI instruments should serve to assist medical professionals instead of replacing them in their tasks. The healthcare industry remains in an ongoing process to establish regulatory guidelines to handle AI in healthcare including data protection measures and requirements for algorithm transparency alongside minimizing AI system biases during model use. Widely used AI systems in clinical practice require testing and design processes to eliminate bias from algorithms and guarantee patient information security.
Advancement in technology necessitates various development research programs targeting AI. AI models at successive levels need two types of data: diagnostic imaging alongside pathology data in combination with full patient medical records and genomic details as well as ongoing healthcare monitoring through wearable medical devices. Collective work between multiple branches of medical science will develop AI systems to achieve stronger diagnostic and predictive strength. Modern healthcare prevention and medical treatment are likely to improve significantly through the creation of real-time monitoring systems. AI technology that analyzes immunotherapy and targeted therapy responses to predict treatment outcomes provides substantial value for maximizing medical results for individual patients.
Table 3.
Overview of pathology imaging datasets.
Table 3.
Overview of pathology imaging datasets.
| Source |
Dataset |
Release Date |
Sample size |
Number of Images |
Image Modality |
Image Dimension |
Image Format |
Ground Truth Availability |
| Nusraat Nawreen et al. [50] |
Cancer
Imaging Archive |
2021 |
NS |
NS |
CT |
256 x 256 |
DICOM |
No |
| Zhang Li et al. [23] |
The ACDC@LungHP |
2021 |
200 |
200 |
WSI |
768 x 768 x 3 |
H&E |
Yes |
| Ahmed S. Sakr [51] |
LC25000 |
2022 |
25 |
25000 |
Histopathological images |
768 x 768 |
JPEG |
Yes |
| Xi Wang et al. [52] |
The Cancer Genome Atlas (TCGA) |
2020 |
112 |
112 |
WSI |
244 x 244 x 3 |
JPEG RGB |
Yes |
| Nowshin Tasnim et al. [53] |
IQ-OTH lung cancer |
2024 |
1097 |
32717 |
CT |
512 × 512, 512×623, 404 × 511 |
NS |
Yes |
| Yoganand Balagurunathan et al. [54] |
ISBI 2018 Lung Nodule Malignancy Prediction test set |
2021 |
1593 |
100 |
LDCT |
3D scans |
DICOM, DICOM-RT, NIfTI |
Yes |
| D. Jayaraj and S. Sathiamoorthy [55] |
Lung Image Database Consortium (LIDC) |
2019 |
NS |
1018 |
CT |
512 x 512 |
JPEG Grayscale (transformed from DICOM format) |
Yes |
| Sayyada Hajera Begum et al. [56] |
Lung Cancer CT Scan Images Dataset |
2022 |
110 |
800 |
CT |
256x256 |
NS |
Yes |
| Worawate Ausawalaithong et al., [57] |
JSRT Dataset |
2018 |
247 |
247 |
X-ray |
2048 x 2048 |
NS |
Yes |
| Worawate Ausawalaithong et al., [57] |
ChestX-ray14 Dataset |
2018 |
112120 |
112120 |
X-ray |
1024 x 1024 |
NS |
Yes |
| Juan Cañada et al. [58] |
LC25000 dataset |
2019 |
25 |
1200 |
Histopathological images |
512 x 512 |
IMG |
Yes |
| Massion, P. P., et al. [59] |
NLST dataset |
2020 |
463 |
14761 |
CT |
0.25x 0.25 x 1 |
NS |
Yes |
| Rohit Y. Bhalerao et al. [60] |
LIDC (Lung Image Database Consortium) |
2019 |
910 |
910 |
CT |
256x256 |
DICOM, JPEG,PNG |
Yes |
5. Conclusions
The proposed review study examined how AI-based approaches can enhance lung cancer diagnosis through digital pathology systems and CT-based imaging analyses. The findings highlight that AI enables more precise diagnoses by delivering standardized evaluations, thereby reducing the variability inherent in traditional medical approaches. Lung cancer screening becomes more effective as deep learning models embedded in AI algorithms demonstrate superior diagnostic performance compared to current medical standards. Given the global shortage of onco-pathologists, technological interventions such as AI-generated diagnoses and reports have the potential to significantly reduce their cognitive workload. In such a framework, the role of pathologists could shift toward overseeing and validating AI-generated outputs, rather than performing every diagnostic task manually. However, several important limitations must be acknowledged. The performance of AI models is directly dependent on the quality and diversity of their training datasets. Many existing studies rely on relatively small datasets derived from homogeneous populations, resulting in limited generalizability to broader and more diverse patient cohorts. Additionally, the standardization of AI system implementation in clinical environments is adversely affected by inconsistencies in performance metrics, imaging protocols, and AI processing techniques across studies. Furthermore, the limited transparency of AI systems, particularly deep learning algorithms, presents challenges for healthcare practitioners, who may hesitate to adopt these “black box” solutions. Finally, the deployment of AI in clinical practice must navigate significant ethical and regulatory challenges, including the protection of patient privacy and the mitigation of algorithmic biases.
The successful integration of AI technology into lung cancer assessment requires addressing several critical factors. Developing comprehensive AI models requires the use of complete and detailed datasets, particularly through federated databases that ensure privacy and security while integrating real-time monitoring data, genomic profiles, and comprehensive clinical histories. Such datasets are essential for enhancing the precision, robustness, and generalizability of AI systems. The implementation of explainable AI is equally vital, as it fosters physician trust and facilitates the safe and effective deployment of these technologies in clinical practice. Furthermore, advancing research on AI’s capacity to predict treatment outcomes and optimize personalized therapeutic strategies will significantly enhance its impact on patient care. Ultimately, realizing the full clinical potential of AI in lung cancer diagnostics depends on systematically addressing its current limitations while pursuing targeted research and technological advancements.
Author Contributions
“Conceptualization, Bharat Jasani and Prashant K. Jamwal.; methodology, Akim Kapsalyamov; formal analysis, Ingkar Chegedekova and Akim Kapsalyamov; investigation, Ingkar Chegedekova; resources, Ingkar Chegedekova; data curation, Ingkar Chegedekova; writing—original draft preparation, Ingkar Chegedekova, Akim Kapsalyamov and Prashant K. Jamwal.; writing—review and editing, Prashant K. Jamwal, Dimitris Parthimos, and Bharat Jasani; supervision, Prashant K. Jamwal, and Bharat Jasani; project administration, Prashant K. Jamwal; funding acquisition, Prashant K. Jamwal. All authors have read and agreed to the published version of the manuscript.”
Funding
This research was funded by the Science Committee of the Ministry of Science and Higher Education of the Republic of Kazakhstan, grant number BR27199433.
Institutional Review Board Statement
The study did not require ethical approval.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors have reviewed and edited the output and take full responsibility for the content of this publication.”
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results”.
References
- C. S. Dela Cruz, L. T. Tanoue, and R. A. Matthay, “Lung cancer: epidemiology, etiology, and prevention,” Clin Chest Med, vol. 32, no. 4, pp. 605-44, Dec, 2011.
- D. Yessenbayev, and Z. Khamidullina, “Epidemiology of Lung Cancer in Kazakhstan: Trends and Geographic Distribution,” vol. 24, no. 5, pp. 1521-1532, May 1, 2023. [CrossRef]
- J. A. Barta, C. A. Powell, and J. P. Wisnivesky, “Global Epidemiology of Lung Cancer,” Ann Glob Health, vol. 85, no. 1, Jan 22, 2019.
- G. J. Amir, and H. P. Lehmann, “After Detection:: The Improved Accuracy of Lung Cancer Assessment Using Radiologic Computer-aided Diagnosis,” Academic Radiology, vol. 23, no. 2, pp. 186-191, 2016/02/01/, 2016.
- M. J. van den Bent, “Interobserver variation of the histopathological diagnosis in clinical trials on glioma: a clinician’s perspective,” Acta Neuropathologica, vol. 120, no. 3, pp. 297-304, 2010/09/01, 2010.
- H.-Y. Chiu, H.-S. Chao, and Y.-M. Chen, "Application of Artificial Intelligence in Lung Cancer," Cancers, 14, 2022]. [CrossRef]
- D. Li, Z. Li, S. Li, H. Zhang, S. Yao, Y. Li, and J. Chen, "Development and Validation of a Prediction Model for Positive Findings of Preoperative Flexible Bronchoscopy in Patients with Peripheral Lung Cancer," Current Oncology, 30, 2023].
- R. S. Winokur, B. B. Pua, B. W. Sullivan, and D. C. Madoff, “Percutaneous lung biopsy: technique, efficacy, and complications,” Semin Intervent Radiol, vol. 30, no. 2, pp. 121-7, Jun, 2013.
- X. Yin, H. Liao, H. Yun, N. Lin, S. Li, Y. Xiang, and X. Ma, “Artificial intelligence-based prediction of clinical outcome in immunotherapy and targeted therapy of lung cancer,” Seminars in Cancer Biology, vol. 86, pp. 146-159, 2022/11/01/, 2022. [CrossRef]
- G. G, J. Thimmiaraja, C. J. Shelke, G. Pavithra, V. K. Sharma, and D. Verma, "Deep Learning with Unsupervised and Supervised Approaches in Medical Image Analysis." pp. 1580-1584.
- M. Šarić, M. Russo, M. Stella, and M. Sikora, "CNN-based Method for Lung Cancer Detection in Whole Slide Histopathology Images." pp. 1-4.
- Y. Wang, Q. Zhang, L. Ying, and C. Zhou, “Deep Reinforcement Learning for Early Diagnosis of Lung Cancer,” Proceedings of the AAAI Conference on Artificial Intelligence, vol. 38, no. 20, pp. 22410-22419, 03/24, 2024.
- D. Moitra, and R. K. Mandal, “Automated AJCC (7th edition) staging of non-small cell lung cancer (NSCLC) using deep convolutional neural network (CNN) and recurrent neural network (RNN),” Health Inf Sci Syst, vol. 7, no. 1, pp. 14, Dec, 2019.
- L. Wang, “Deep Learning Techniques to Diagnose Lung Cancer,” Cancers (Basel), vol. 14, no. 22, Nov 13, 2022. [CrossRef]
- J. P. Matthew, E. M. Joanne, M. B. Patrick, B. Isabelle, C. H. Tammy, D. M. Cynthia, S. Larissa, M. T. Jennifer, A. A. Elie, E. B. Sue, C. Roger, G. Julie, M. G. Jeremy, H. Asbjørn, M. L. Manoj, L. Tianjing, W. L. Elizabeth, M.-W. Evan, M. Steve, A. M. Luke, A. S. Lesley, T. James, C. T. Andrea, A. W. Vivian, W. Penny, and M. David, “The PRISMA 2020 statement: an updated guideline for reporting systematic reviews,” BMJ, vol. 372, pp. n71, 2021.
- J. R. Molina, P. Yang, S. D. Cassivi, S. E. Schild, and A. A. Adjei, “Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship,” Mayo Clin Proc, vol. 83, no. 5, pp. 584-94, May, 2008.
- S. Blandin Knight, P. A. Crosbie, H. Balata, J. Chudziak, T. Hussell, and C. Dive, “Progress and prospects of early detection in lung cancer,” Open Biol, vol. 7, no. 9, Sep, 2017.
- I. Castiglioni, F. Gallivanone, P. Soda, M. Avanzo, J. Stancanello, M. Aiello, M. Interlenghi, and M. Salvatore, “AI-based applications in hybrid imaging: how to build smart and truly multi-parametric decision models for radiomics,” European Journal of Nuclear Medicine and Molecular Imaging, vol. 46, no. 13, pp. 2673-2699, 2019/12/01, 2019.
- A. Teramoto, T. Tsukamoto, Y. Kiriyama, and H. Fujita, “Automated Classification of Lung Cancer Types from Cytological Images Using Deep Convolutional Neural Networks,” BioMed Research International, vol. 2017, pp. 4067832, 2017/08/13, 2017. [CrossRef]
- T. Kim, H. Chang, B. Kim, J. Yang, D. Koo, J. Lee, J. W. Chang, G. Hwang, G. Gong, N. H. Cho, C. W. Yoo, J. Y. Pyo, and Y. Chong, “Deep learning-based diagnosis of lung cancer using a nationwide respiratory cytology image set: improving accuracy and inter-observer variability,” Am J Cancer Res, vol. 13, no. 11, pp. 5493-5503, 2023.
- N. Coudray, P. S. Ocampo, T. Sakellaropoulos, N. Narula, M. Snuderl, D. Fenyö, A. L. Moreira, and N. Razavian, “Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning,” vol. 24, no. 10, pp. 1559-1567, Oct, 2018.
- W. Choi, J. H. Oh, S. Riyahi, C. J. Liu, F. Jiang, W. Chen, C. White, A. Rimner, J. G. Mechalakos, J. O. Deasy, and W. Lu, “Radiomics analysis of pulmonary nodules in low-dose CT for early detection of lung cancer,” Med Phys, vol. 45, no. 4, pp. 1537-1549, Apr, 2018.
- Z. Li, J. Zhang, T. Tan, X. Teng, X. Sun, H. Zhao, L. Liu, Y. Xiao, B. Lee, Y. Li, Q. Zhang, S. Sun, Y. Zheng, J. Yan, N. Li, Y. Hong, J. Ko, H. Jung, Y. Liu, Y. c. Chen, C. w. Wang, V. Yurovskiy, P. Maevskikh, V. Khanagha, Y. Jiang, L. Yu, Z. Liu, D. Li, P. J. Schüffler, Q. Yu, H. Chen, Y. Tang, and G. Litjens, “Deep Learning Methods for Lung Cancer Segmentation in Whole-Slide Histopathology Images—The ACDC@LungHP Challenge 2019,” IEEE Journal of Biomedical and Health Informatics, vol. 25, no. 2, pp. 429-440, 2021. [CrossRef]
- Y. Wu, Y. Wu, J. Wang, Z. Yan, L. Qu, B. Xiang, and Y. Zhang, “An optimal tumor marker group-coupled artificial neural network for diagnosis of lung cancer,” Expert Systems with Applications, vol. 38, no. 9, pp. 11329-11334, 2011/09/01/, 2011.
- P. Khosravi, E. Kazemi, M. Imielinski, O. Elemento, and I. Hajirasouliha, “Deep Convolutional Neural Networks Enable Discrimination of Heterogeneous Digital Pathology Images,” EBioMedicine, vol. 27, pp. 317-328, 2018/01/01/, 2018.
- M. Alexander, B. Solomon, D. L. Ball, M. Sheerin, I. Dankwa-Mullan, A. M. Preininger, G. P. Jackson, and D. M. Herath, “Evaluation of an artificial intelligence clinical trial matching system in Australian lung cancer patients,” JAMIA Open, vol. 3, no. 2, pp. 209-215, 2020. [CrossRef]
- N. Nasrullah, J. Sang, M. S. Alam, M. Mateen, B. Cai, and H. Hu, "Automated Lung Nodule Detection and Classification Using Deep Learning Combined with Multiple Strategies," Sensors, 19, 2019].
- S. Cui, S. Ming, Y. Lin, F. Chen, Q. Shen, H. Li, G. Chen, X. Gong, and H. Wang, “Development and clinical application of deep learning model for lung nodules screening on CT images,” Scientific Reports, vol. 10, no. 1, pp. 13657, 2020/08/12, 2020.
- S. Duan, H. Cao, H. Liu, L. Miao, J. Wang, X. Zhou, W. Wang, P. Hu, L. Qu, and Y. Wu, “Development of a machine learning-based multimode diagnosis system for lung cancer,” Aging (Albany NY), vol. 12, no. 10, pp. 9840-9854, May 23, 2020. [CrossRef]
- G. M. Kalkan, B. Dönmez, Y. C. Tok, S. Gürel, E. AkdaĞliŞ, H. Çapkan, M. Z. Kaya, D. Kandaz, M. K. Uçar, and S. N. Şenol, "Diagnosis of Lung Cancer with Hybrid Artificial Intelligence Method." pp. 1-4.
- S. Das, G. L, S. J. Prakash, N. S. Dey, J. Panuganti, and R. Poojitha, "Lung Cancer Detection and Classification using Transfer Learning with Pre-trained VGG19 Convolutional Neural Networks." pp. 1-6.
- A. Alomar, M. Alazzam, H. Mustafa, and A. Mustafa, "Lung Cancer Detection Using Deep Learning and Explainable Methods." pp. 1-4.
- B. Gajera, S. R. Kapil, D. Ziaei, J. Mangalagiri, E. Siegel, and D. Chapman, “CT-Scan Denoising Using a Charbonnier Loss Generative Adversarial Network,” IEEE Access, vol. 9, pp. 84093-84109, 2021. [CrossRef]
- D. Hişam, and E. Hişam, "Deep learning models for classifying cancer and COVID-19 lung diseases." pp. 1-4.
- M. S. Ahmed, K. N. Iqbal, and M. G. R. Alam, "Interpretable Lung Cancer Detection using Explainable AI Methods." pp. 1-6.
- M. Mamun, A. Farjana, M. A. Mamun, and M. S. Ahammed, "Lung cancer prediction model using ensemble learning techniques and a systematic review analysis." pp. 187-193.
- R. P. R. Kumar, S. Polepaka, D. Likithasree, and S. Keerthika, "An Investigation on CNN-based Lung Cancer Prediction Method." pp. 1-5.
- P. S. Bharathi, and T. P. K. R, "Neural Network based Earlier Stage Lung Cancer Prediction Scheme with Differential Learning Assistance." pp. 01-06.
- F. Ciompi, K. Chung, S. J. van Riel, A. A. A. Setio, P. K. Gerke, C. Jacobs, E. T. Scholten, C. Schaefer-Prokop, M. M. W. Wille, A. Marchianò, U. Pastorino, M. Prokop, and B. van Ginneken, “Towards automatic pulmonary nodule management in lung cancer screening with deep learning,” Scientific Reports, vol. 7, no. 1, pp. 46479, 2017/04/19, 2017.
- C. Zhang, X. Sun, K. Dang, K. Li, X. w. Guo, J. Chang, Z. q. Yu, F. y. Huang, Y. s. Wu, Z. Liang, Z. y. Liu, X. g. Zhang, X. l. Gao, S. h. Huang, J. Qin, W. n. Feng, T. Zhou, Y. b. Zhang, W. j. Fang, M. f. Zhao, X. n. Yang, Q. Zhou, Y. l. Wu, and W. z. Zhong, “Toward an Expert Level of Lung Cancer Detection and Classification Using a Deep Convolutional Neural Network,” The Oncologist, vol. 24, no. 9, pp. 1159-1165, 2019.
- D. Ardila, A. P. Kiraly, S. Bharadwaj, B. Choi, J. J. Reicher, L. Peng, D. Tse, M. Etemadi, W. Ye, G. Corrado, D. P. Naidich, and S. Shetty, “End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography,” Nature Medicine, vol. 25, no. 6, pp. 954-961, 2019/06/01, 2019.
- P. Petousis, A. Winter, W. Speier, D. R. Aberle, W. Hsu, and A. A. T. Bui, “Using Sequential Decision Making to Improve Lung Cancer Screening Performance,” IEEE Access, vol. 7, pp. 119403-119419, 2019. [CrossRef]
- M. Koizumi, K. Motegi, M. Koyama, M. Ishiyama, T. Togawa, T. Makino, Y. Arisaka, and T. Terauchi, “Diagnostic performance of a computer-assisted diagnostic system: sensitivity of BONENAVI for bone scintigraphy in patients with disseminated skeletal metastasis is not so high,” Annals of Nuclear Medicine, vol. 34, no. 3, pp. 200-211, 2020/03/01, 2020.
- J. Li, D. Dong, M. Fang, R. Wang, J. Tian, H. Li, and J. Gao, “Dual-energy CT–based deep learning radiomics can improve lymph node metastasis risk prediction for gastric cancer,” European Radiology, vol. 30, no. 4, pp. 2324-2333, 2020/04/01, 2020. [CrossRef]
- X. Yang, L. Wu, W. Ye, K. Zhao, Y. Wang, W. Liu, J. Li, H. Li, Z. Liu, and C. Liang, “Deep Learning Signature Based on Staging CT for Preoperative Prediction of Sentinel Lymph Node Metastasis in Breast Cancer,” Academic Radiology, vol. 27, no. 9, pp. 1226-1233, 2020/09/01/, 2020.
- Y. Gao, Z.-D. Zhang, S. Li, Y.-T. Guo, Q.-Y. Wu, S.-H. Liu, S.-J. Yang, L. Ding, B.-C. Zhao, S. Li, Y. Lu, and Y.-Y. Ji, “Deep neural network-assisted computed tomography diagnosis of metastatic lymph nodes from gastric cancer,” Chinese Medical Journal, vol. 132, no. 23, pp. 2804-2811, 2019/12/05, 2019. [CrossRef]
- M. Dohopolski, L. Chen, D. J. Sher, and J. Wang, “Predicting Lymph Node Metastasis in Patients with Oropharyngeal Cancer by Convolutional Neural Networks with associated Epistemic Uncertainty,” International Journal of Radiation Oncology, Biology, Physics, vol. 105, no. 1, pp. S122, 2019.
- Y. Ariji, M. Fukuda, Y. Kise, M. Nozawa, Y. Yanashita, H. Fujita, A. Katsumata, and E. Ariji, “Contrast-enhanced computed tomography image assessment of cervical lymph node metastasis in patients with oral cancer by using a deep learning system of artificial intelligence,” Oral Surgery, Oral Medicine, Oral Pathology and Oral Radiology, vol. 127, no. 5, pp. 458-463, 2019/05/01/, 2019. [CrossRef]
- Y. P. Zhou, S. Li, X. X. Zhang, Z. D. Zhang, Y. X. Gao, L. Ding, and Y. Lu, “[High definition MRI rectal lymph node aided diagnostic system based on deep neural network],” Zhonghua wai ke za zhi [Chinese journal of surgery], vol. 57, no. 2, pp. 108-113, 2019/02//, 2019.
- N. Nawreen, U. Hany, and T. Islam, "Lung Cancer Detection and Classification using CT Scan Image Processing." pp. 1-6.
- A. S. Sakr, "Automatic Detection of Various Types of Lung Cancer Based on Histopathological Images Using a Lightweight End-to-End CNN Approach." pp. 141-146.
- X. Wang, H. Chen, C. Gan, H. Lin, Q. Dou, E. Tsougenis, Q. Huang, M. Cai, and P. A. Heng, “Weakly Supervised Deep Learning for Whole Slide Lung Cancer Image Analysis,” IEEE Transactions on Cybernetics, vol. 50, no. 9, pp. 3950-3962, 2020. [CrossRef]
- N. Tasnim, K. R. Noor, M. Islam, M. N. Huda, and I. H. Sarker, "A Deep Learning Based Image Processing Technique for Early Lung Cancer Prediction." pp. 1060-1064.
- Y. Balagurunathan, A. Beers, M. Mcnitt-Gray, L. Hadjiiski, S. Napel, D. Goldgof, G. Perez, P. Arbelaez, A. Mehrtash, T. Kapur, E. Yang, J. W. Moon, G. B. Perez, R. Delgado-Gonzalo, M. M. Farhangi, A. A. Amini, R. Ni, X. Feng, A. Bagari, K. Vaidhya, B. Veasey, W. Safta, H. Frigui, J. Enguehard, A. Gholipour, L. S. Castillo, L. A. Daza, P. Pinsky, J. Kalpathy-Cramer, and K. Farahani, “Lung Nodule Malignancy Prediction in Sequential CT Scans: Summary of ISBI 2018 Challenge,” IEEE Transactions on Medical Imaging, vol. 40, no. 12, pp. 3748-3761, 2021. [CrossRef]
- D. Jayaraj, and S. Sathiamoorthy, "Random Forest based Classification Model for Lung Cancer Prediction on Computer Tomography Images." pp. 100-104.
- S. H. Begum, M. I. Baig, M. A. Hussain, and M. A. Muqeet, "A Lightweight Deep Learning Model for Automatic Diagnosis of Lung Cancer." pp. 1-5.
- W. Ausawalaithong, A. Thirach, S. Marukatat, and T. Wilaiprasitporn, "Automatic Lung Cancer Prediction from Chest X-ray Images Using the Deep Learning Approach." pp. 1-5.
- J. Cañada, E. Cuello, L. Téllez, J. M. García, F. J. Velasco, and J. Cabrera, "Assistance to lung cancer detection on histological images using Convolutional Neural Networks." pp. 1-4.
- P. P. Massion, S. Antic, S. Ather, C. Arteta, J. Brabec, H. Chen, J. Declerck, D. Dufek, W. Hickes, T. Kadir, J. Kunst, B. A. Landman, R. F. Munden, P. Novotny, H. Peschl, L. C. Pickup, C. Santos, G. T. Smith, A. Talwar, and F. Gleeson, “Assessing the Accuracy of a Deep Learning Method to Risk Stratify Indeterminate Pulmonary Nodules,” Am J Respir Crit Care Med, vol. 202, no. 2, pp. 241-249, Jul 15, 2020.
- R. Y. Bhalerao, H. P. Jani, R. K. Gaitonde, and V. Raut, "A novel approach for detection of Lung Cancer using Digital Image Processing and Convolution Neural Networks." pp. 577-583.
- D. R. Baldwin, J. Gustafson, L. Pickup, C. Arteta, P. Novotny, J. Declerck, T. Kadir, C. Figueiras, A. Sterba, A. Exell, V. Potesil, P. Holland, H. Spence, A. Clubley, E. O'Dowd, M. Clark, V. Ashford-Turner, M. E. Callister, and F. V. Gleeson, “External validation of a convolutional neural network artificial intelligence tool to predict malignancy in pulmonary nodules,” Thorax, vol. 75, no. 4, pp. 306-312, Apr, 2020. [CrossRef]
- P. Rajpurkar, J. Irvin, R. L. Ball, K. Zhu, B. Yang, H. Mehta, T. Duan, D. Ding, A. Bagul, C. P. Langlotz, B. N. Patel, K. W. Yeom, K. Shpanskaya, F. G. Blankenberg, J. Seekins, T. J. Amrhein, D. A. Mong, S. S. Halabi, E. J. Zucker, A. Y. Ng, and M. P. Lungren, “Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists,” PLOS Medicine, vol. 15, no. 11, pp. e1002686, 2018.
- S. Ashhar, S. Mokri, A. A. Abd. Rahni, A. Huddin, N. Zulkarnain, N. Azmi, and T. Mahaletchumy, “Comparison of deep learning convolutional neural network (CNN) architectures for CT lung cancer classification,” International Journal of Advanced Technology and Engineering Exploration, vol. 8, pp. 126-134, 01/31, 2021.
- F. Essaf, Improved Convolutional Neural Network for Lung Cancer Detection, 2020.
- K. Yang, J. Liu, W. Tang, H. Zhang, R. Zhang, J. Gu, R. Zhu, J. Xiong, X. Ru, and J. Wu, “Identification of benign and malignant pulmonary nodules on chest CT using improved 3D U-Net deep learning framework,” European Journal of Radiology, vol. 129, pp. 109013, 2020/08/01/, 2020.
- M. Ye, L. Tong, X. Zheng, H. Wang, H. Zhou, X. Zhu, C. Zhou, P. Zhao, Y. Wang, Q. Wang, L. Bai, Z. Cai, F.-M. Kong, Y. Wang, Y. Li, M. Feng, X. Ye, D. Yang, Z. Liu, Q. Zhang, Z. Wang, S. Han, L. Sun, N. Zhao, Z. Yu, J. Zhang, X. Zhang, R. L. Katz, J. Sun, and C. Bai, “A Classifier for Improving Early Lung Cancer Diagnosis Incorporating Artificial Intelligence and Liquid Biopsy,” Frontiers in Oncology, vol. 12, 2022. [CrossRef]
- D. Ueda, A. Yamamoto, A. Shimazaki, S. L. Walston, T. Matsumoto, N. Izumi, T. Tsukioka, H. Komatsu, H. Inoue, D. Kabata, N. Nishiyama, and Y. Miki, “Artificial intelligence-supported lung cancer detection by multi-institutional readers with multi-vendor chest radiographs: a retrospective clinical validation study,” BMC Cancer, vol. 21, no. 1, pp. 1120, Oct 18, 2021.
- Y. Chen, X. Tian, K. Fan, Y. Zheng, N. Tian, and K. Fan, “The Value of Artificial Intelligence Film Reading System Based on Deep Learning in the Diagnosis of Non-Small-Cell Lung Cancer and the Significance of Efficacy Monitoring: A Retrospective, Clinical, Nonrandomized, Controlled Study,” vol. 2022, pp. 2864170, 2022.
- J. Chamberlin, M. R. Kocher, J. Waltz, M. Snoddy, N. F. C. Stringer, J. Stephenson, P. Sahbaee, P. Sharma, S. Rapaka, U. J. Schoepf, A. F. Abadia, J. Sperl, P. Hoelzer, M. Mercer, N. Somayaji, G. Aquino, and J. R. Burt, “Automated detection of lung nodules and coronary artery calcium using artificial intelligence on low-dose CT scans for lung cancer screening: accuracy and prognostic value,” BMC Medicine, vol. 19, no. 1, pp. 55, 2021/03/04, 2021. [CrossRef]
- J. G. Nam, H. J. Kim, E. H. Lee, W. Hong, J. Park, E. J. Hwang, C. M. Park, and J. M. Goo, “Value of a deep learning-based algorithm for detecting Lung-RADS category 4 nodules on chest radiographs in a health checkup population: estimation of the sample size for a randomized controlled trial,” European Radiology, vol. 32, no. 1, pp. 213-222, 2022/01/01, 2022.
- M. M. Naeem Abid, T. Zia, M. Ghafoor, and D. Windridge, “Multi-view Convolutional Recurrent Neural Networks for Lung Cancer Nodule Identification,” Neurocomputing, vol. 453, pp. 299-311, 2021/09/17/, 2021. [CrossRef]
- A. Gürsoy Çoruh, B. Yenigün, Ç. Uzun, Y. Kahya, E. U. Büyükceran, A. Elhan, K. Orhan, and A. Kayı Cangır, “A comparison of the fusion model of deep learning neural networks with human observation for lung nodule detection and classification,” British Journal of Radiology, vol. 94, no. 1123, pp. 20210222, 2021.
- S. Trajanovski, D. Mavroeidis, C. L. Swisher, B. G. Gebre, B. S. Veeling, R. Wiemker, T. Klinder, A. Tahmasebi, S. M. Regis, C. Wald, B. J. McKee, S. Flacke, H. MacMahon, and H. Pien, “Towards radiologist-level cancer risk assessment in CT lung screening using deep learning,” Computerized Medical Imaging and Graphics, vol. 90, pp. 101883, 2021/06/01/, 2021.
- Y.-C. Hsu, Y.-H. Tsai, H.-H. Weng, L.-S. Hsu, Y.-H. Tsai, Y.-C. Lin, M.-S. Hung, Y.-H. Fang, and C.-W. Chen, “Artificial neural networks improve LDCT lung cancer screening: a comparative validation study,” BMC Cancer, vol. 20, no. 1, pp. 1023, 2020/10/22, 2020. [CrossRef]
- J. Li, and J. P. Fine, “Assessing the dependence of sensitivity and specificity on prevalence in meta-analysis,” Biostatistics, vol. 12, no. 4, pp. 710-722, 2011.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).