1. Introduction
Allogenic hematopoietic stem cell transplantation (allo-HSCT) is a curative therapy for several malignant and nonmalignant hematological disorders. The matching status of human leukocyte antigens (HLA) between donors and recipients is a major factor for a successful transplantation. Mismatches in classical HLA genes (HLA-A, B, C, DRB1, DQB1, and DPB1) are associated with adverse HSCT outcomes such as non-engraftment, acute or chronic graft versus host disease (aGVHD and cGVHD, respectively), reduced overall survival, and reduced disease-free survival after HSCT [
1]. Therefore, an HLA-identical sibling donor (ISD) transplant is the best choice of donor if available, as it is associated with reduced risk of GVHD, increased overall survival (OS), and enhanced engraftment [
2]. Nevertheless, adverse HSCT outcomes could also occur in HLA-ISD allo-HSCT that were attributed to genetic contributors within or outside the HLA genomic cluster [
3].
Several studies reported significant associations between HSCT outcomes and HLAs in HLA-ISD HSCT. Two studies from Switzerland and the UK, showed that HLA-DR*15 is significantly associated with reduced relapse rate and improved survival in patients with leukemia or non-Hodgkin lymphoma after HLA-ISD HSCT [
4,
5]. Similarly, A large study (n=1204) reported a significant impact of HLA DR15 in reducing secondary graft failure after HLA-ISD HSCT [
6]. An Iranian study found several HLA associations. HLA-B*07 was linked to a higher incidence of acute graft-versus-host disease (aGVHD) (grades II-IV), while HLA-A*01, B*40, B*41, and B*57 significantly increased the risk of chronic GVHD (cGVHD). Regarding mortality, HLA-DRB1*02, B*13, B*40, and DRB1*04 were significantly associated with an increased death rate. HLA-DRB1*15 showed a non-significant trend towards a decreased incidence of aGVHD [
7]. In a Slovakian study, recipients carrying the HLA-A*01, -DRB1*03 and -DQB1*03 alleles showed lower incidence of aGVHD, while HLA-DQB1*06 allele was associated with a higher incidence of cGVHD [
8]. Another study on Japanese patients with leukemia or myelodysplastic reported an association of HLA-B*60 with grades II-IV aGVHD increased risk, and HLA B*62 with decreased grades II-IV [
3]. Consistently a recent report from a dataset of Japanese patients with adult T-cell leukemia/lymphoma (ATL) revealed that HLA-B60 was associated with an increased risk of acute GVHD and overall mortality, while HLA-B62 was associated with a decreased risk of overall and transplant related mortality [
9]. Based on existing research, it's evident that replication of findings may be observed only when studies are conducted within the same ethnic group. This may indicate that the HLA contribution to unfavorable HSCT outcomes may differ globally due to variations in HLA genomics, specifically linkage disequilibrium blocks, and the unique distribution of HLA alleles across distinct ethnicities.
Currently, there is a gap in research from Arab countries regarding the impact of HLA alleles on HSCT outcomes. Our study aims to address this by investigating an Omani cohort, marking the first such report from the Arab population. Allogenic HSCT is conducted in Oman in a single center at Sultan Qaboos University Hospital (SQUH), with 84% of procedures utilizing HLA-ISDs and a reported 18% risk aGVHD (Grades II-IV) and an 8% risk of cGVHD following transplants [
10]. The current study is to evaluate the influence of HLA alleles on HLA-ISD HSCT outcomes, which could refine the risk assessment and outcome prediction for HSCT recipients and improve post-HSCT management.
2. Materials and Methods
Patients
Data was collected retrospectively for 153 registered patients, who underwent allo-HSCT from fully HLA-matched related donors in the period from 2012 to 2022. All donors were siblings except one, who was an HLA identical mother. The baseline parameters collected were the date of HSCT, donor and recipient demographics (age, gender, blood group), CMV status, stem cell characteristics (source and dose), conditioning regimen, and GVHD prophylaxis.
Inclusion and Exclusion Criteria
The study included both male and female patients who received matched sibling transplants, either at SQUH or elsewhere if their early post-transplant care was managed at SQUH. The inclusion period covered Jan 2012 to Dec 2022. Patients with HLA reports missing, incomplete clinical data or with less than seven days survival after HSCT were excluded. The characteristics of patients are shown in
Table 1.
Outcomes
We assessed several HSCT outcomes, including aGVHD, an inflammatory condition occur -ing within 100 days of transplantation, and cGVHD, which develops after 100 days and exhibits autoimmune-like characteristics [
11,
12]. We also analyzed patient chimerism status (complete or mixed) at 6 to 12 months post-HSCT. Chimerism refers to the proportion of donor and recipient cells in the recipient's blood or bone marrow after HSCT. Complete chimerism indicates only donor cells are present, while mixed chimerism shows both donor and recipient cells, with 5-95% of cells being of donor origin [
13].
Moreover, the study examined clinical parameters including neutrophil and platelet engraftment times following HSCT, and five-year overall patient survival. Neutrophil engraftment was defined as a sustained neutrophil count exceeding 500 × 10^6/L over three consecutive days [
14], while platelet engraftment required seven days of transfusion independence, and a platelet count above 20 × 10^9/L [
15].
Data was retrieved from the HLA laboratory at Sultan Qaboos University (SQU). For samples collected between 2012 and 2014, serological techniques were used for HLA class I typing HLA-A, B and C) while PCR-SSP was employed for HLA class II genotyping (HLA-DRB1 and DQB1). From 2012-2014, available genotypes were at low-resolution level for HLA class I (serological methods) and HLA class (PCR-SSP technique) for patients and donors. We converted class I serotypes to the most probable molecular HLA alleles according to WHO-HLA Nomenclature Committee [
16]. Genotypes after 2014 were typed for class I and II on the Luminex platform (Luminex Corp., USA) which is based on sequence-specific oligonucleotide (SSO) method. For all patients, five HLA loci: HLA-A, B, C, DRB1 and DQB1 were reported at low-resolution level (two digits).
3. Results
3.1. Baseline Characteristics
This study involved 153 patients, categorized as having either malignant (62 patients) or non-malignant (91 patients) hematological disorders. Most of the patients were males (61%) and pediatric cases (below 18 years old) comprised 37.9% of the cohort (
Table 1).
At the time of transplant, the patients' median age was 21 years. Peripheral blood was the primary source of stem cells (68.6%). Over half the patients (55.8%) received fludarabine, busulfan, and anti-thymocyte globulin (Flu/Bu/ATG) for conditioning, and most (64.7%) were given cyclosporine (CSA) for GVHD prevention. Neutrophil and platelet engraftment occurred at a median of 14 and 15 days, respectively. The incidence of acute and chronic GVHD (all grades) was 16% and 15%, respectively. Within 6 to 12 months post-HSCT, mixed chimerism was observed in 20.3% of patients, while 2% experienced complete rejection.
3.2. GVHD, Engraftment and Chimerism
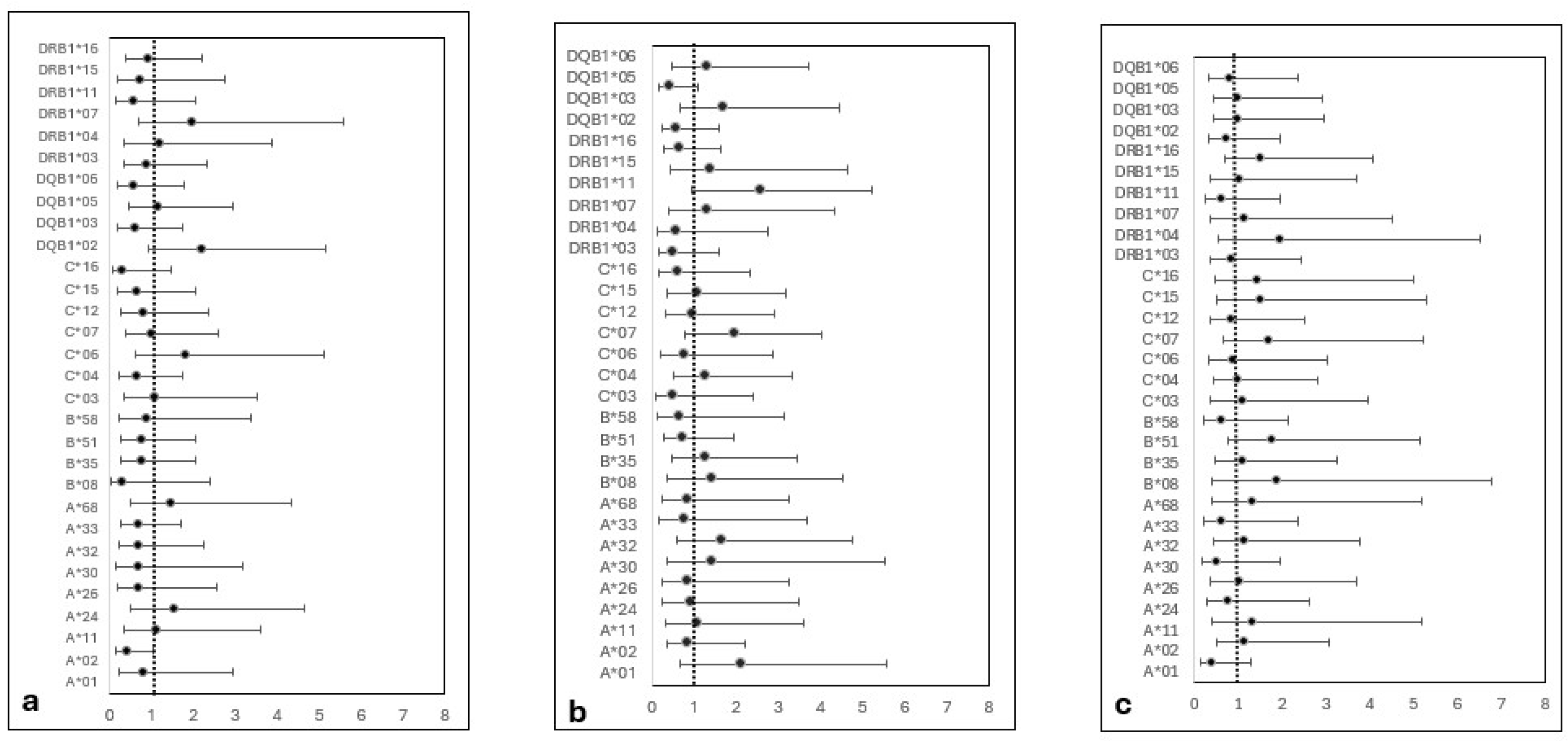
Univariate regression analysis revealed no significant association between the selected HLA antigens and the risk of aGVHD, cGVHD, chimerism status, or engraftment (
Figure 2). However, HLA DQB1*05 allele was associated with a trend of decreased cGVHD risk but did not reach significance (OR 0.43, CI (0.17-1.09) and P=0.07). Detailed ratios and p-values are available in supplementary table S1.
Also, there was no significant association between HLA and neutrophil or platelet engraftment times (
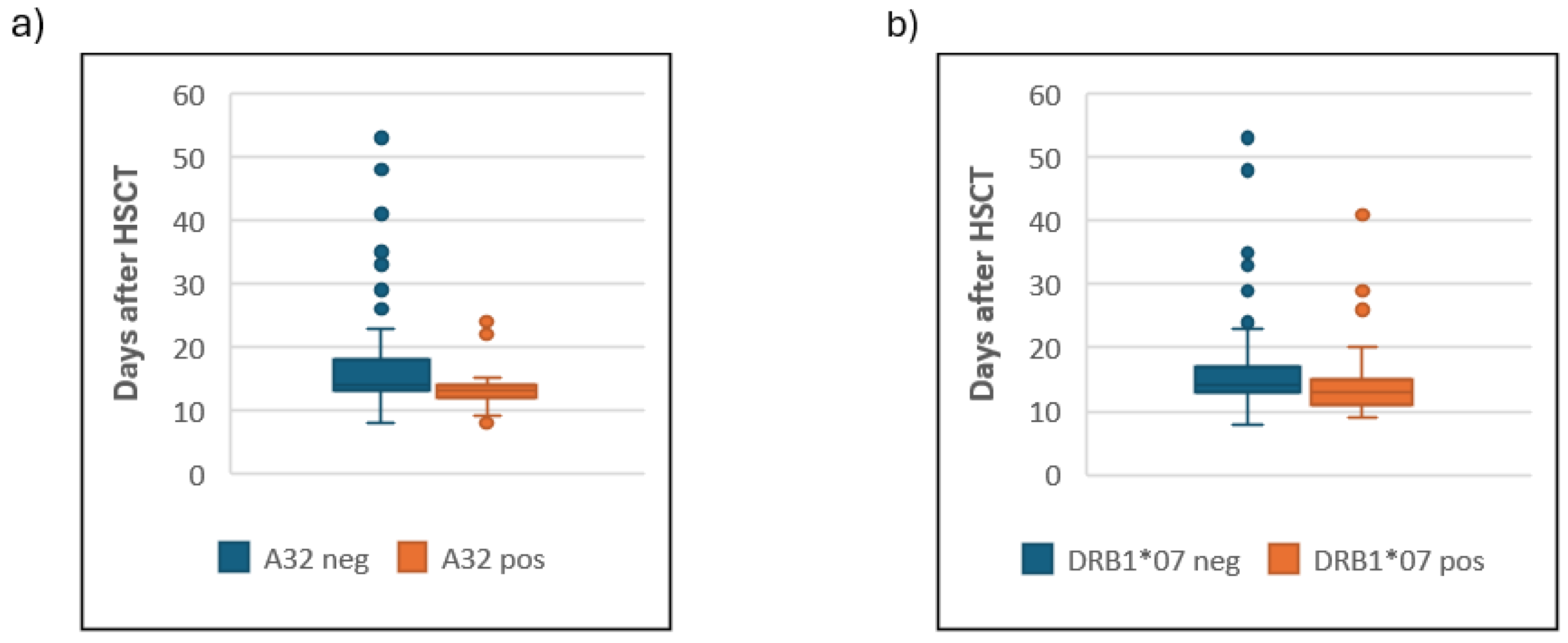
Table S2). Although trends of earlier neutrophil engraftment in HLA A*
32 and DRB1*07 carriers (medians 13 vs 14 days) were observed (
Figure 3), these did not achieve statistical significance after correction (
Pc = 0.23 and 0.18 respectively).
3.3. Overall Survival
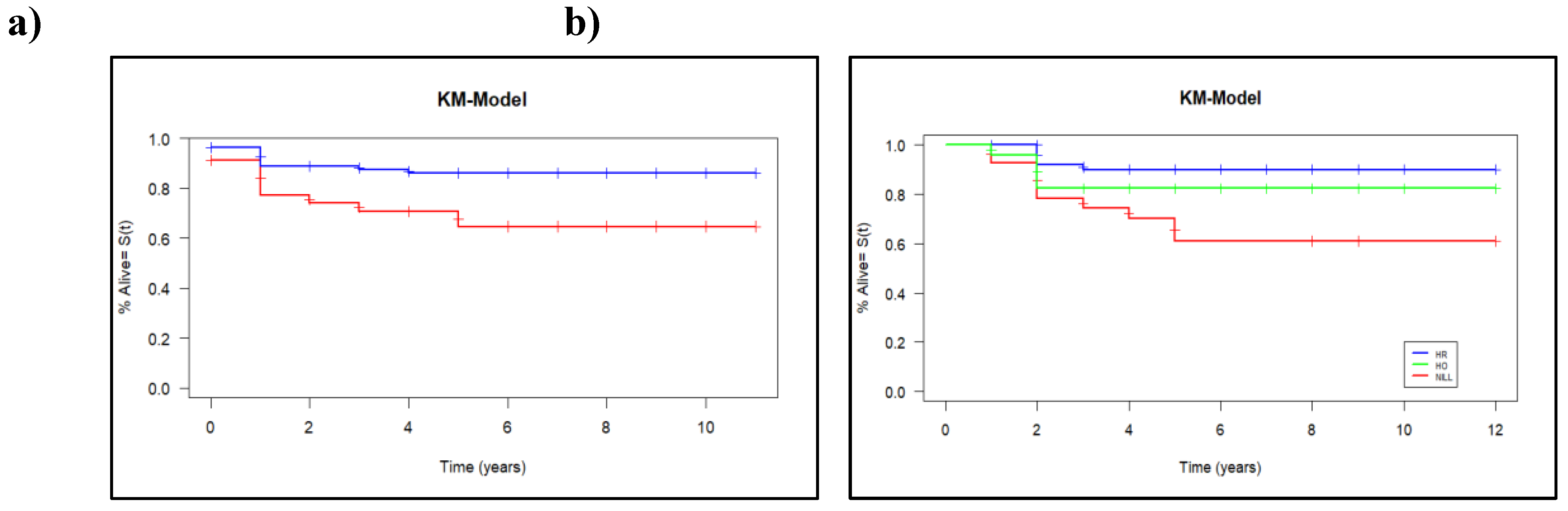
With a median follow-up of five years, 126 patients (82.4%) were alive, and statistically significant better survivals were associated with the presence of HLA-DQB1*05 (P = 0.010, Pc = 0.04). The presence of DQB1*05 correlated with improvement in estimated 5-year overall survival, with rates of 90% (95% CI: 88-93%) compared to 68% (95% CI: 65-73%) in its absence, as illustrated in
Figure 4a. Using Cox proportional hazard model, the hazard ratio for carrying DQB1*05 was 0.39 (95% CI, 0.18 to 0.84, P
-value = 0.01, Pc= 0.05). The Cox model revealed also that age, gender and source of stem cell were not statistically significant predictors of the overall survival (
P-value = 0.52, 0.20 and 0.69, respectively).
We analyzed the effect of DQB1*05 genotype (heterozygous, homozygous, or non-carrier) on overall survival. The log-rank test revealed a significant difference (P = 0.01), driven by a significant decrease in survival among non-carriers compared to both heterozygous and homozygous carriers. However, no significant difference in survival was observed between DQB1*05 heterozygous and homozygous patients (
Figure 4b).
We found also a trend of better survival in HLA-DRB1*16 carriers, but it did not remain significant after multiple testing correction (
P-value=0.05,
Pc=0.3). No significant differences in survival curves were observed between HLA-positive and HLA-negative patients for the remaining HLA alleles (
Table S3).
4. Discussion
The current study is the first to analyze HLA-ISD HSCT outcomes in Oman and, uniquely, explores the influence of HLA on HSCT outcomes. Despite the small cohort size, our patient’s recovery pattern agreed almost with the published reported ranges and have lower incidences of both aGVHD and cGVHD (
Table 2).
Given the heterogeneity of the study population, we compared HSCT outcomes within the cohort subgroups based on age, disease type, sex match and source of stem cell (
Table S4). Values were comparable to the existing literature, with very low cGVHD incidence in children (7.40%) and in bone marrow transplanted patients (4.65%).
Among 30 HLA alleles assessed, HLA-DQB1*05 was the only significant predictor (Pc=0.048), associated with improved 5-year OS. Although, the study small sample size may have limited the ability to achieve high statistical significance, DQB1*05 represents 46% of the DQB1 antigens in Oman, with three alleles DQB1*05:02 (0.80), 05:01 (0.16) and 05:03 (0.04) (unpublished data). Possibly, its high frequency may have enhanced the detection of the association [29].
Further, we postulated that this protective effect might be mediated through lowering cGVHD incidence, as DQB1*05 exhibited the lowest odds ratio (RR: 0.87, p= 0.07,
Figure 2b). Notably, cGVHD have an autoimmune-like underlying pattern [
22] and DQB1*05 and its alleles were shown to confer protection against autoimmune diseases like type 1 diabetes in Arabs [
23,30]. Thus, DQB1*05 alleles may modulate post-HSCT immune responses and decrease cGVHD incidence.
In this study, the HLA-DQB1*05 allele showed similar frequencies in children, present in 75% of those with chronic graft-versus-host disease (cGVHD) and 74% of those without it. In contrast, among adults, the allele was more common in patients who did not develop cGVHD (74%) compared to those who did (50%) suggesting that this allele have more protective effect in adults.
The underlaying mechanisms for HLA association with HCST outcomes remain to be elucidated. Clearly, different outcomes, such as aGVHD and cGVHD are associated with different immunological patterns and thus different HLA Class I and or II alleles. Also, potentially the association might not be only due to antigen presentation, as some studies have associated differential HLA allele-specific expression with HSCT outcomes. For example, aGVHD incidence was greater in patients with highly expressed HLA-DPB1 alleles who received an HLA-DPB1 mismatched transplant from a donor with low expression HLA-DPB1 alleles [31].
The main finding of this study was the association between HLA-DQB1*05 and improved five-year overall survival in Omani patients undergoing HLA-ISD HSCT, given the high prevalence of HLA-DQB1*05 in Oman (37.56% in the general population [
17], 48.7% in our study). Consistent with other studies, we found that HSCT outcomes likely depend not only on HLA allele matching but also on the HLA allele itself and possibly genetic background.
It is worth mentioning here that HLA A*32 and DRB1*07, showed association with a trend of faster neutrophil engraftment that became insignificant after correction. This weak association might be attributed to its low frequency in the population (4%), and the relatively small cohort size.
As the first retrospective study to evaluate the association of HLA alleles with HSCT outcomes, several limitations were identified that should be addressed in future research. First, a small sample size potentially influenced the identification of association, especially with relatively uncommon alleles. Second, heterogeneous cohort of adult and pediatric patients with various diseases that may differentially influence HSCT outcome and HLA effect on malignant conditions is not as in heritable and hyperinflammatory disorders. Third, currently most transplants are from HLA identical sibling or related donors that do not demand high resolution typing. However, low resolution typing groups multiple alleles that can differ significantly at the amino acid level, while 4-digit typing distinguishes between alleles that may have distinct peptide-binding profiles and immune responses. Moreover, new approaches such as RNA-based next generation sequencing (NGS) enable us to quantify HLA allele-specific expression differences between individuals with different phenotypes.
5. Conclusion
In conclusion, from the dataset of a heterogenous cohort of Omani patients who received HLA-ISD HSCT, we have detected a decreased incidence of cGVHD and a significant association with five-year OS with HLA DQB1*05. This could be of a predictive value for long-term HSCT outcomes and contribute to precision medicine. Comprehensive investigation facilitated by collaborative efforts among neighboring countries and across diverse ethnicities, is needed to explore validate detected associations and there underlaying mechanisms.
Ethical Approval
The study was reviewed and accepted by the Medical Research Ethic Committee (MREC) at the College of Medicine and Health Sciences/Sultan Qaboos University (MREC # 2924).
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
For this work, Conceptualization, was by Aliya Al Ansari. and Fatma Al Lawati.; methodology was proposed by Murtadha Al Kabori who also provided the software for data collection, Data collection, formal analysis and statistical investigation was done by Fatma Al Lawati, who also wrote the original draft. Aliya, Murtadha, and Salma reviewed the drafts. There was no funding allocated for this work.
Funding
“This research received no external funding”
Institutional Review Board Statement
“The study was conducted in accordance with the Declaration of Helsinki, and approved by Medical Research Ethic Committee (MREC) at the College of Medicine and Health Sciences/Sultan Qaboos University (MREC # 2924).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
We encourage all authors of articles published in MDPI journals to share their research data. In this section, please provide details regarding where data supporting reported results can be found, including links to publicly archived datasets analyzed or generated during the study. Where no new data was created, or where data is unavailable due to privacy or ethical restrictions, a statement is still required. Suggested Data Availability Statements are available in section “MDPI Research Data Policies” at
https://www.mdpi.com/ethics.
Acknowledgments
We extend our thanks to the nursing team in the hematology department for their help in retrieving HSCT patient information from the SQUH system. We're also especially grateful to Dr. Najmus Sehar for her assistance with additional clinical information.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| Allo-HSCT |
Allogenic Hematopoietic Stem Cell Transplantation |
| ISD |
Identical Sibling Donor |
| HLA |
Human Leukocyte Antigen |
| SQUH |
Sultan Qaboos University Hospital |
| GVHD |
Graft versus Host disease |
| cGVHD |
Chronic Graft versus Host Disease |
| aGVHD |
Acute Graft versus Host Disease |
| OS |
Overall Survival |
| ATL |
Adult T cell Leukemia/Lymphoma |
| CMV |
Cytomegalo virus |
| WHO |
World Health Organisation |
| SSO |
Single strand oligonucleotide |
| CSA |
Cyclosporin A |
| OR |
Odd Ratio |
References
- Bertaina, A.; Andreani, M. Major Histocompatibility Complex and Hematopoietic Stem Cell Transplantation: Beyond the Classical HLA Polymorphism. Int J Mol Sci. 2018, 19, 2. [Google Scholar] [CrossRef] [PubMed]
- Níchonghaile, M. Donor Selection. 2018.
- Morishima, S.; Fukuda, T.; Doki, N.; Mori, T.; Onizuka, M.; Kawakita, T.; et al. Individual HLAs influence immunological events in allogeneic stem cell transplantation from HLA-identical sibling donors. Bone Marrow Transplant. 2021, 56, 646–54. [Google Scholar] [CrossRef]
- Cruz-Tapias, P.; Castiblanco, J.; Anaya, J.M. Chapter 10 Major histocompatibility complex: Antigen processing and presentation. In: Anaya JM, Shoenfeld Y, Rojas-Villarraga A, Levy RA, Cervera R, editors. Autoimmunity: From Bench to Bedside. 1st ed. 2013. p. 169–84.
- Shiina, T.; Kulski, J.K. HLA Genetics for the Human Diseases. Adv Exp Med Biol. 2024, 1444, 237–258. [Google Scholar]
- Trowsdale, J. The MHC, disease and selection. Immunol Lett. 2011, 137, 1–8. [Google Scholar] [CrossRef]
- Battiwalla, M.; Hahn, T.; Radovic, M.; Roy, H.; Wahab, A.; Duman, E.; et al. Human leukocyte antigen (HLA) DR15 is associated with reduced incidence of acute GVHD in HLA-matched allogeneic transplantation but does not impact chronic GVHD incidence. Blood. 2006, 107, 1970–3. [Google Scholar] [CrossRef] [PubMed]
- Davidson, J.A.; Tate, D.G.; Poulton, K.V.; Lucas, G.S.; Yin, J.L.; Liakopoulou, E.F.; et al. HLA-DR15, reduced relapse rate and improved survival after HLA identical sibling hemopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2007, 13, 493–4. [Google Scholar] [CrossRef]
- Stern, M.; Passweg, J.; Tiercy, J.M.; Genitsch, A.; Meyer-Monard, S.; Heim, D.; et al. Human leukocyte antigen DR15 is associated with reduced relapse rate and improved survival after human leukocyte antigen-identical sibling hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2006, 12, 1169–75. [Google Scholar] [CrossRef]
- Al-Khabori, M.; Al-Huneini, M. Hematopoietic stem cell transplantation in the Sultanate of Oman. Hematol Oncol Stem Cell Ther. 2017, 10, 305–7. [Google Scholar] [CrossRef]
- Toubai, T.; Sun, Y.; Reddy, P. GVHD pathophysiology: is acute different from chronic? Best Pract Res Clin Haematol. 2008, 21, 101–17. [Google Scholar] [CrossRef]
- Olivieri, A.; Mancini, G. Current Approaches for the Prevention and Treatment of Acute and Chronic GVHD. Cells. 2024, 13, 18. [Google Scholar] [CrossRef]
- Baron, F.; Sandmaier, B.M. Chimerism and outcomes after allogeneic hematopoietic cell transplantation following nonmyeloablative conditioning. Leukemia. 2006, 20, 1690–700. [Google Scholar] [CrossRef] [PubMed]
- Wolff, S.N. Second hematopoietic stem cell transplantation for the treatment of graft failure, graft rejection or relapse after allogeneic transplantation. Bone Marrow Transplant. 2002, 29, 545–52. [Google Scholar] [CrossRef] [PubMed]
- Teltschik, H.M.; Heinzelmann, F.; Gruhn, B.; Feuchtinger, T.; Schlegel, P.; Schumm, M.; et al. Treatment of graft failure with TNI-based reconditioning and haploidentical stem cells in paediatric patients. Br J Haematol. 2016, 175, 115–22. [Google Scholar] [CrossRef] [PubMed]
- Bodmer, J.G.; Marsh, S.G.; Albert, E.D.; Bodmer, W.F.; Bontrop, R.E.; Dupont, B.; et al. Nomenclature for factors of the HLA system, 1998. Tissue Antigens. 1999, 53, 407–46. [Google Scholar] [CrossRef]
- Al Hudhili, I.; Al Zadjali, F.; Al Khabori, M. HLA Alleles and Haplotypes Frequencies in Omani Population and Validation of Maximum Likelihood Estimation Predictive Algorithm. [Muscat]: Sultan Qaboos University; 2020. [Google Scholar]
- Battiwalla, M.; Wang, T.; Carreras, J.; Deeg, H.J.; Ayas, M.; Bajwa, R.P.S.; et al. HLA-matched sibling transplantation for severe aplastic anemia: impact of HLA DR15 antigen status on engraftment, graft-versus-host disease, and overall survival. Biol Blood Marrow Transplant. 2012, 18, 1401–6. [Google Scholar] [CrossRef]
- Justiz Vaillant, A.A.; Modi, P.; Mohammadi, O. Graft-Versus-Host Disease. 2024.
- Spierings, E. Minor histocompatibility antigens: past, present, and future. Tissue Antigens. 2014, 84, 374–60. [Google Scholar] [CrossRef]
- Laurin, D.; Spierings, E.; van der Veken, L.T.; Hamrouni, A.; Falkenburg, J.H.F.; Souillet, G.; et al. Minor histocompatibility antigen DDX3Y induces HLA-DQ5-restricted T cell responses with limited TCR-Vbeta usage both in vivo and in vitro. Biol Blood Marrow Transplant. 2006, 12, 1114–24. [Google Scholar] [CrossRef]
- Nilsson, J.B.; Kaabinejadian, S.; Yari, H.; Kester, M.G.D.; van Balen, P.; Hildebrand, W.H.; et al. Accurate prediction of HLA class II antigen presentation across all loci using tailored data acquisition and refined machine learning. Sci Adv. 2023, 9, eadj6367. [Google Scholar] [CrossRef]
- Thibodeau, J.; Moulefera, M.A.; Balthazard, R. On the structure-function of MHC class II molecules and how single amino acid polymorphisms could alter intracellular trafficking. Hum Immunol. 2019, 80, 15–31. [Google Scholar] [CrossRef]
- Fleury, S.; Thibodeau, J.; Croteau, G.; Labrecque, N.; Aronson, H.E.; Cantin, C.; et al. HLA-DR polymorphism affects the interaction with CD4. J Exp Med. 1995, 182, 733–41. [Google Scholar] [CrossRef]
- Dendrou, C.A.; Petersen, J.; Rossjohn, J.; Fugger, L. HLA variation and disease. Nat Rev Immunol. 2018, 18, 325–39. [Google Scholar] [CrossRef]
- Petersdorf, E.W.; Malkki, M.; O’hUigin, C.; Carrington, M.; Gooley, T.; Haagenson, M.D.; et al. High HLA-DP Expression and Graft-versus-Host Disease. N Engl J Med. 2015, 373, 599–609. [Google Scholar] [CrossRef]
- van den, E. lsen PJ. Expression regulation of major histocompatibility complex class I and class II encoding genes. Front Immunol. 2011, 2, 48. [Google Scholar]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).