Submitted:
23 July 2025
Posted:
24 July 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Database
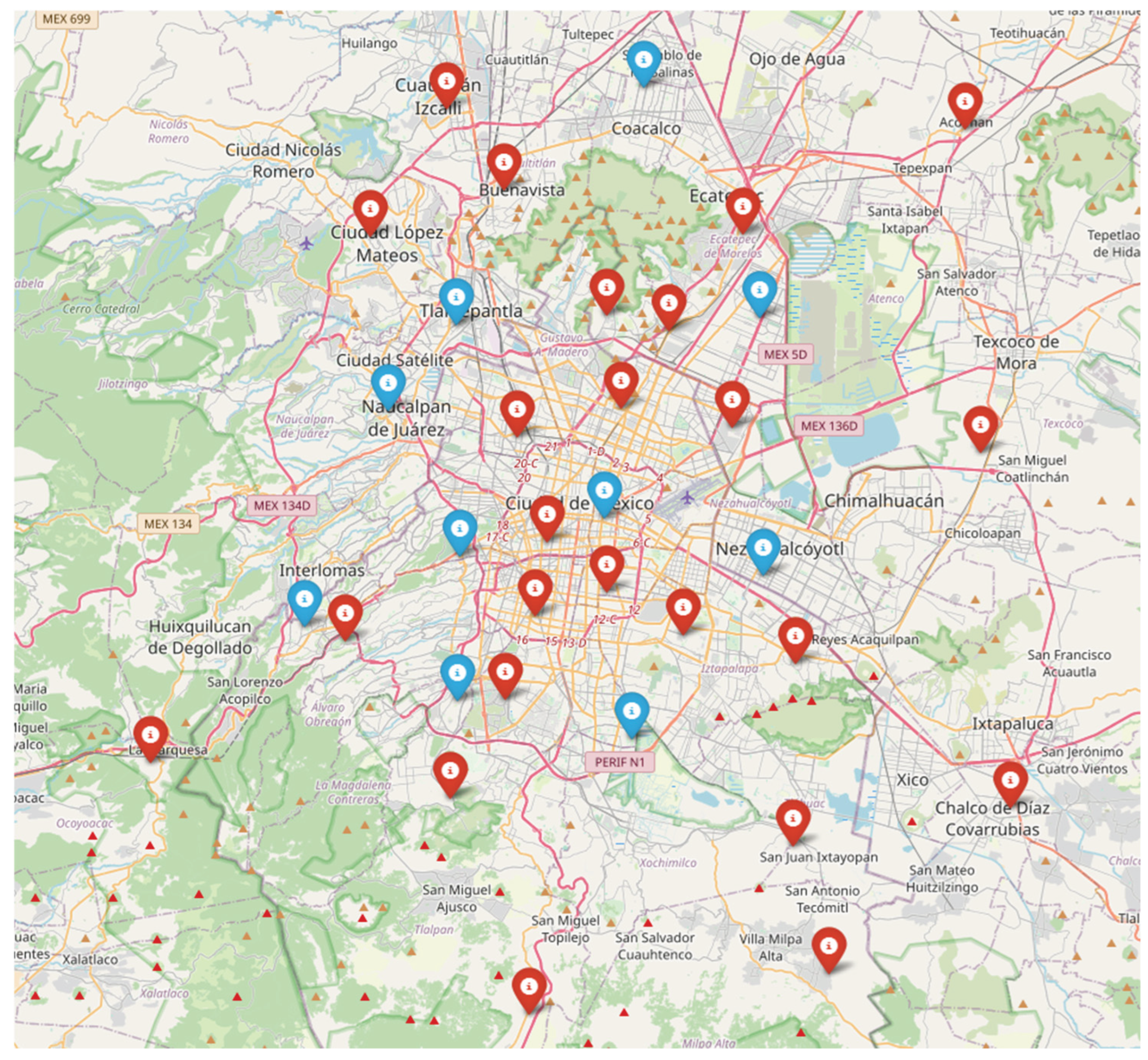
2.1.1. Study Area
2.1.2. Data Collection and Integration
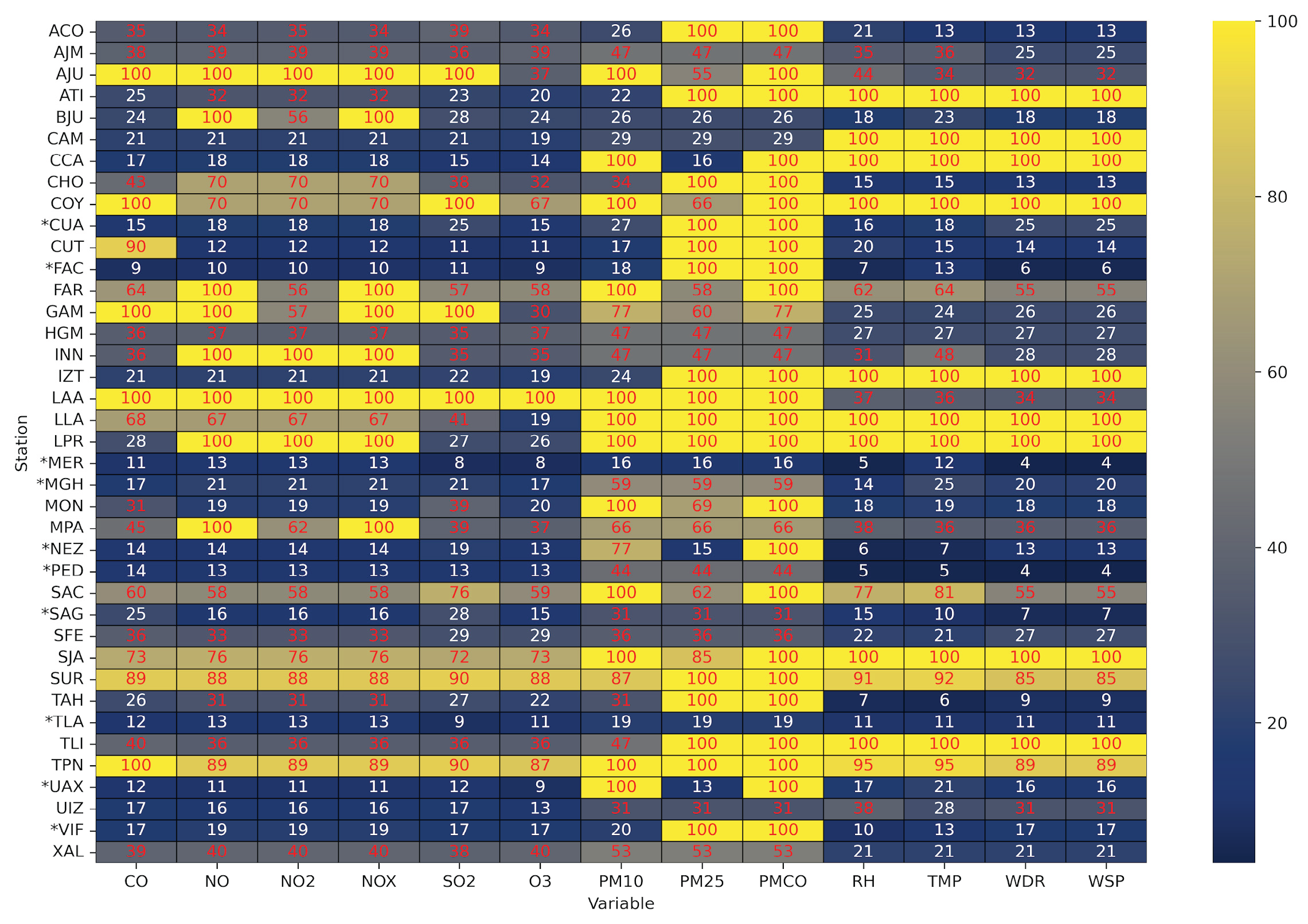
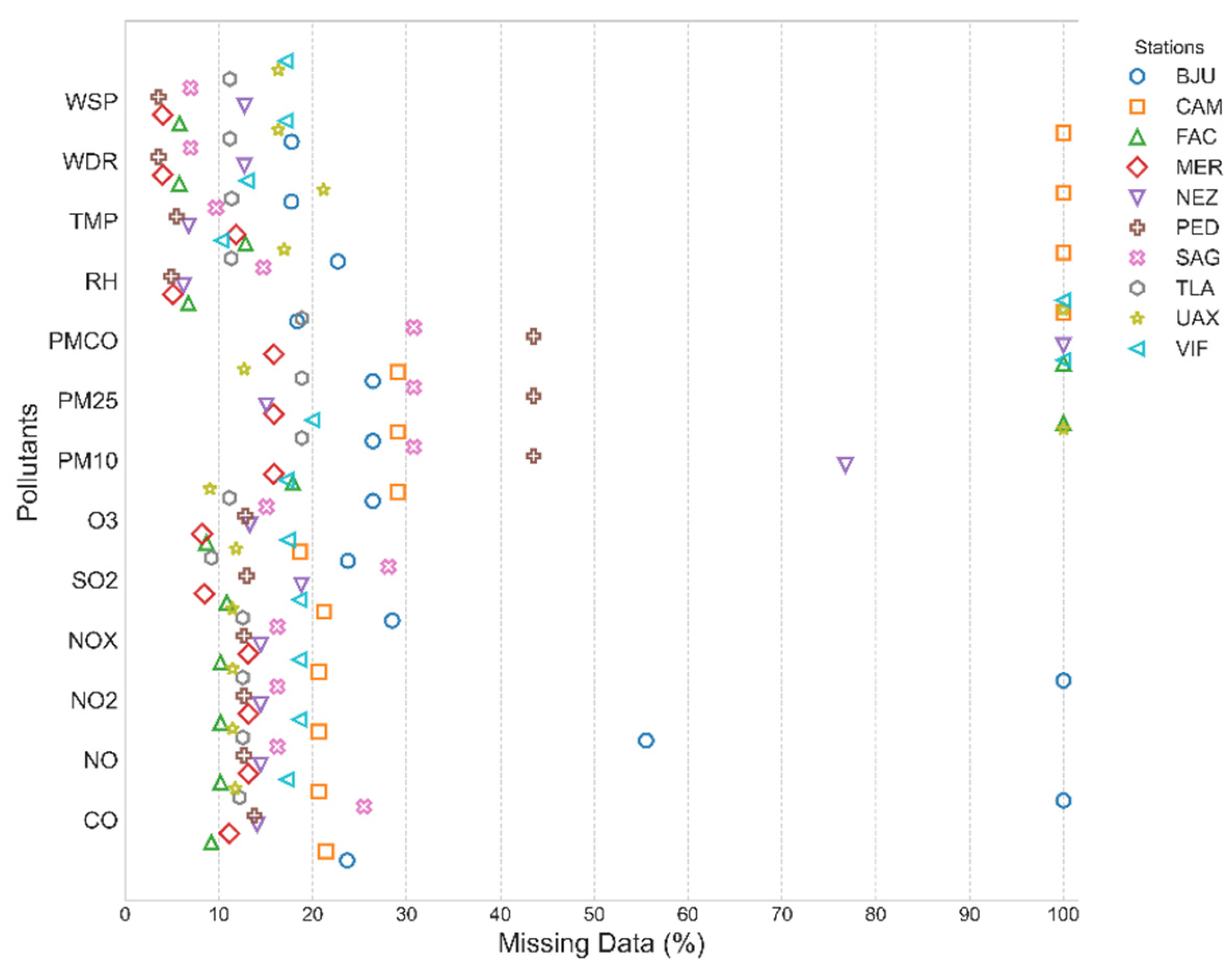
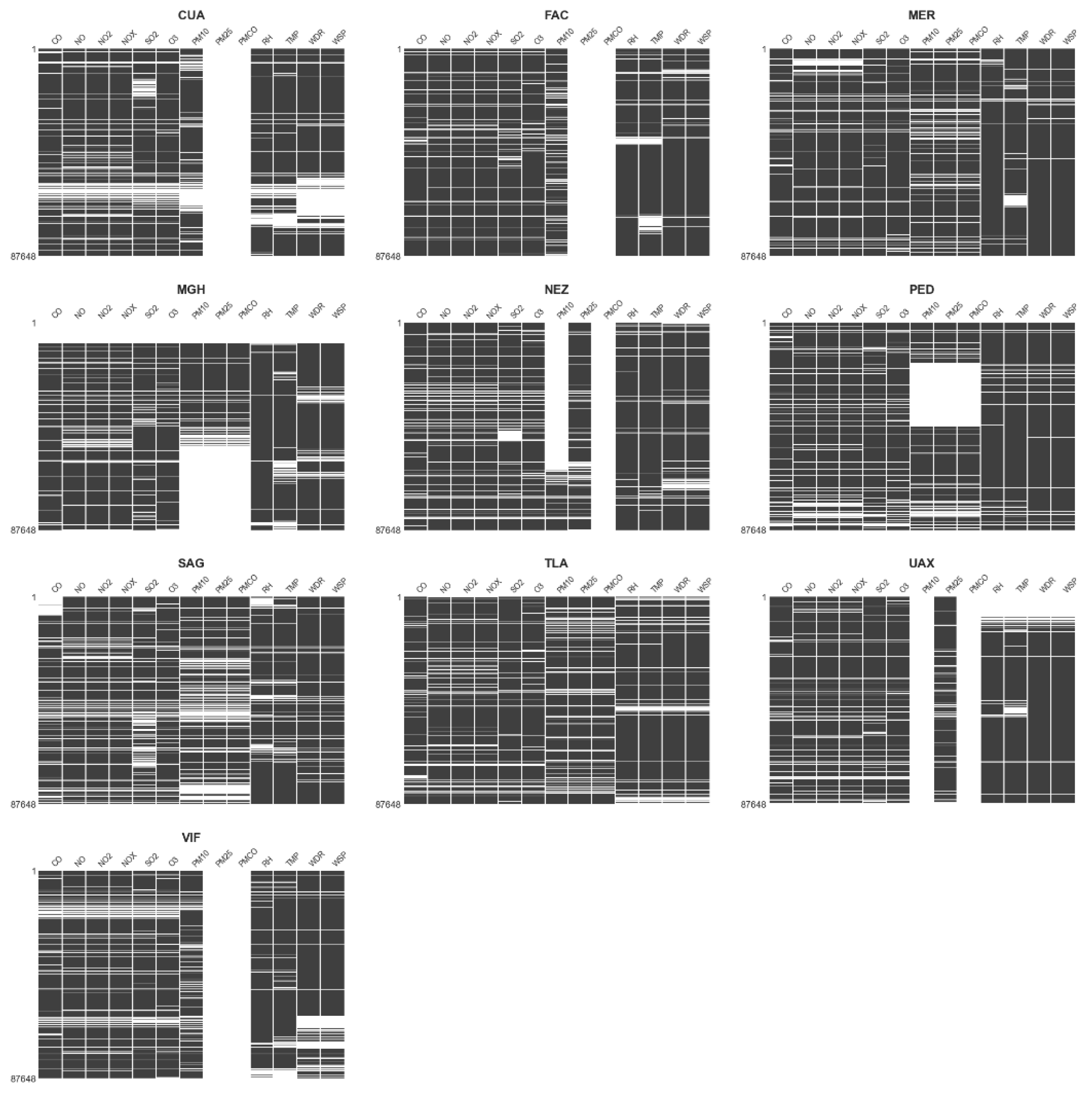
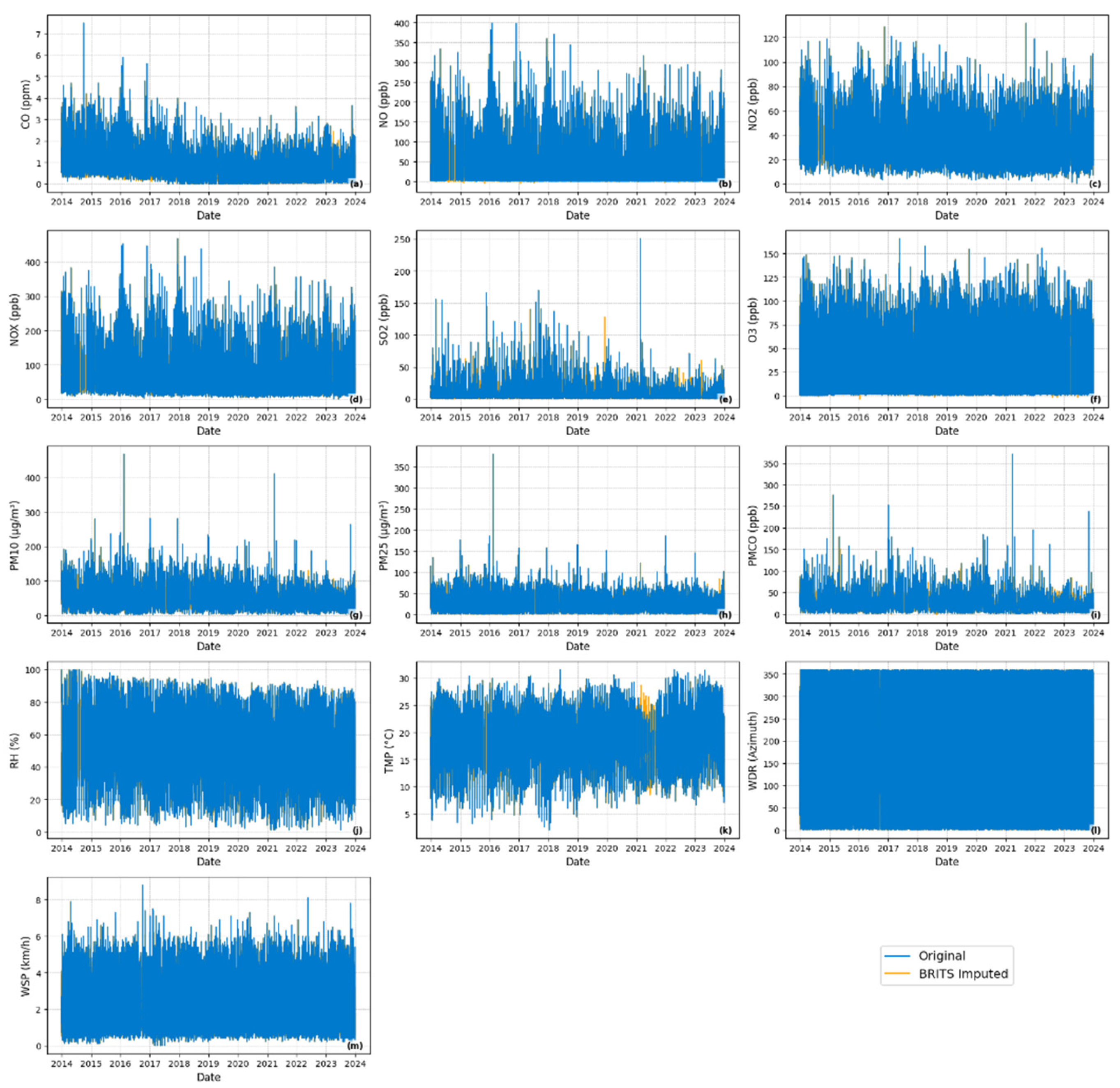
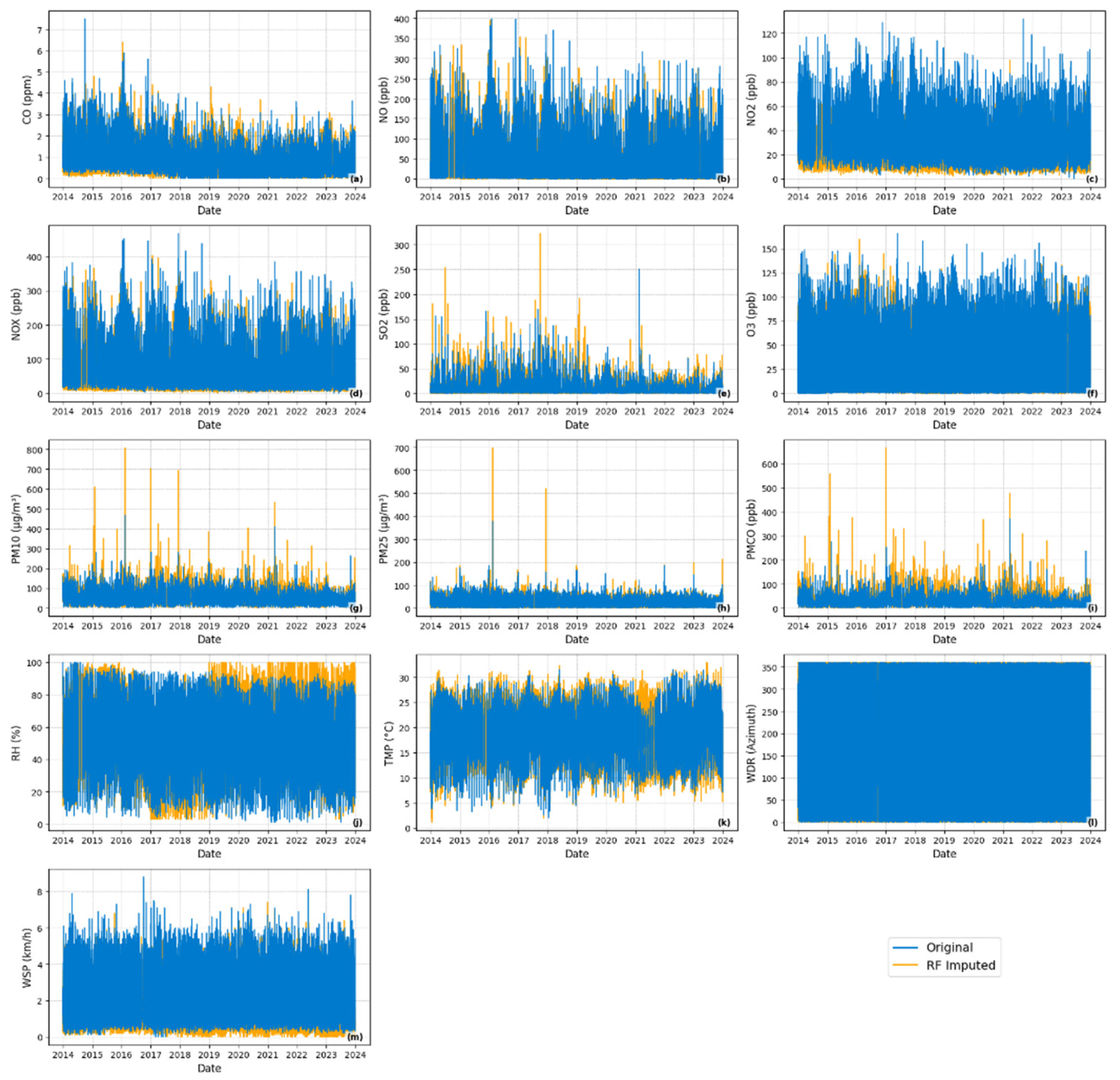
2.1.3. Missing Data Patterns Analysis and Station Filtering
2.1.4. Missing Data Patterns in Selected Monitoring Station
2.2. Model Training and Evaluation Pipeline
- Missing data identification: Binary masks were created to identify real missing values in the dataset.
- Data splitting: Each station’s dataset was divided into 80% for training and 20% for testing.
- Hyperparameter optimization: Hyperparameter tuning was performed in sequential steps due to computational constraints.
- Artificial missingness for evaluation: To enable controlled performance assessment, 20% of the observed values in the test set were randomly removed under a Missing Completely At Random (MCAR) assumption. This masking allowed direct comparison between imputed and ground-truth values.
- Normalization: For the BRITS method, all variables were standardized using MinMaxScaler normalization, applied separately to the training and test datasets.
- Model Training and Evaluation: After hyperparameters optimization, models were retrained using the full training set and evaluated on the masked test set. Imputation performance was assessed using several evaluation metrics including MAE, RMSE, Wasserstein Distance, and TOST equivalence tests.
2.3. Model Training
2.3.1. Random Forest (RF) Hyperparameter Optimization
- n_estimators: Number of trees in the ensemble.
- max_depth: Maximum depth of individual trees.
- max_iter: Number of iterations in the iterative imputation process (specific to IterativeImputer).
- min_samples_split: Minimum number of samples required to split an internal node.
- min_samples_leaf: Minimum number of samples required to be at a leaf node.
2.3.2. BRITS Hyperparameter Optimization
- Stage 1: RNN units and subsequence length were tuned, with learning rate = 0.005, batch size = 64, use_regularization = False, and dropout_rate = 0.2 held constant.
- Stage 2: learning rate and batch size were tuned, keeping use_regularization = False and dropout_rate = 0.2 fixed.
- Stage 3: use_regularization and dropout_rate were jointly optimized.
- RNN units: Number of hidden units in the RNN layers. Higher values improve model capacity to learn temporal patterns.
- subsequence length: Input window size used during training.
- learning rate: Controls the optimizer’s step size.
- batch size: Number of samples per training batch.
- use_regularization: Whether to apply dropout/L2 to prevent overfitting.
- dropout_rate: Proportion of neurons randomly deactivated during training.
2.4. Model Evaluation
- Mean Absolute Error (MAE) quantifies the average absolute difference between imputed values ŷₜ and observed values yₜ:
- 2.
- Root Mean Squared Error (RMSE) measures the square root of the average squared differences between imputed and observed values:
- 3.
- Wasserstein Distance, also known as Earth Mover’s Distance (EMD), measures the dissimilarity between two probability distributions by calculating the minimum effort required to transform one distribution into another [23]. In the context of air quality time series, it is useful for comparing distributions of imputed and observed values. Given two cumulative distribution functions, and , the first-order Wasserstein distance is defined as:
- 4.
- The Two One-Sided Test (TOST [22]) is a statistical procedure used to assess the equivalence between the mean of imputed and observed values. Unlike conventional statistical tests that seek to detect significant differences, TOST is specifically designed to demonstrate equivalence, that is, to confirm that the difference is small enough to be considered negligible within a predefined tolerance margin. Let μₒ and μᵢ be the means of observed and imputed values, respectively. According to Santamaría-Bonfil et al. [2], TOST is implemented as two one-sided t-tests:
3. Results
3.1. Results of Hyperparameter Optimization
3.2. Results of Imputation Models
3.2.1. Performance Evaluation on Masked Subset
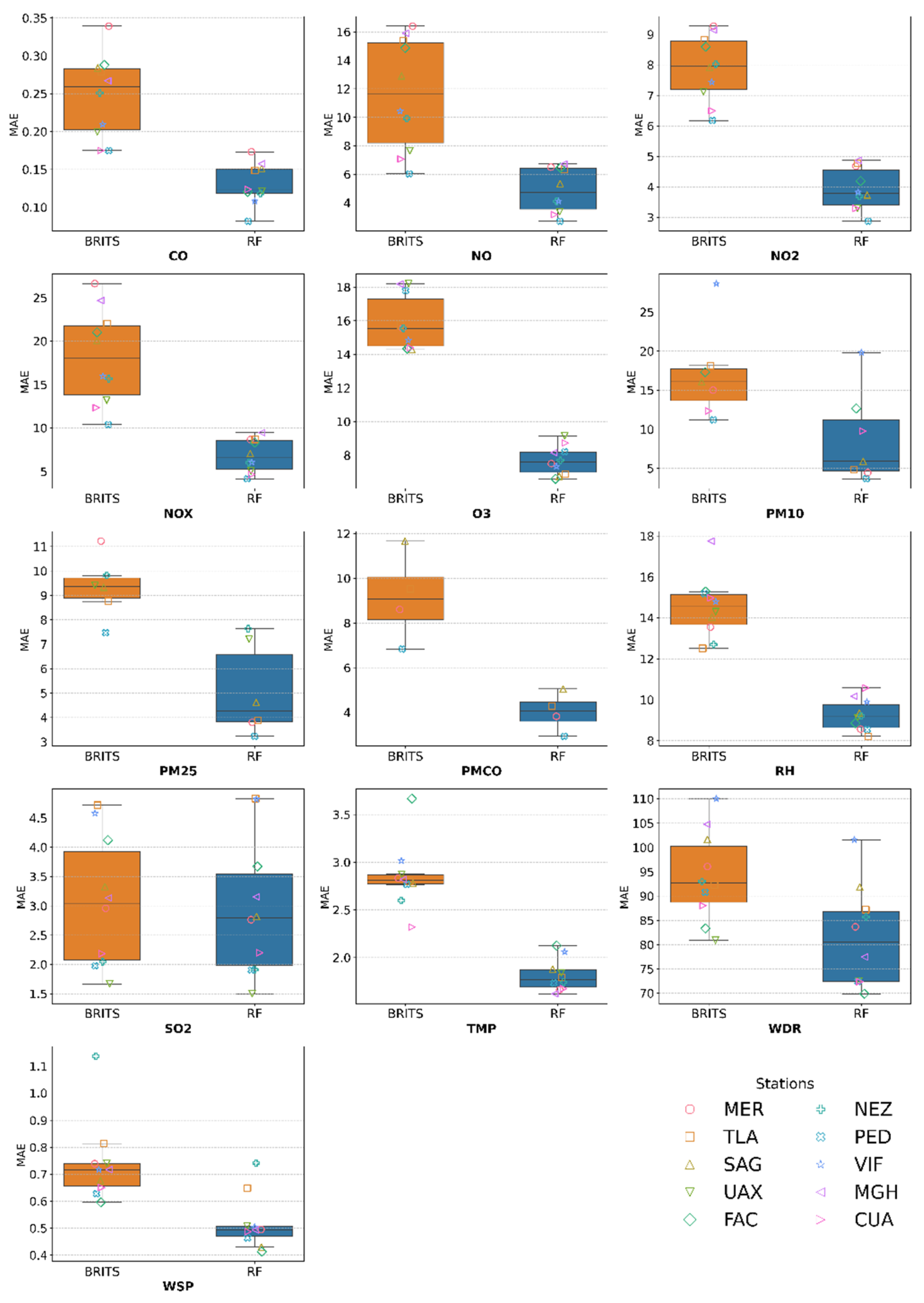
MAE Results
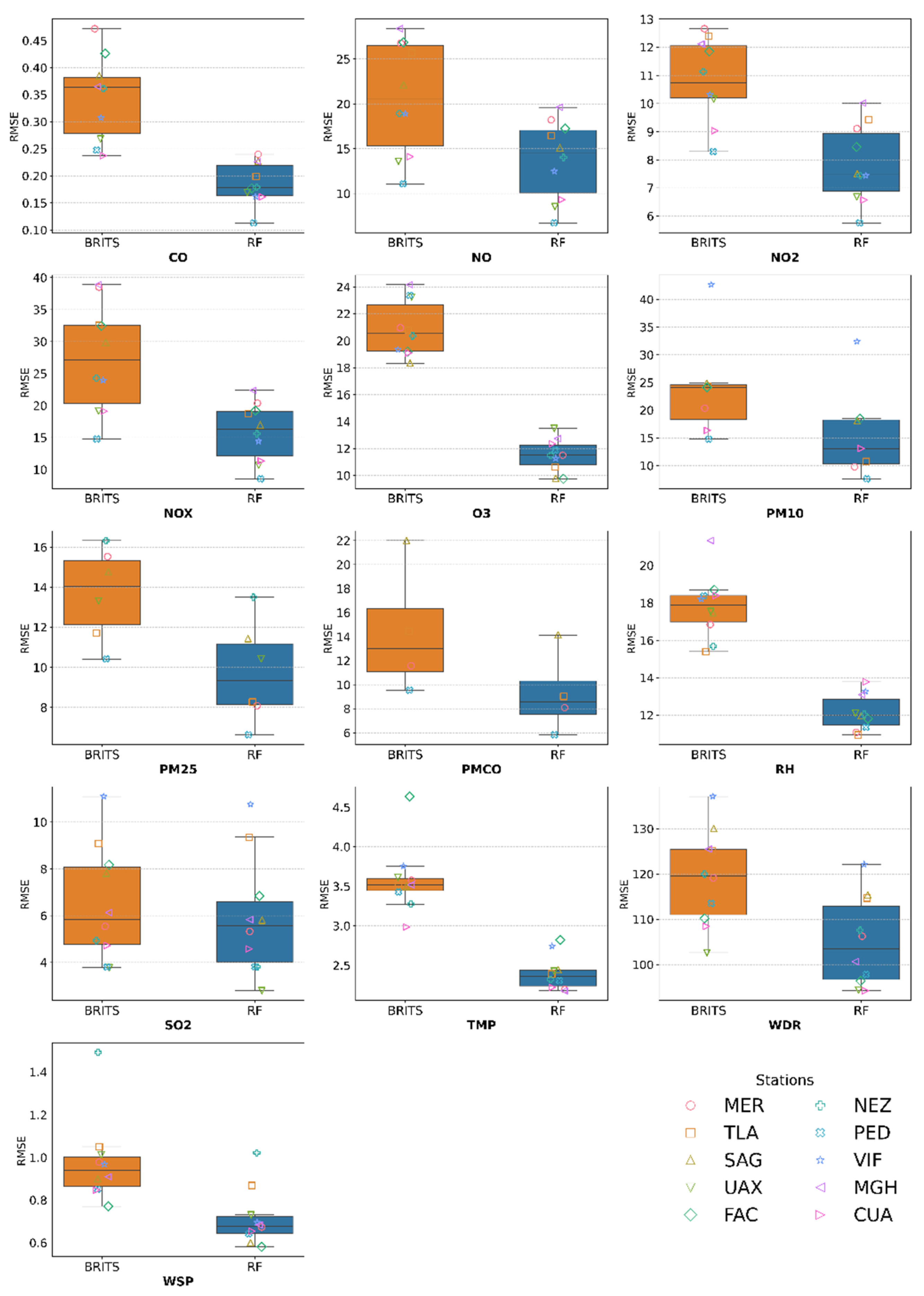
RMSE Results
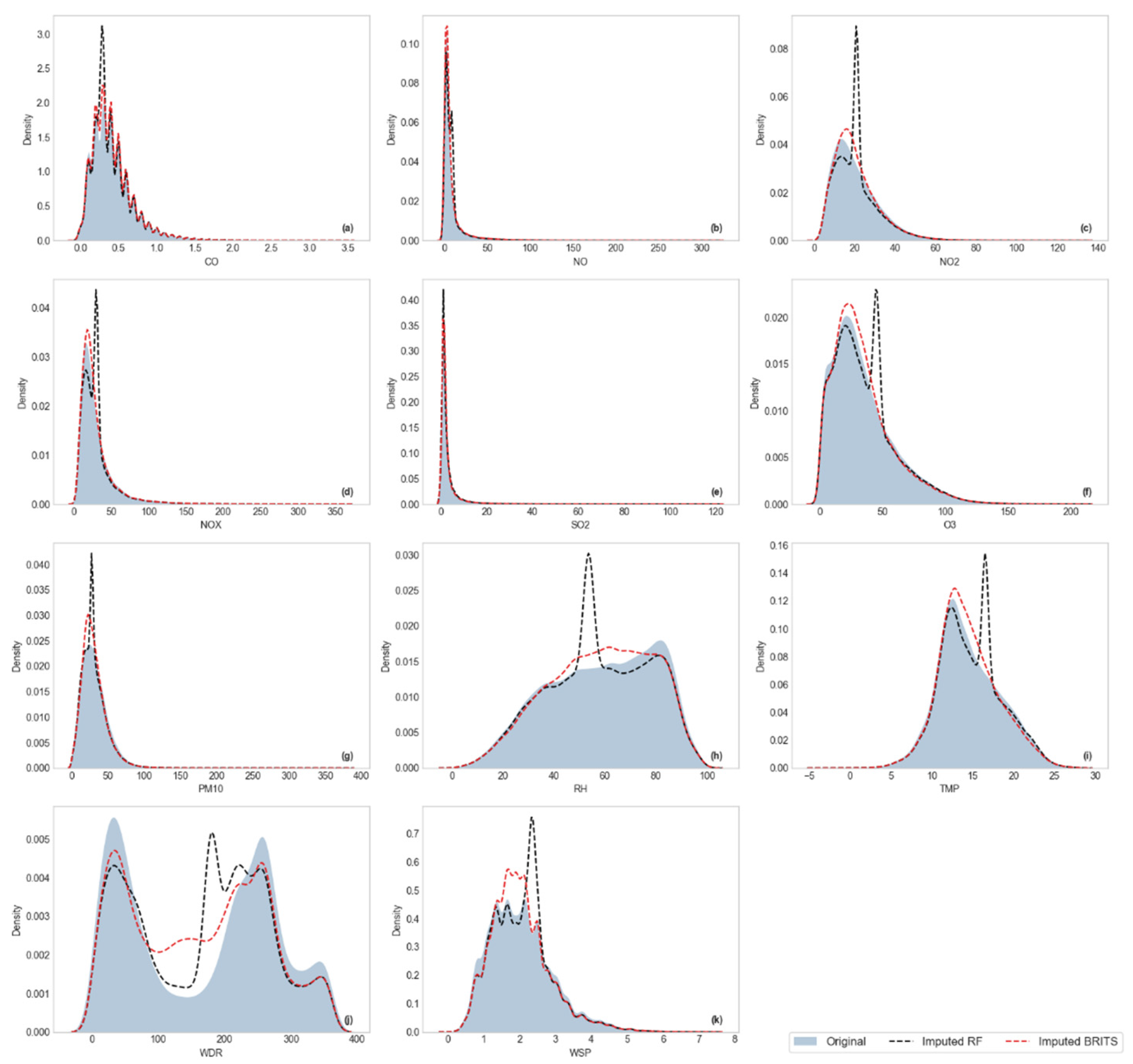
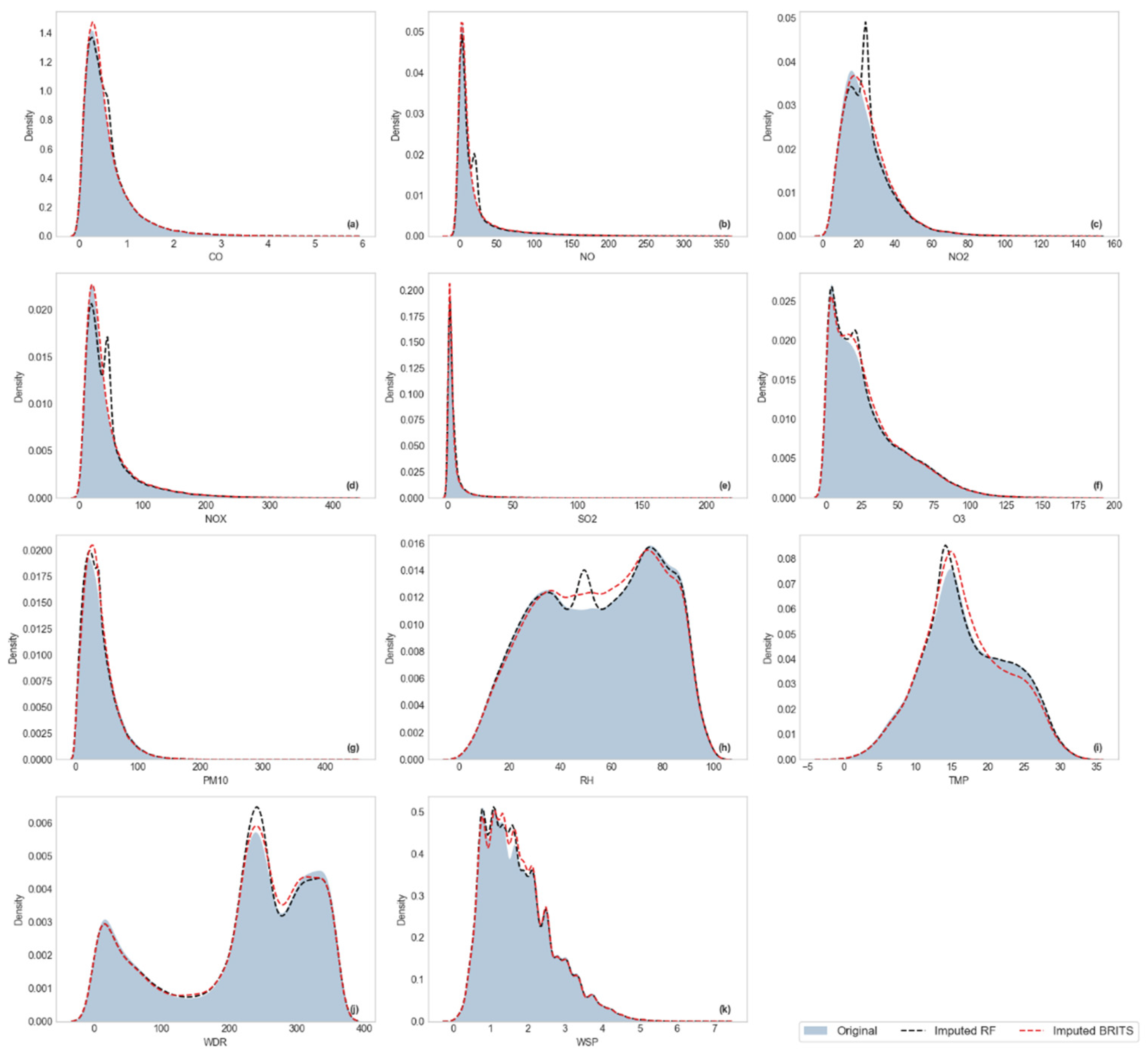
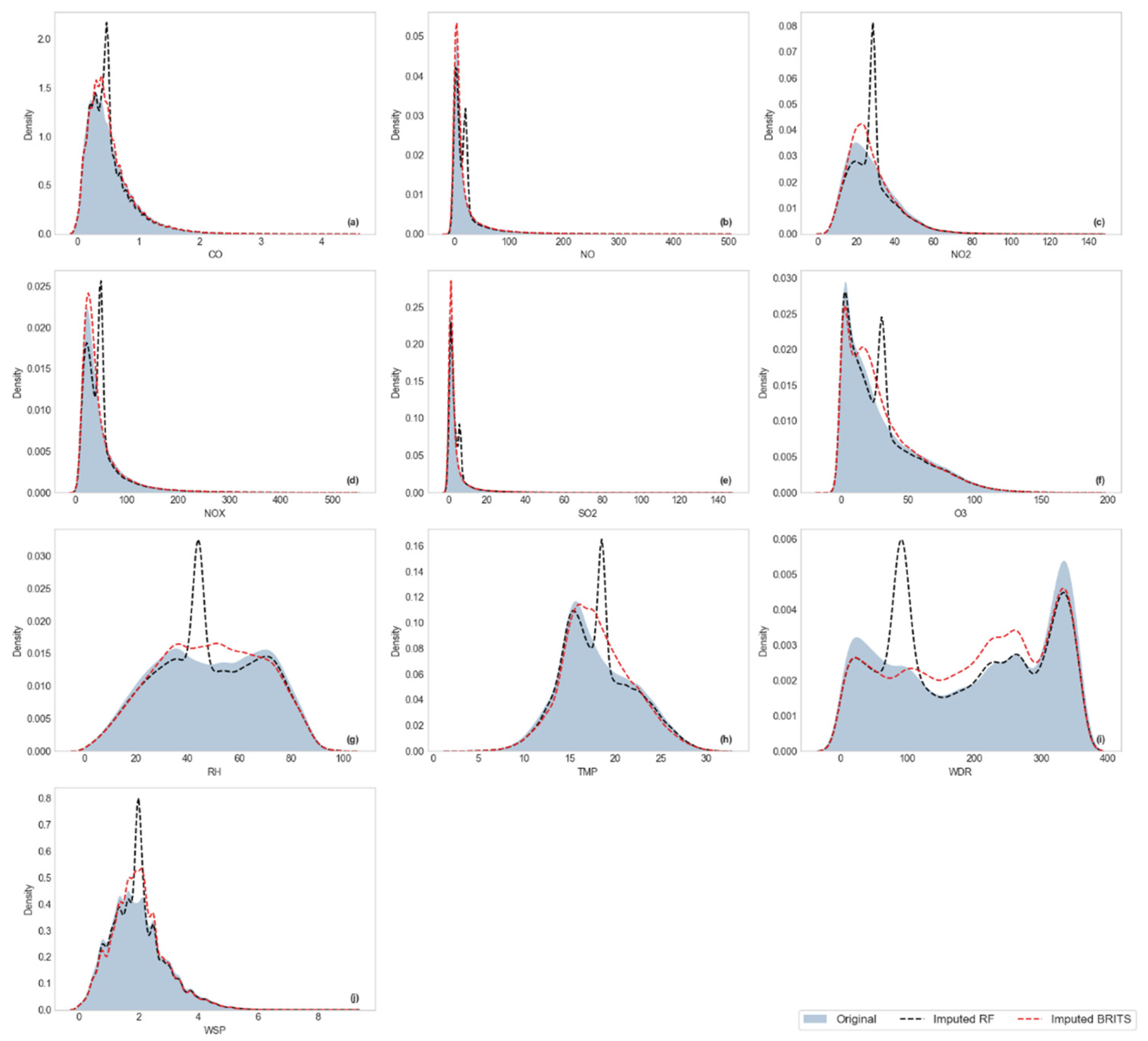
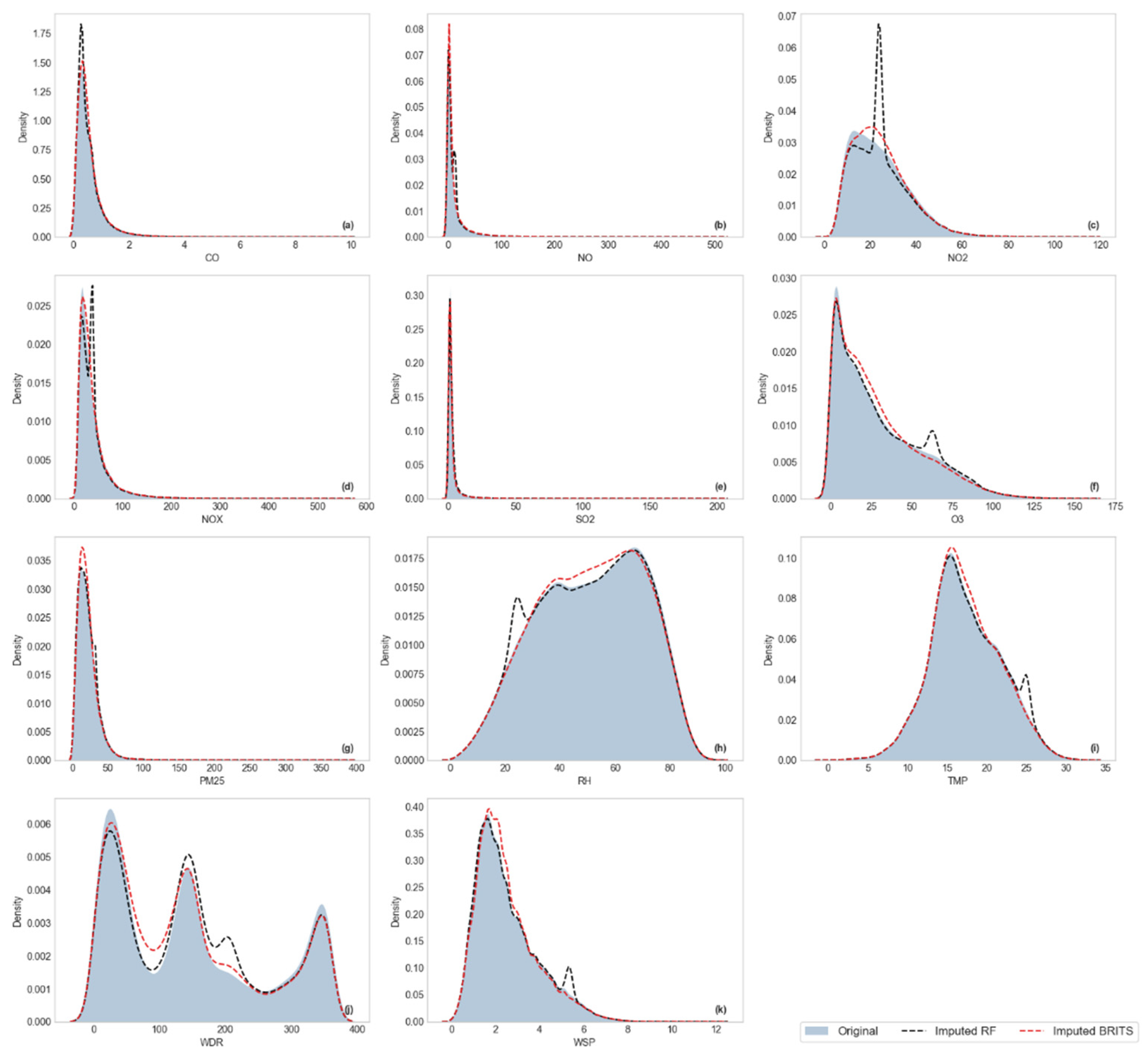
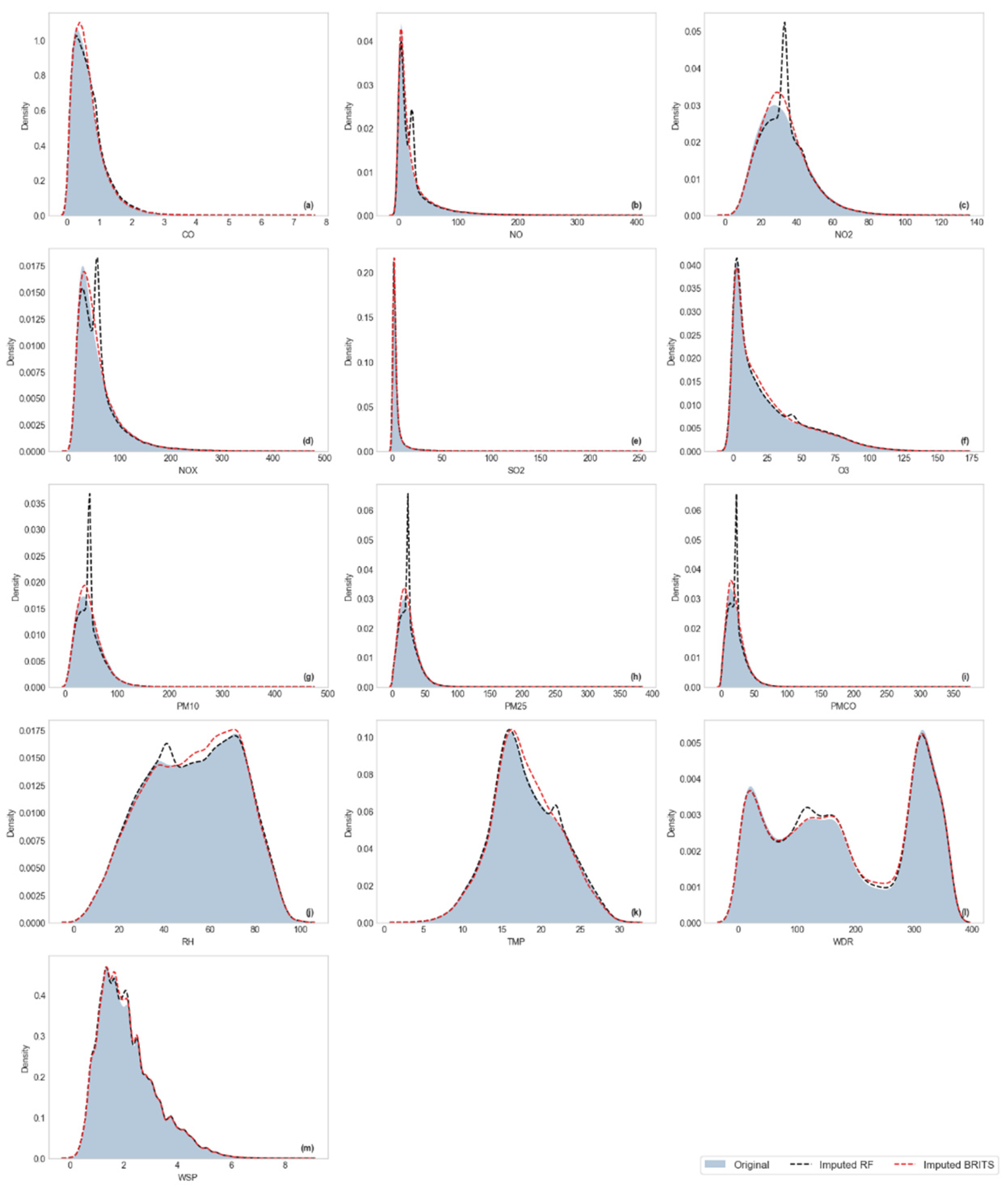
3.2.2. Distributional Similarity Assessment
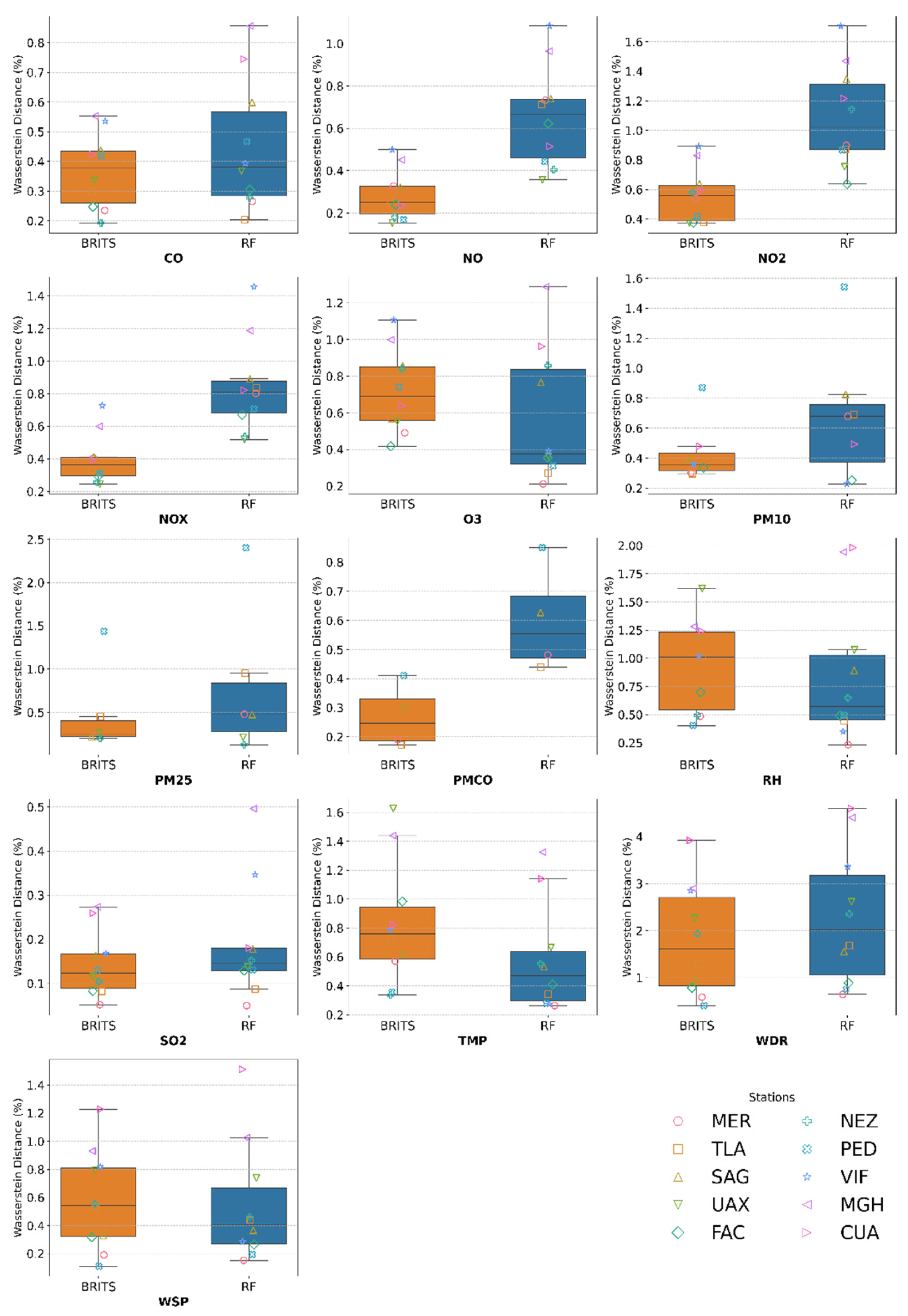
Results of Wasserstein Distance
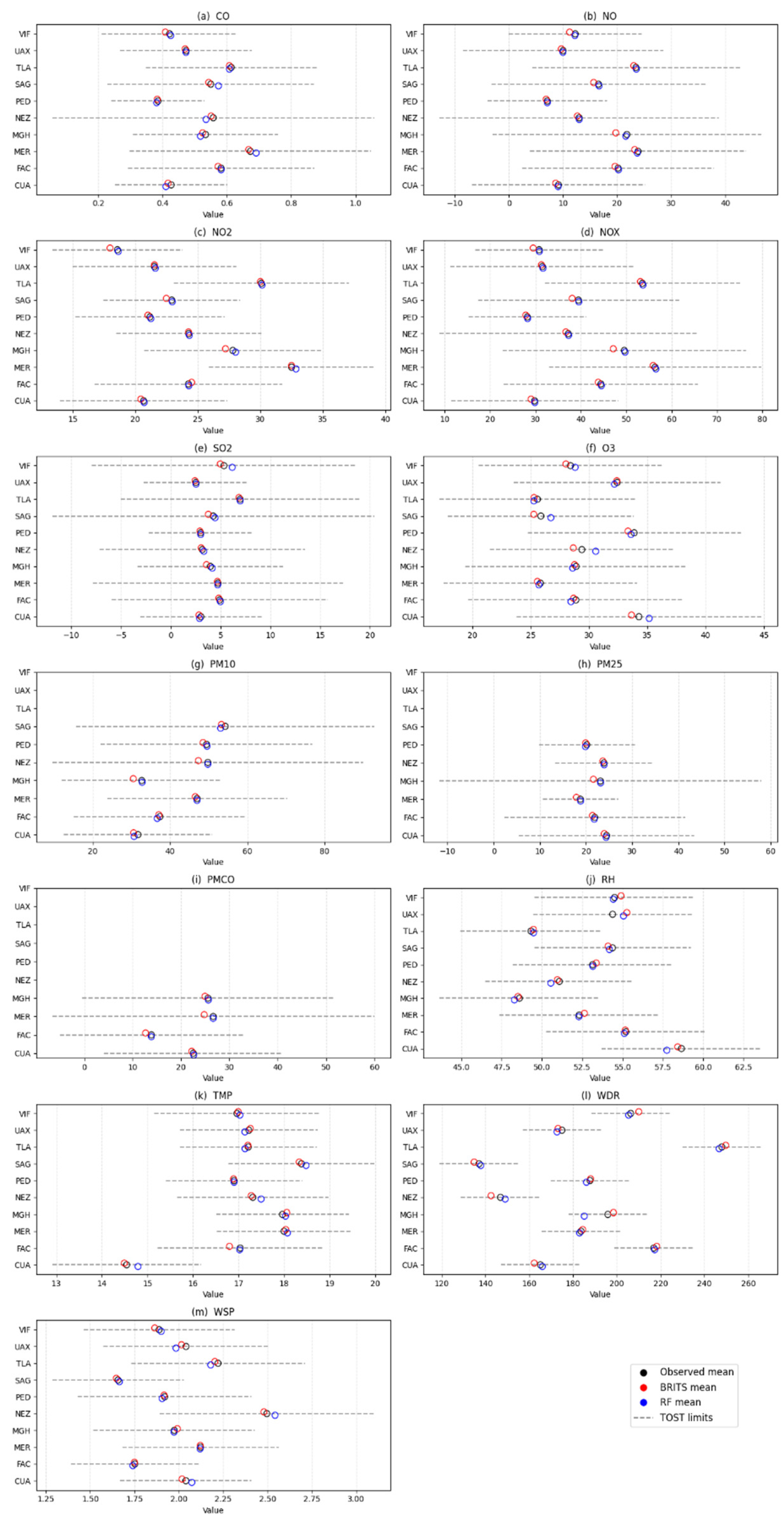
Results of TOST
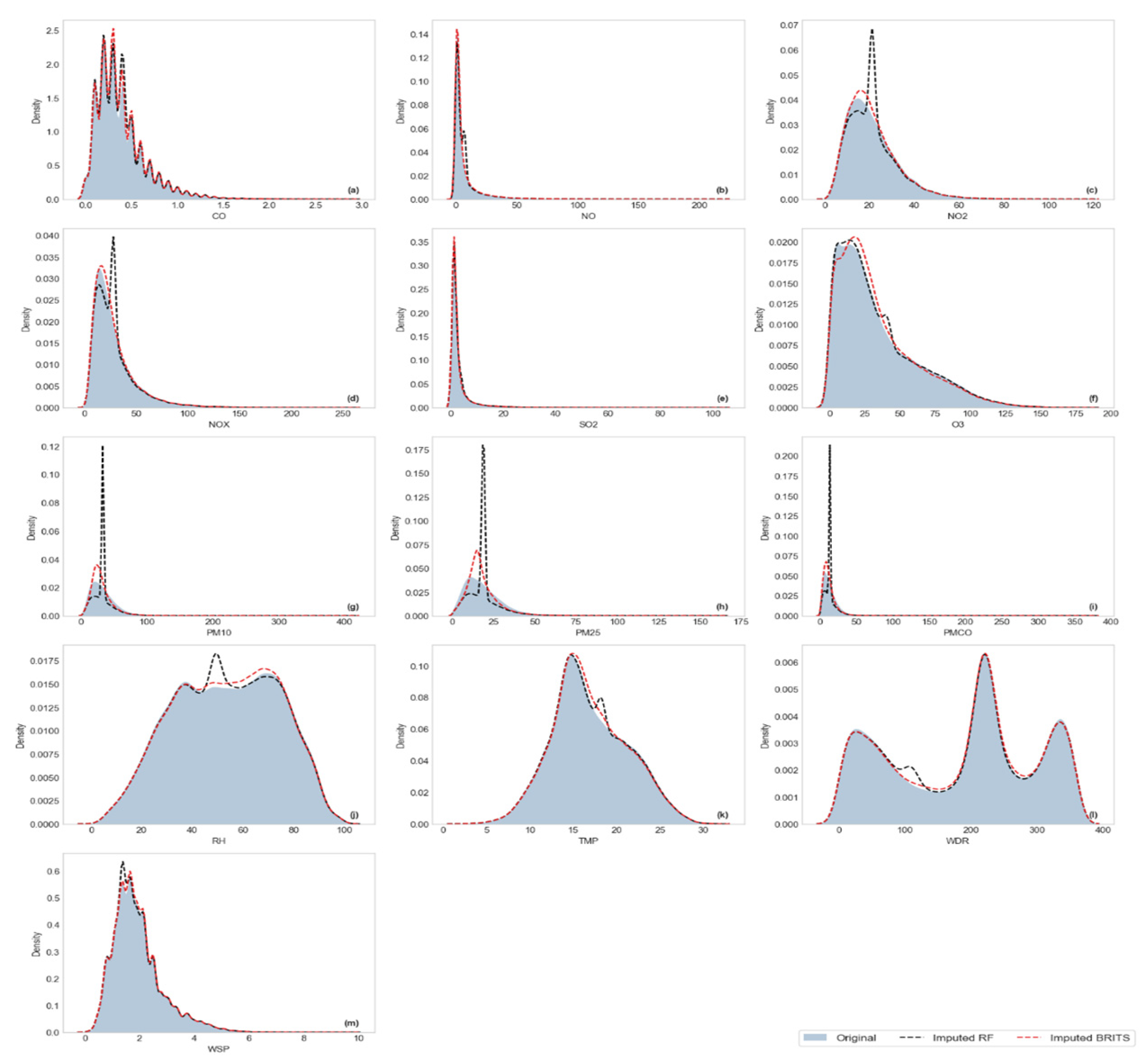
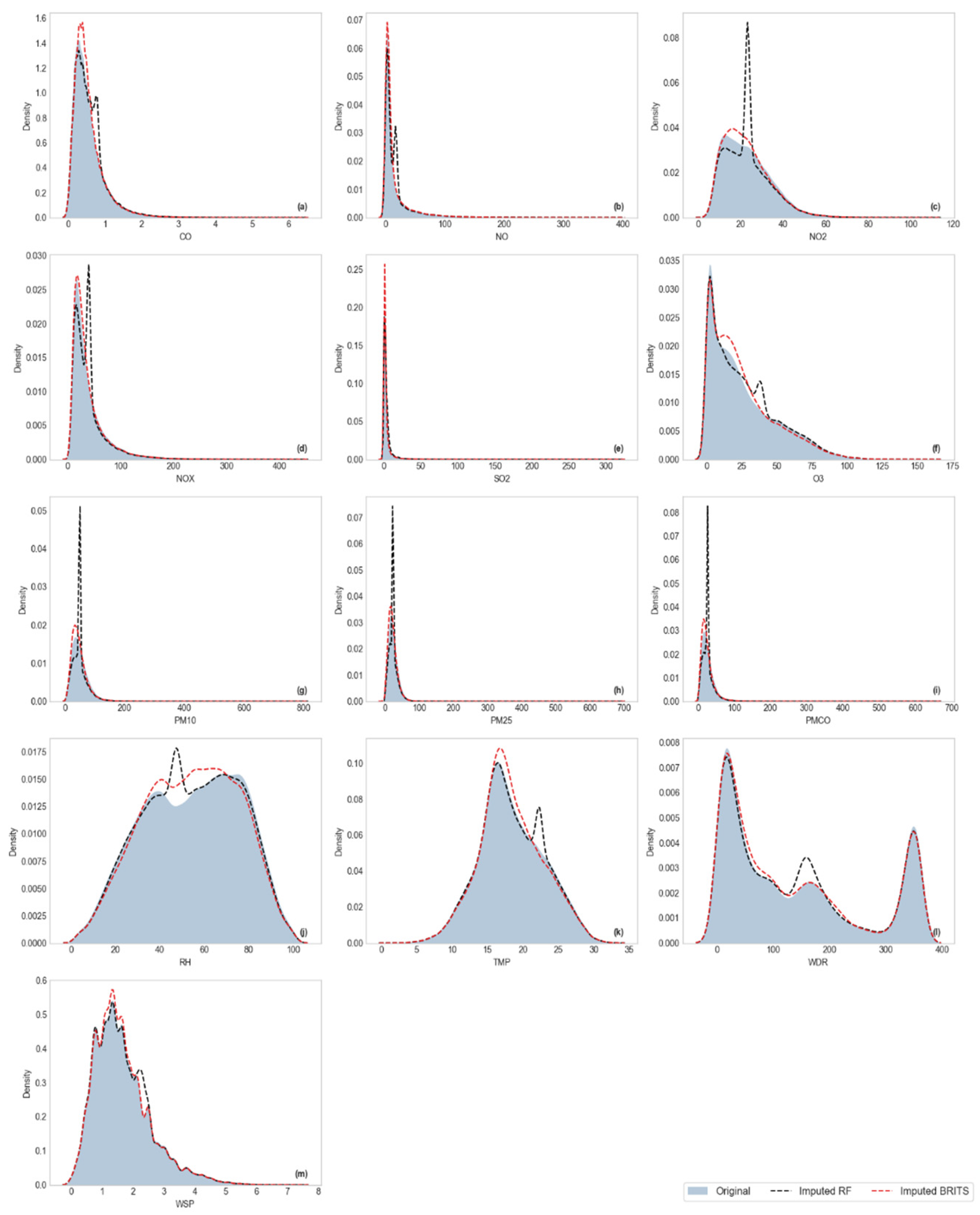
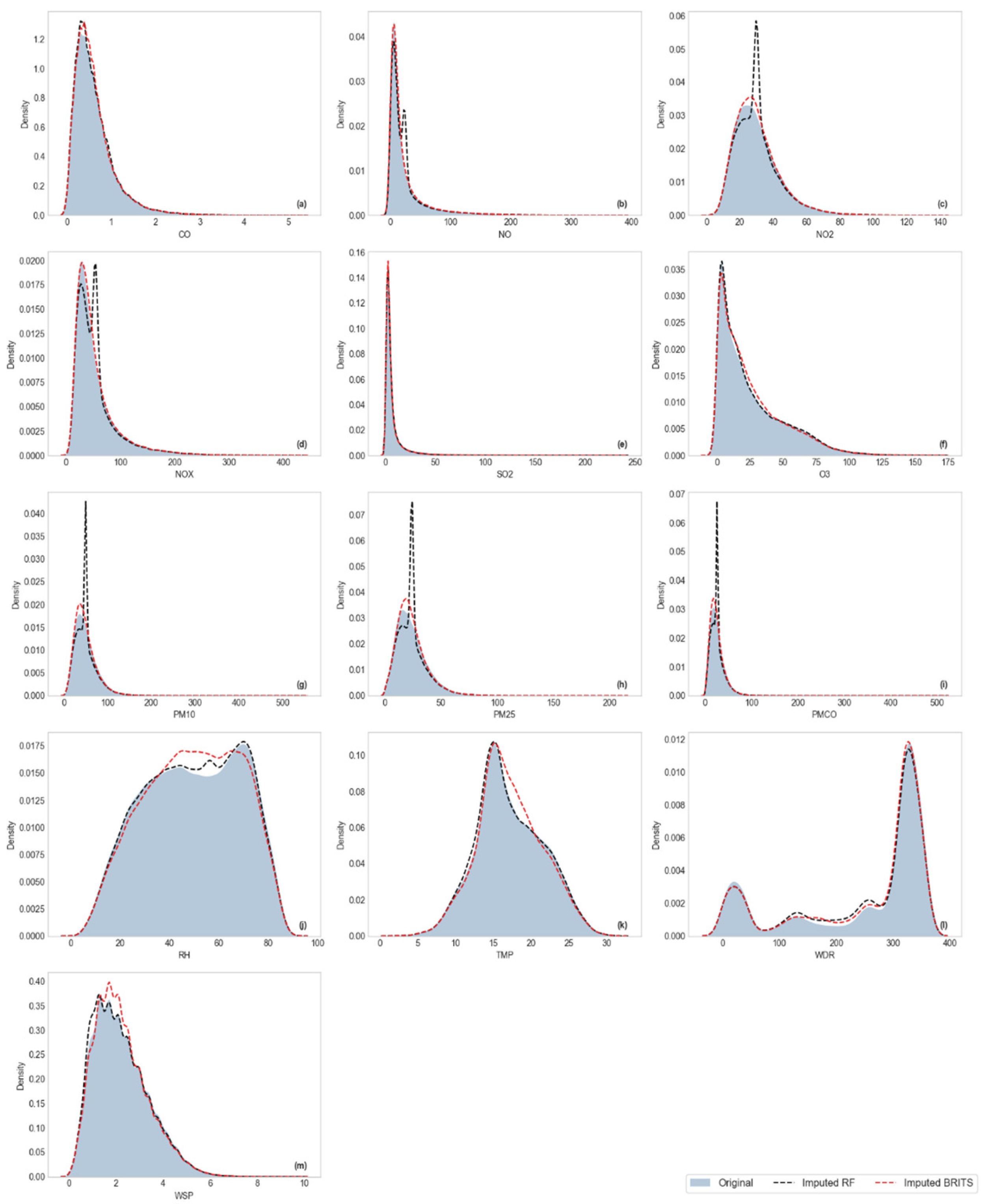
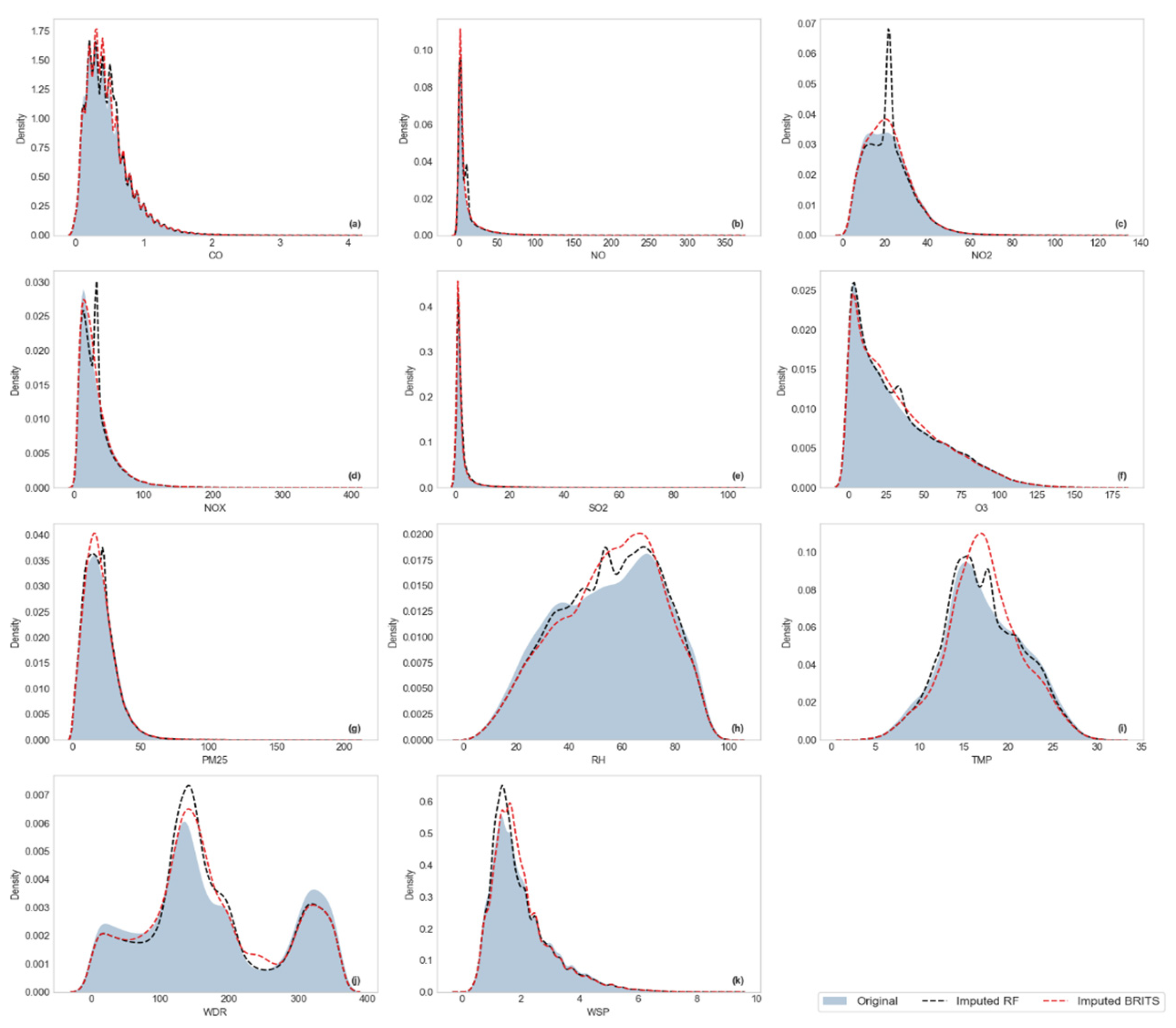
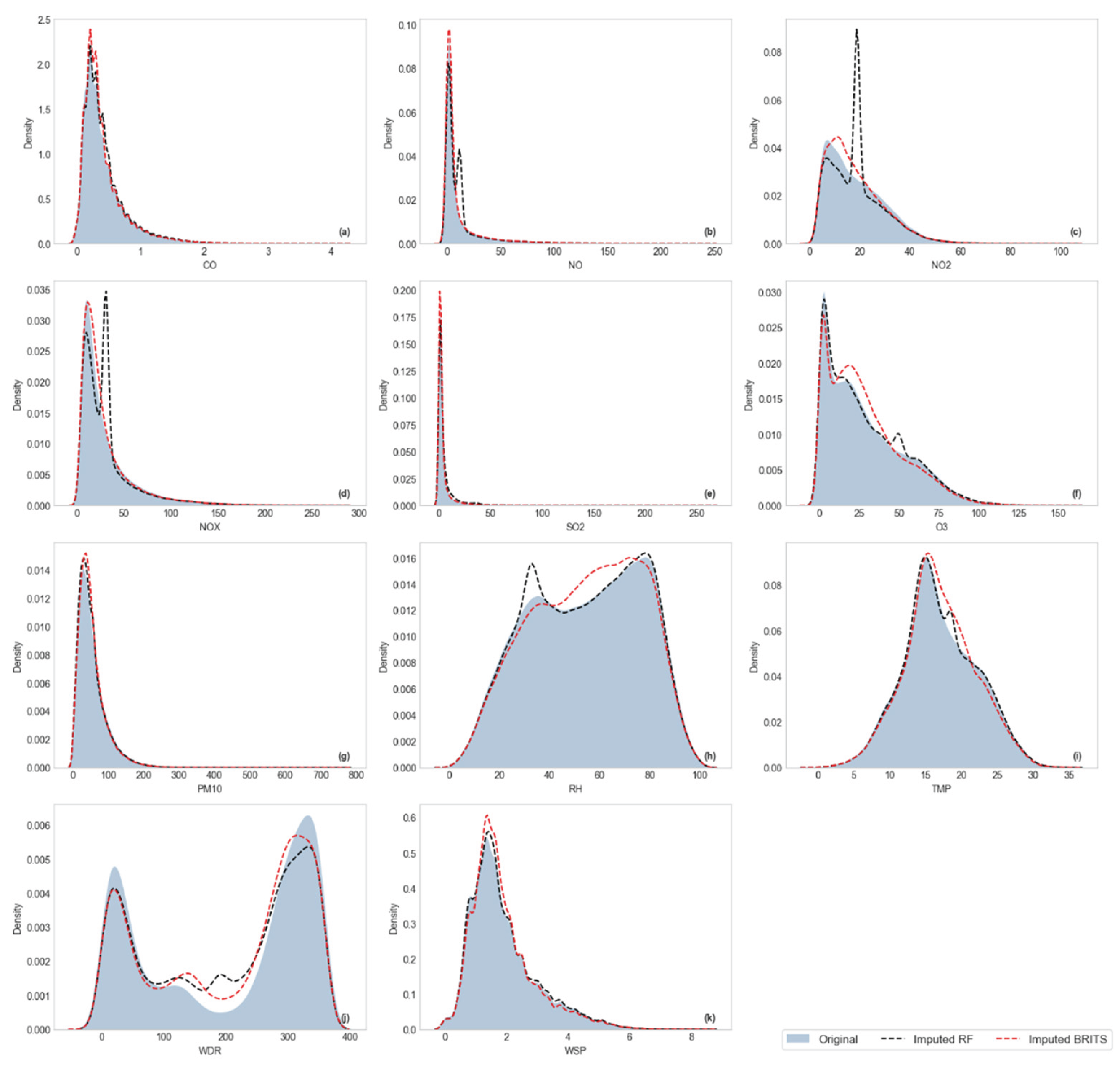
Visualization of Kernel Density Estimation (KDE)
4. Discussion
5. Conclusions
Author Contributions
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| # | Station | Variable | Masked Subset | Complete distribution dataset | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAE | RMSE | Wasserstein Distance (%) | Original Mean | Imputed mean | TOST Interval ± | |||||||
| RF | BRITS | RF | BRITS | RF | BRITS | RF | BRITS | |||||
| 1 | MER | CO | 0.17 | 0.34 | 0.24 | 0.47 | 0.26 | 0.24 | 0.7 | 0.7 | 0.7 | 0.4 |
| 2 | NO | 6.5 | 16.4 | 18.2 | 26.75 | 0.73 | 0.33 | 23.9 | 23.7 | 23.2 | 20 | |
| 3 | NO2 | 4.68 | 9.27 | 9.11 | 12.66 | 0.9 | 0.54 | 32.5 | 32.9 | 32.5 | 6.6 | |
| 4 | NOX | 8.66 | 26.62 | 20.36 | 38.5 | 0.8 | 0.4 | 56.4 | 56.5 | 55.9 | 23.4 | |
| 5 | O3 | 7.5 | 15.54 | 11.51 | 20.96 | 0.21 | 0.49 | 25.8 | 25.7 | 25.6 | 8.3 | |
| 6 | PM10 | 4.43 | 15 | 9.77 | 20.37 | 0.68 | 0.3 | 47 | 47 | 46.5 | 23.3 | |
| 7 | PM25 | 3.8 | 11.22 | 8.08 | 15.54 | 0.48 | 0.22 | 24.4 | 24.4 | 23.9 | 19 | |
| 8 | PMCO | 3.82 | 8.61 | 8.09 | 11.57 | 0.48 | 0.19 | 22.6 | 22.6 | 22.2 | 18.5 | |
| 9 | RH | 8.56 | 13.55 | 11.08 | 16.84 | 0.23 | 0.49 | 52.3 | 52.3 | 52.6 | 5 | |
| 10 | SO2 | 2.76 | 2.96 | 5.33 | 5.54 | 0.05 | 0.05 | 4.7 | 4.7 | 4.7 | 12.6 | |
| 11 | TMP | 1.66 | 2.83 | 2.2 | 3.57 | 0.26 | 0.57 | 18 | 18.1 | 18 | 1.5 | |
| 12 | WDR | 83.67 | 96.06 | 106.24 | 119.12 | 0.65 | 0.59 | 183.7 | 183 | 184.4 | 18 | |
| 13 | WSP | 0.49 | 0.74 | 0.67 | 0.98 | 0.15 | 0.19 | 2.1 | 2.1 | 2.1 | 0.4 | |
| 14 | TLA | CO | 0.15 | 0.28 | 0.2 | 0.37 | 0.2 | 0.3 | 0.6 | 0.6 | 0.6 | 0.3 |
| 15 | NO | 6.35 | 15.37 | 16.47 | 25.7 | 0.71 | 0.26 | 23.5 | 23.5 | 23.1 | 19.2 | |
| 16 | NO2 | 4.78 | 8.83 | 9.42 | 12.39 | 0.87 | 0.38 | 30.1 | 30.2 | 30 | 7 | |
| 17 | NOX | 8.67 | 21.97 | 18.76 | 32.52 | 0.83 | 0.33 | 53.6 | 53.7 | 53.1 | 21.5 | |
| 18 | O3 | 6.87 | 15.75 | 10.63 | 20.8 | 0.27 | 0.57 | 25.6 | 25.3 | 25.3 | 8.4 | |
| 19 | PM10 | 4.85 | 18.14 | 10.77 | 24.25 | 0.69 | 0.3 | 49.5 | 49.6 | 48.6 | 27.5 | |
| 20 | PM25 | 3.89 | 8.75 | 8.27 | 11.72 | 0.95 | 0.45 | 23.9 | 23.9 | 23.6 | 10.6 | |
| 21 | PMCO | 4.27 | 9.52 | 9.03 | 14.44 | 0.44 | 0.17 | 25.6 | 25.7 | 25 | 26 | |
| 22 | RH | 8.21 | 12.51 | 10.96 | 15.4 | 0.44 | 1.01 | 49.3 | 49.5 | 49.5 | 4.4 | |
| 23 | SO2 | 4.83 | 4.72 | 9.36 | 9.09 | 0.09 | 0.08 | 7 | 7 | 6.8 | 12 | |
| 24 | TMP | 1.79 | 2.8 | 2.39 | 3.52 | 0.34 | 0.73 | 17.2 | 17.1 | 17.2 | 1.5 | |
| 25 | WDR | 87.19 | 92.27 | 114.68 | 125.17 | 1.68 | 1.29 | 248 | 246.7 | 249.7 | 18 | |
| 26 | WSP | 0.65 | 0.81 | 0.87 | 1.05 | 0.44 | 0.54 | 2.2 | 2.2 | 2.2 | 0.5 | |
| 27 | SAG | CO | 0.15 | 0.28 | 0.23 | 0.39 | 0.6 | 0.44 | 0.6 | 0.6 | 0.5 | 0.3 |
| 28 | NO | 5.33 | 12.91 | 15.1 | 22.09 | 0.74 | 0.32 | 16.5 | 16.6 | 15.7 | 19.8 | |
| 29 | NO2 | 3.73 | 7.91 | 7.5 | 10.35 | 1.35 | 0.64 | 22.9 | 23 | 22.5 | 5.5 | |
| 30 | NOX | 7.05 | 20.04 | 16.98 | 29.87 | 0.89 | 0.41 | 39.5 | 39.6 | 38.1 | 22.2 | |
| 31 | O3 | 6.75 | 14.29 | 9.78 | 18.33 | 0.77 | 0.86 | 25.9 | 26.7 | 25.3 | 8 | |
| 32 | PM10 | 5.9 | 16.08 | 18.06 | 24.86 | 0.83 | 0.39 | 49.8 | 49.8 | 47.3 | 40.3 | |
| 33 | PM25 | 4.61 | 9.33 | 11.43 | 14.77 | 0.47 | 0.22 | 23.1 | 23.2 | 21.6 | 34.9 | |
| 34 | PMCO | 5.04 | 11.67 | 14.14 | 21.99 | 0.63 | 0.3 | 26.7 | 26.7 | 24.8 | 33.3 | |
| 35 | RH | 9.35 | 14.04 | 11.98 | 17.46 | 0.89 | 1.2 | 54.4 | 54.2 | 54.1 | 4.9 | |
| 36 | SO2 | 2.82 | 3.33 | 5.8 | 7.81 | 0.18 | 0.16 | 4.3 | 4.4 | 3.8 | 16.2 | |
| 37 | TMP | 1.88 | 2.78 | 2.44 | 3.5 | 0.53 | 0.62 | 18.4 | 18.5 | 18.3 | 1.6 | |
| 38 | WDR | 91.83 | 101.65 | 115.36 | 130.03 | 1.56 | 0.95 | 137.1 | 137.9 | 134.8 | 18 | |
| 39 | WSP | 0.43 | 0.68 | 0.6 | 0.9 | 0.37 | 0.33 | 1.7 | 1.7 | 1.6 | 0.4 | |
| 40 | UAX | CO | 0.12 | 0.2 | 0.17 | 0.27 | 0.37 | 0.33 | 0.5 | 0.5 | 0.5 | 0.2 |
| 41 | NO | 3.37 | 7.64 | 8.56 | 13.59 | 0.36 | 0.15 | 10 | 10 | 9.7 | 18.6 | |
| 42 | NO2 | 3.3 | 7.12 | 6.69 | 10.16 | 0.75 | 0.37 | 21.5 | 21.6 | 21.5 | 6.6 | |
| 43 | NOX | 5.05 | 13.2 | 10.68 | 19.12 | 0.52 | 0.25 | 31.5 | 31.7 | 31.3 | 20.4 | |
| 44 | O3 | 9.15 | 18.21 | 13.5 | 23.24 | 0.36 | 0.55 | 32.4 | 32.2 | 32.4 | 8.9 | |
| 45 | PM25 | 7.21 | 9.4 | 10.42 | 13.31 | 0.21 | 0.26 | 20.2 | 19.9 | 19.9 | 10.4 | |
| 46 | RH | 9.13 | 14.3 | 12.11 | 17.54 | 1.07 | 1.62 | 54.4 | 55.1 | 55.3 | 4.9 | |
| 47 | SO2 | 1.5 | 1.67 | 2.79 | 3.78 | 0.14 | 0.11 | 2.5 | 2.5 | 2.4 | 5.3 | |
| 48 | TMP | 1.84 | 2.87 | 2.43 | 3.61 | 0.66 | 1.63 | 17.2 | 17.1 | 17.3 | 1.5 | |
| 49 | WDR | 72.5 | 80.91 | 94.34 | 102.6 | 2.62 | 2.26 | 175 | 172.7 | 173.1 | 18 | |
| 50 | WSP | 0.51 | 0.74 | 0.73 | 1.01 | 0.74 | 0.79 | 2 | 2 | 2 | 0.5 | |
| 51 | FAC | CO | 0.12 | 0.29 | 0.18 | 0.43 | 0.3 | 0.25 | 0.6 | 0.6 | 0.6 | 0.3 |
| 52 | NO | 6.46 | 14.86 | 17.24 | 26.84 | 0.62 | 0.24 | 20.2 | 20.2 | 19.6 | 17.7 | |
| 53 | NO2 | 4.19 | 8.59 | 8.46 | 11.85 | 0.64 | 0.37 | 24.3 | 24.3 | 24.5 | 7.5 | |
| 54 | NOX | 8.33 | 21.02 | 19.2 | 32.45 | 0.67 | 0.29 | 44.4 | 44.5 | 43.8 | 21.5 | |
| 55 | O3 | 6.6 | 14.33 | 9.75 | 19.2 | 0.36 | 0.42 | 28.9 | 28.4 | 28.7 | 9.3 | |
| 56 | PM10 | 12.67 | 17.31 | 18.44 | 24.11 | 0.25 | 0.34 | 37.4 | 36.7 | 37.1 | 22.4 | |
| 57 | RH | 8.85 | 15.28 | 11.82 | 18.7 | 0.49 | 0.7 | 55.2 | 55.1 | 55.2 | 5 | |
| 58 | SO2 | 3.67 | 4.12 | 6.84 | 8.17 | 0.13 | 0.08 | 4.9 | 5 | 4.8 | 10.9 | |
| 59 | TMP | 2.12 | 3.67 | 2.82 | 4.63 | 0.41 | 0.98 | 17 | 17 | 16.8 | 1.8 | |
| 60 | WDR | 69.89 | 83.32 | 96.36 | 110.18 | 0.89 | 0.78 | 216.9 | 217.3 | 218.2 | 18 | |
| 61 | WSP | 0.41 | 0.6 | 0.58 | 0.77 | 0.27 | 0.32 | 1.8 | 1.7 | 1.7 | 0.4 | |
| 62 | NEZ | CO | 0.12 | 0.25 | 0.18 | 0.36 | 0.28 | 0.19 | 0.6 | 0.5 | 0.6 | 0.5 |
| 63 | NO | 4.1 | 9.93 | 14.03 | 18.93 | 0.4 | 0.18 | 13 | 13 | 12.6 | 25.8 | |
| 64 | NO2 | 3.69 | 8.02 | 7.44 | 11.14 | 1.14 | 0.58 | 24.3 | 24.3 | 24.3 | 5.8 | |
| 65 | NOX | 5.99 | 15.67 | 15.6 | 24.28 | 0.54 | 0.26 | 37.2 | 37.3 | 36.7 | 28.4 | |
| 66 | O3 | 7.69 | 15.55 | 11.49 | 20.37 | 0.86 | 0.84 | 29.4 | 30.6 | 28.7 | 7.9 | |
| 67 | PM25 | 7.64 | 9.81 | 13.51 | 16.34 | 0.12 | 0.2 | 21.9 | 21.8 | 21.4 | 19.6 | |
| 68 | RH | 9.21 | 12.7 | 12.05 | 15.69 | 0.65 | 0.49 | 51.1 | 50.5 | 51 | 4.6 | |
| 69 | SO2 | 1.91 | 2.05 | 3.82 | 4.93 | 0.15 | 0.1 | 3.2 | 3.3 | 3 | 10.3 | |
| 70 | TMP | 1.74 | 2.6 | 2.32 | 3.27 | 0.55 | 0.34 | 17.3 | 17.5 | 17.3 | 1.7 | |
| 71 | WDR | 85.72 | 93.01 | 107.58 | 119.99 | 2.35 | 1.94 | 146.9 | 149 | 142.6 | 18 | |
| 72 | WSP | 0.74 | 1.14 | 1.02 | 1.49 | 0.46 | 0.55 | 2.5 | 2.5 | 2.5 | 0.6 | |
| 73 | PED | CO | 0.08 | 0.17 | 0.11 | 0.25 | 0.47 | 0.42 | 0.4 | 0.4 | 0.4 | 0.1 |
| 74 | NO | 2.7 | 6.03 | 6.72 | 11.08 | 0.44 | 0.17 | 7.1 | 7.1 | 6.9 | 11.1 | |
| 75 | NO2 | 2.87 | 6.17 | 5.75 | 8.29 | 0.87 | 0.42 | 21.2 | 21.3 | 21 | 6 | |
| 76 | NOX | 4.15 | 10.39 | 8.54 | 14.76 | 0.71 | 0.31 | 28.2 | 28.3 | 27.9 | 13 | |
| 77 | O3 | 8.2 | 17.79 | 11.87 | 23.37 | 0.31 | 0.74 | 33.9 | 33.6 | 33.3 | 9.2 | |
| 78 | PM10 | 3.66 | 11.2 | 7.6 | 14.77 | 1.54 | 0.87 | 32.7 | 32.8 | 30.5 | 20.8 | |
| 79 | PM25 | 3.23 | 7.47 | 6.63 | 10.42 | 2.4 | 1.44 | 18.8 | 18.8 | 17.9 | 8.2 | |
| 80 | PMCO | 2.92 | 6.83 | 5.83 | 9.54 | 0.85 | 0.41 | 13.9 | 13.9 | 12.7 | 19 | |
| 81 | RH | 8.54 | 15.18 | 11.36 | 18.37 | 0.5 | 0.4 | 53.1 | 53.2 | 53.4 | 5 | |
| 82 | SO2 | 1.91 | 1.98 | 3.81 | 3.8 | 0.13 | 0.13 | 3 | 3 | 2.9 | 5.3 | |
| 83 | TMP | 1.73 | 2.77 | 2.31 | 3.43 | 0.28 | 0.36 | 16.9 | 16.9 | 16.9 | 1.5 | |
| 84 | WDR | 72.41 | 90.79 | 97.81 | 113.5 | 0.75 | 0.41 | 187.7 | 186.1 | 188 | 18 | |
| 85 | WSP | 0.46 | 0.63 | 0.64 | 0.85 | 0.19 | 0.11 | 1.9 | 1.9 | 1.9 | 0.5 | |
| 86 | VIF | CO | 0.11 | 0.21 | 0.16 | 0.31 | 0.39 | 0.54 | 0.4 | 0.4 | 0.4 | 0.2 |
| 87 | NO | 4.1 | 10.42 | 12.47 | 18.89 | 1.08 | 0.5 | 12.2 | 12.2 | 11.2 | 12.3 | |
| 88 | NO2 | 3.83 | 7.43 | 7.44 | 10.32 | 1.71 | 0.89 | 18.6 | 18.7 | 18 | 5.2 | |
| 89 | NOX | 6.05 | 15.96 | 14.41 | 23.93 | 1.45 | 0.73 | 30.8 | 30.9 | 29.5 | 14.1 | |
| 90 | O3 | 7.34 | 14.84 | 11.27 | 19.34 | 0.39 | 1.11 | 28.4 | 28.8 | 28 | 7.9 | |
| 91 | PM10 | 19.81 | 28.63 | 32.39 | 42.66 | 0.23 | 0.36 | 54.3 | 53.1 | 53.3 | 38.6 | |
| 92 | RH | 9.89 | 14.8 | 13.27 | 18.2 | 0.35 | 1.02 | 54.5 | 54.4 | 54.9 | 5 | |
| 93 | SO2 | 4.82 | 4.58 | 10.75 | 11.09 | 0.35 | 0.17 | 5.3 | 6.2 | 5 | 13.3 | |
| 94 | TMP | 2.06 | 3.01 | 2.74 | 3.75 | 0.27 | 0.79 | 17 | 17 | 17 | 1.8 | |
| 95 | WDR | 101.58 | 110.04 | 122.12 | 137.12 | 3.36 | 2.85 | 206.3 | 205.4 | 210 | 18 | |
| 96 | WSP | 0.5 | 0.72 | 0.7 | 0.97 | 0.28 | 0.82 | 1.9 | 1.9 | 1.9 | 0.4 | |
| 97 | MGH | CO | 0.16 | 0.27 | 0.23 | 0.37 | 0.86 | 0.55 | 0.5 | 0.5 | 0.5 | 0.2 |
| 98 | NO | 6.72 | 15.9 | 19.61 | 28.37 | 0.96 | 0.45 | 21.8 | 21.6 | 19.7 | 24.9 | |
| 99 | NO2 | 4.87 | 9.14 | 10.02 | 12.12 | 1.47 | 0.83 | 27.8 | 28 | 27.2 | 7.1 | |
| 100 | NOX | 9.46 | 24.66 | 22.37 | 38.89 | 1.19 | 0.6 | 49.6 | 49.7 | 47.1 | 26.8 | |
| 101 | O3 | 8.15 | 18.19 | 12.73 | 24.17 | 1.29 | 1 | 28.9 | 28.6 | 28.8 | 9.5 | |
| 102 | RH | 10.16 | 17.75 | 13.09 | 21.33 | 1.94 | 1.28 | 48.6 | 48.3 | 48.5 | 5 | |
| 103 | SO2 | 3.15 | 3.13 | 5.83 | 6.13 | 0.5 | 0.27 | 4 | 4.2 | 3.6 | 7.3 | |
| 104 | TMP | 1.62 | 2.82 | 2.18 | 3.52 | 1.32 | 1.44 | 18 | 18 | 18.1 | 1.5 | |
| 105 | WDR | 77.46 | 104.76 | 100.65 | 125.51 | 4.4 | 2.9 | 195.9 | 185.1 | 198.5 | 18 | |
| 106 | WSP | 0.49 | 0.72 | 0.68 | 0.91 | 1.02 | 0.93 | 2 | 2 | 2 | 0.5 | |
| 107 | CUA | CO | 0.12 | 0.17 | 0.16 | 0.24 | 0.74 | 0.42 | 0.4 | 0.4 | 0.4 | 0.2 |
| 108 | NO | 3.16 | 7.06 | 9.31 | 14.1 | 0.51 | 0.23 | 9.2 | 9.1 | 8.6 | 16.1 | |
| 109 | NO2 | 3.29 | 6.49 | 6.57 | 9.03 | 1.22 | 0.59 | 20.7 | 20.7 | 20.5 | 6.7 | |
| 110 | NOX | 4.64 | 12.33 | 11.34 | 19.12 | 0.82 | 0.39 | 29.8 | 29.9 | 29 | 18.4 | |
| 111 | O3 | 8.73 | 14.38 | 12.36 | 19.1 | 0.96 | 0.64 | 34.3 | 35.2 | 33.6 | 10.5 | |
| 112 | PM10 | 9.76 | 12.33 | 13.09 | 16.34 | 0.49 | 0.48 | 31.7 | 30.7 | 30.5 | 19.2 | |
| 113 | RH | 10.58 | 14.97 | 13.78 | 18.39 | 1.98 | 1.24 | 58.6 | 57.7 | 58.4 | 5 | |
| 114 | SO2 | 2.2 | 2.19 | 4.58 | 4.72 | 0.18 | 0.26 | 3 | 2.9 | 2.8 | 6.1 | |
| 115 | TMP | 1.67 | 2.32 | 2.22 | 2.99 | 1.14 | 0.82 | 14.5 | 14.8 | 14.5 | 1.6 | |
| 116 | WDR | 72.39 | 88.03 | 94.23 | 108.43 | 4.59 | 3.92 | 165.1 | 166 | 162.3 | 18 | |
| 117 | WSP | 0.49 | 0.65 | 0.65 | 0.85 | 1.51 | 1.23 | 2 | 2.1 | 2 | 0.4 | |
References
- World Health Organization (WHO). Ambient (Outdoor) Air Pollution. Available online: https://www.who.int/news-room/fact-sheets/detail/ambient- (accessed on day month year).
- State of Global Air (SoGA). State of Global Air Report 2024. Available online: https://www.stateofglobalair.org/hap (accessed on 19 July 2025).
- World Health Organization (WHO). Air Pollution. Available online: https://www.who.int/health-topics/air-pollution#tab=tab_1 (accessed on 19 July 2025).
- Kim, T.; Kim, J.; Yang, W.; Lee, H.; Choo, J. Missing Value Imputation of Time-Series Air-Quality Data via Deep Neural Networks. Int. J. Environ. Res. Public Health 2021, 18, 12213. [Google Scholar] [CrossRef] [PubMed]
- He, M.Z.; Yitshak-Sade, M.; Just, A.C.; Gutiérrez-Avila, I.; Dorman, M.; de Hoogh, K.; Kloog, I. Predicting Fine-Scale Daily NO₂ over Mexico City Using an Ensemble Modeling Approach. Atmos. Pollut. Res. 2023, 14, 101763. [Google Scholar] [CrossRef] [PubMed]
- Gobierno de México. Programa de Gestión para Mejorar la Calidad del Aire (PROAIRE). Available online: https://www.gob.mx/semarnat/acciones-y-programas/programas-de-gestion-para-mejorar-la-calidad-del-aire (accessed on 19 July 2025).
- Gobierno de la Ciudad de México. Mexico City Atmospheric Monitoring System (SIMAT). Available online: http://www.aire.cdmx.gob.mx/default.php (accessed on 19 July 2025).
- Zhang, X.; Zhou, P. A Transferred Spatio-Temporal Deep Model Based on Multi-LSTM Auto-Encoder for Air Pollution Time Series Missing Value Imputation. Future Gener. Comput. Syst. 2024, 156, 325–338. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, K.; He, Y.; Fu, Q.; Luo, W.; Li, W.; Xiao, S. Research on Missing Value Imputation to Improve the Validity of Air Quality Data Evaluation on the Qinghai-Tibetan Plateau. Atmosphere 2023, 14, 1821. [Google Scholar] [CrossRef]
- Hua, V.; Nguyen, T.; Dao, M.S.; Nguyen, H.D.; Nguyen, B.T. The Impact of Data Imputation on Air Quality Prediction Problem. PLoS ONE 2024, 19, e0306303. [Google Scholar] [CrossRef] [PubMed]
- Alkabbani, H.; Ramadan, A.; Zhu, Q.; Elkamel, A. An Improved Air Quality Index Machine Learning-Based Forecasting with Multivariate Data Imputation Approach. Atmosphere 2022, 13, 1144. [Google Scholar] [CrossRef]
- Camastra, F.; Capone, V.; Ciaramella, A.; Riccio, A.; Staiano, A. Prediction of Environmental Missing Data Time Series by Support Vector Machine Regression and Correlation Dimension Estimation. Environ. Model. Softw. 2022, 150, 105343. [Google Scholar] [CrossRef]
- Che, Z.; Purushotham, S.; Cho, K.; Sontag, D.; Liu, Y. Recurrent Neural Networks for Multivariate Time Series with Missing Values. Sci. Rep. 2018, 8, 6085. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Wang, D.; Li, J.; Zhou, H.; Li, L.; Li, Y. BRITS: Bidirectional Recurrent Imputation for Time Series. Adv. Neural Inf. Process. Syst. 2018, 31, 6775–6785. [Google Scholar]
- Yoon, J.; Jordon, J.; Schaar, M. GAIN: Missing Data Imputation Using Generative Adversarial Nets. In Proceedings of the 35th International Conference on Machine Learning, Stockholm, Sweden, 10–15 July 2018; pp. 5689–5698. [Google Scholar]
- Shahbazian, R.; Greco, S. Generative Adversarial Networks Assist Missing Data Imputation: A Comprehensive Survey and Evaluation. IEEE Access 2023, 11, 88908–88928. [Google Scholar] [CrossRef]
- Cini, A.; Marisca, I.; Alippi, C. Filling the Gaps: Multivariate Time Series Imputation by Graph Neural Networks. arXiv 2021, arXiv:2108.00298. [Google Scholar] [CrossRef]
- Du, W.; Côté, D.; Liu, Y. SAITS: Self-Attention-Based Imputation for Time Series. Expert Syst. Appl. 2023, 219, 119619. [Google Scholar] [CrossRef]
- Colorado Cifuentes, G.U.; Flores Tlacuahuac, A. A Short-Term Deep Learning Model for Urban Pollution Forecasting with Incomplete Data. Can. J. Chem. Eng. 2021, 99, S417–S431. [Google Scholar] [CrossRef]
- Alahamade, M.; Lake, A. Handling Missing Data in Air Quality Time Series: Evaluation of Statistical and Machine Learning Approaches. Atmosphere 2021, 12, 1130. [Google Scholar] [CrossRef]
- World Population Review. Mexico City Population. Available online: https://worldpopulationreview.com/cities/mexico/mexico-city (accessed on 19 July 2025).
- Santamaría-Bonfil, G.; Santoyo, E.; Díaz-González, L.; Arroyo-Figueroa, G. Equivalent Imputation Methodology for Handling Missing Data in Compositional Geochemical Databases of Geothermal Fluids. Geothermics 2022, 104, 102440. [Google Scholar] [CrossRef]
- Farjallah, R.; Selim, B.; Jaumard, B.; Ali, S.; Kaddoum, G. Evaluation of Missing Data Imputation for Time Series without Ground Truth. arXiv, 2503. [Google Scholar] [CrossRef]
| Reference | Datasets and Missing Rate | Input Variables and Missing Rate | Techniques | Target | Key Findings |
|---|---|---|---|---|---|
| Kim et al. (2021) [4] | Minute-level air quality data (2020–2021) from South Korea: Guro-gu (24 stations) and Dangjin-si (42 stations). | PM₂.₅ and PM₁₀ concentrations. Missing Rate (Real / Artificial): 7.91–16.1% / 20%. |
A novel N-BEATS deep learning model with interpretable blocks (trend, seasonality, residual) was compared with baseline methods (mean, spatial average, MICE). | Imputation of missing PM₂.₅ and PM₁₀ values. | N-BEATS model outperforms traditional methods and allows interpretability via component decomposition but struggles with long-term seasonal patterns due to fixed-period Fourier terms. |
| Alahamade & Lake (2021) [20] | Hourly pollutant data (PM₁₀, PM₂.₅, O₃, NO₂) of the Automatic Urban and Rural Network (AURN) from UK (2015-2017), covering 167 station types (urban, suburban, rural, roadside, industrial). | Concentrations of PM₁₀, PM₂.₅, O₃ and NO₂. Missing Rate (Real / Artificial): Not quantified (some pollutants entirely missing at certain stations) / Not applied |
Evaluated models: (1) Clustering based on multivariate time series similarity (CA, CA+ENV, CA+REG); (2) Spatial methods using the 1 or 2 nearest stations (1NN, 2NN); (3) Ensemble method: median of all five approaches. | Imputation of missing PM₁₀, PM₂.₅, O₃ and NO₂ using temporal and spatial similarity. | The ensemble method performed best for O₃, PM₁₀, and PM₂.₅, CA+ENV for NO₂. Performance depended on the pollutant and station type. MVTS clustering allowed full imputation; extremes were slightly under- or overestimated. |
| Wang et al. (2023) [9] | Hourly air quality data (2019–2022) from 16 stations in Qinghai Province and Haidong City, China. | Multivariate time series of six pollutants (PM2.5, PM10, O3, NO2, SO2, and CO). Missing Rate (Real / Artificial): 5–22% / 30%. | BRITS-ALSTM (BRITS encoder + LSTM with attention) was compared to Mean, KNN, MICE, MissForest, M-RNN, BRITS, and BRITS-LSTM. | Imputation of missing pollutants (PM2.5, PM10, O3, NO2, SO2, and CO) data with high and irregular missing rates. | BRITS-ALSTM outperformed baselines for all pollutants and missing patterns. |
| He et al. (2023) [5] | Air quality and meteorological data from Mexico City (2005–2019; 42 stations), combined with satellite data from OMI, TROPOMI, and reanalysis data from CAMS. #stations? |
Daily NO₂ concentrations, wind speed/direction, temperature, cloud coverage, and satellite-based NO₂ columns. Missing Rate (Real / Artificial): Not quantified / Not simulated. |
Comparative modeling using RF, XGBoost, and GAM. Missing values were imputed using Random Forest. | Predict daily NO₂ surface concentrations in Mexico City. | XGBoost and RF outperformed GAM. The model integrated hybrid sources (ground, satellite, and meteorological) to improve NO₂ prediction. |
| Colorado Cifuentes and Flores Tlacuahua (2020) [19] | Hourly air quality and meteorological data (2012-2017) from 13 monitoring stations in the Monterrey Metropolitan area, Mexico. | Pollutants and meteorological variables over a 24-hour window. Missing Rate (Real / Artificial): <25% / Not simulated. |
A deep neural network (DNN). Missing values were imputed using interpolation for non-seasonal time series. | 24-hour ahead prediction of O₃, PM₂.₅, and PM₁₀. |
The DNN model achieved good predictive accuracy for all target pollutants, and the imputation process preserved model performance. |
| Hua et al. (2024) [10] | Six real-world hourly datasets from Germany, China, Taiwan, and Vietnam. The datasets include ~15,000 to 1.2 million samples. | Pollutant and meteorological variables. Missing Rate (Real / Artificial): 0-42% / 10-80%. |
Mean, Median, KNN, MICE, SAITS, BRITS, MRNN, and Transformer. | Evaluate the impact of different imputation strategies on the performance of air quality forecasting models, aimed at predicting 24-hour concentrations of AQI, PM₂.₅, PM₁₀, CO, SO₂, and O₃. | SAITS achieved the highest accuracy, followed by BRITS. KNN performed well on large datasets with high missing rates. MICE was effective on smaller datasets but was slower. Transformer model performed worse than the top methods. |
| Zhang and Zhou (2024) [8] | PM2.5 time series (2018-2020; 225 stations) from Xi’an, China, with single, block, and long-interval missing patterns. | PM2.5 data from multiple stations, with spatial-temporal dependencies. Missing Rate (Real / Artificial): <1% / [10%, 20%, 30%, 50%] | TMLSTM-AE was compared to KNN, SVD, ST-MVL, LSTM, and DAE | Impute complex missing patterns in single-feature PM2.5 time series using spatial-temporal dependencies. | TMLSTM-AE outperforms traditional and baselines methods, especially on long and block missing data. |
| Hyperparameter | default value | values evaluated |
|---|---|---|
| (a) RandomForest | ||
| n_estimators | 100 | [10, 20, 50, 80] |
| max_depth | None | [10, 20, None] |
| max_iter* | 10 | [5, 10, 15, 20] |
| min_samples_split | 2 | [2, 5, 10, 15] |
| min_samples_leaf | 1 | [1, 2, 4, 6] |
| (a) BRITS | ||
| RNN units | 64 | [64, 128, 256, 512] |
| subsequence length | 24 | [16, 32, 64, 128, 168] |
| learning rate | 0.001 | [0.001, 0.005, 0.009] |
| batch size | 64 | [16, 32, 64, 128, 168] |
| use regularization | False | [False, True] |
| dropout rate | 0 | [0.10, 0.2, 0.30] |
| Hyperparameter | default value | values evaluated | Best parameters for each station | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PED | MER | SAG | TLA | FAC | NEZ | UAX | CUA | VIF | MGH | |||
| n_estimators | 100 | [10, 20, 50, 80] | 50 | 20 | 50 | 20 | 50 | 20 | 20 | 10 | 20 | 10 |
| max_depth | None | [10, 20, None] | 20 | 20 | 20 | None | None | 20 | 10 | 10 | 20 | 10 |
| max_iter* | 10 | [5, 10, 15, 20] | 10 | 15 | 15 | 15 | 5 | 10 | 20 | 15 | 15 | 10 |
| min_samples_split | 2 | [2, 5, 10, 15] | 5 | 10 | 2 | 2 | 2 | 10 | 5 | 15 | 15 | 10 |
| min_samples_leaf | 1 | [1, 2, 4, 6] | 1 | 4 | 4 | 4 | 1 | 6 | 4 | 4 | 6 | 4 |
| Hyperparameter | default value | values evaluated | Best parameters for each station | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PED | MER | SAG | TLA | FAC | NEZ | UAX | CUA | VIF | MGH | |||
| RNN units | 64 | [64, 128, 256, 512] | 512 | 512 | 512 | 512 | 256 | 512 | 512 | 256 | 256 | 512 |
| subsequence length | 24 | [16, 32, 64, 128, 168] | 32 | 64 | 64 | 32 | 128 | 64 | 64 | 32 | 64 | 168 |
| learning rate | 0.001 | [0.001, 0.005, 0.009] | 0.005 | 0.005 | 0.005 | 0.005 | 0.005 | 0.005 | 0.005 | 0.005 | 0.005 | 0.009 |
| batch size | 64 | [16, 32, 64, 128, 168] | 32 | 32 | 32 | 32 | 32 | 32 | 32 | 32 | 32 | 128 |
| use regularization | False | [False, True] | True | False | True | False | False | True | False | False | True | False |
| dropout rate | 0 | [0.10, 0.2, 0.30] | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | 0.3 | 0.1 | 0.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





