Submitted:
18 July 2025
Posted:
21 July 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
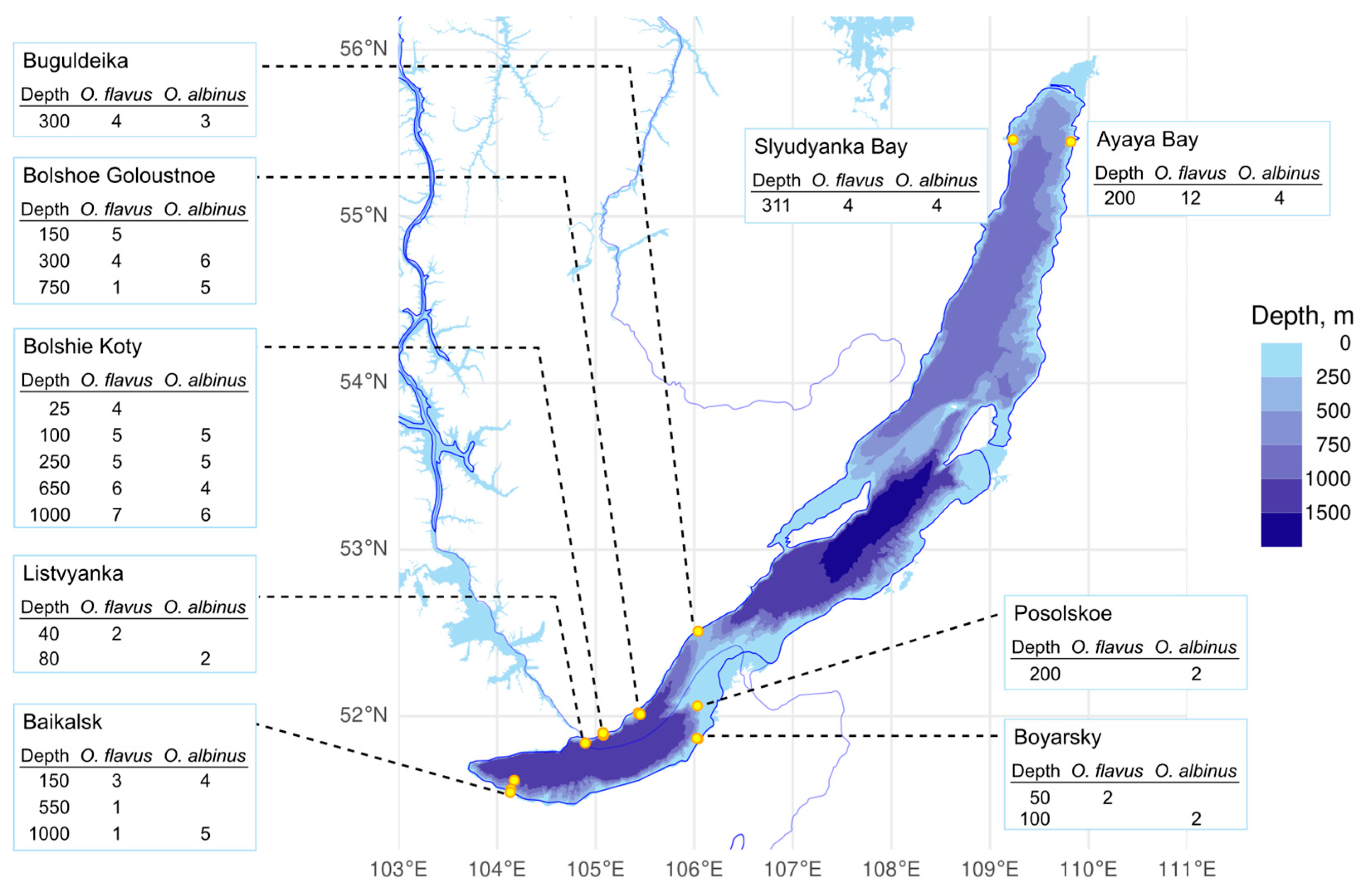
2.1. Sampling, Study Sites and Fixation
2.2. Nucleic Acid Extraction, PCR and Sequencing
2.3. Extraction and Estimation of Carotenoid Concentration
2.4. Data Analysis
3. Results
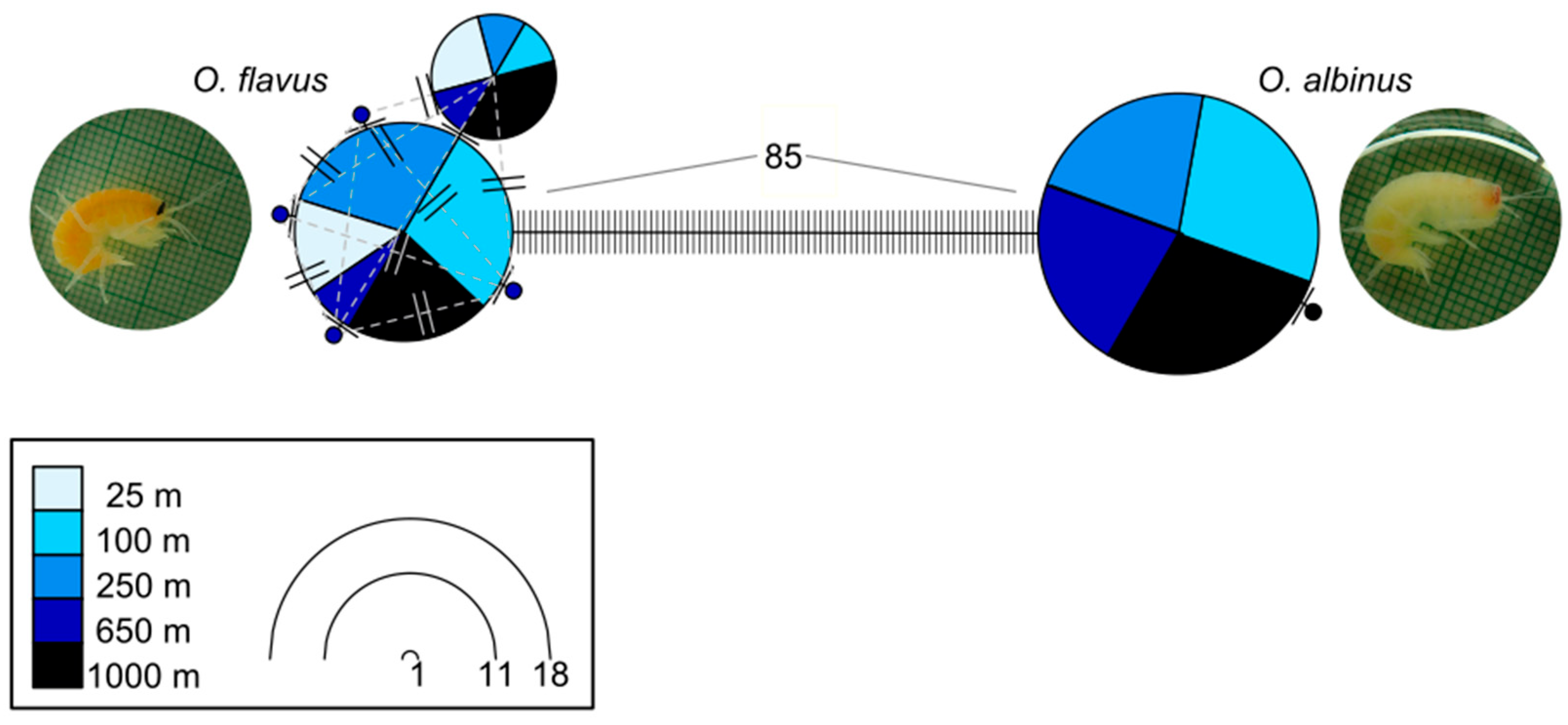
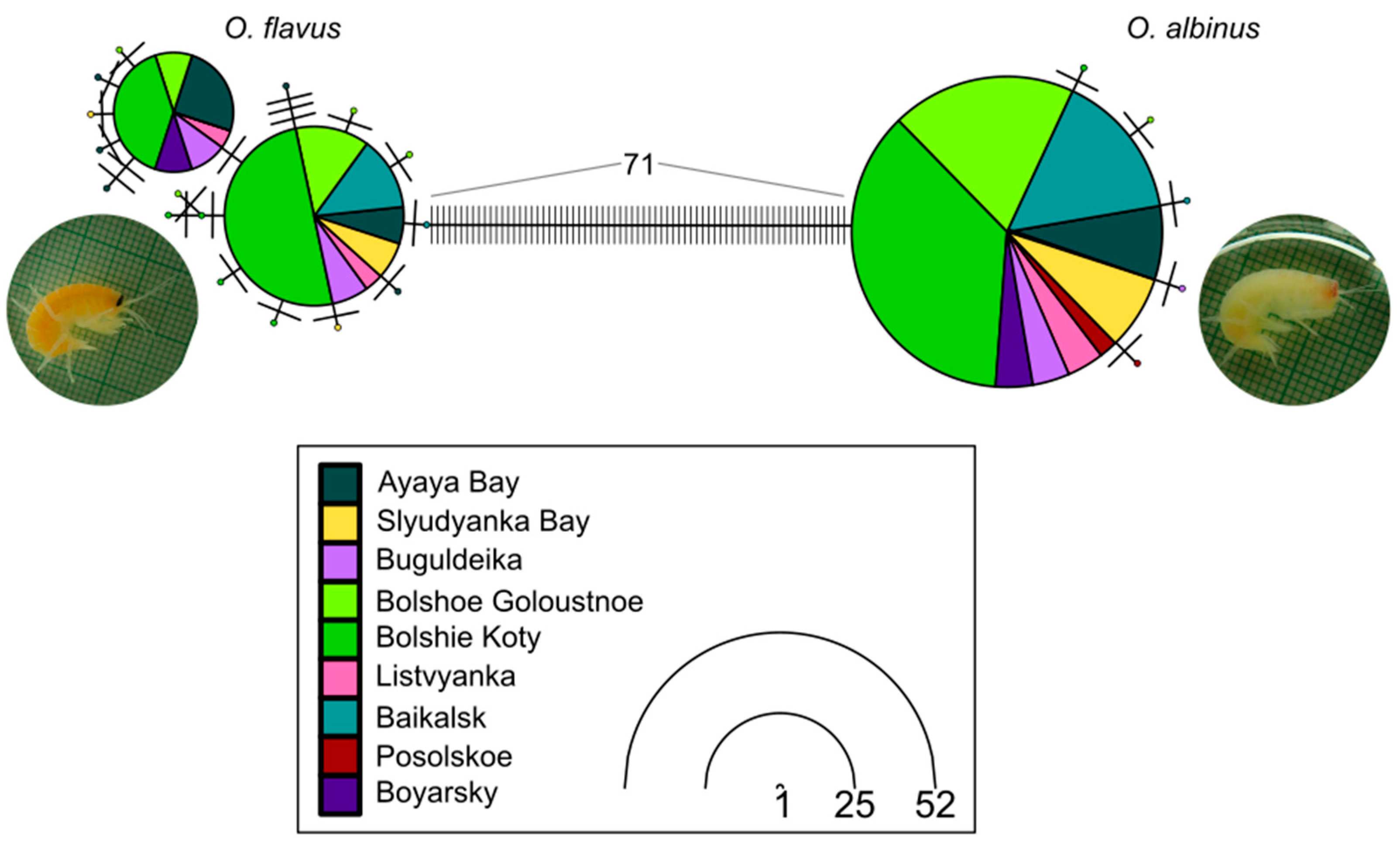
3.1. Intraspecific Genetic Diversity Is Low and Similar Across Locations and Depths
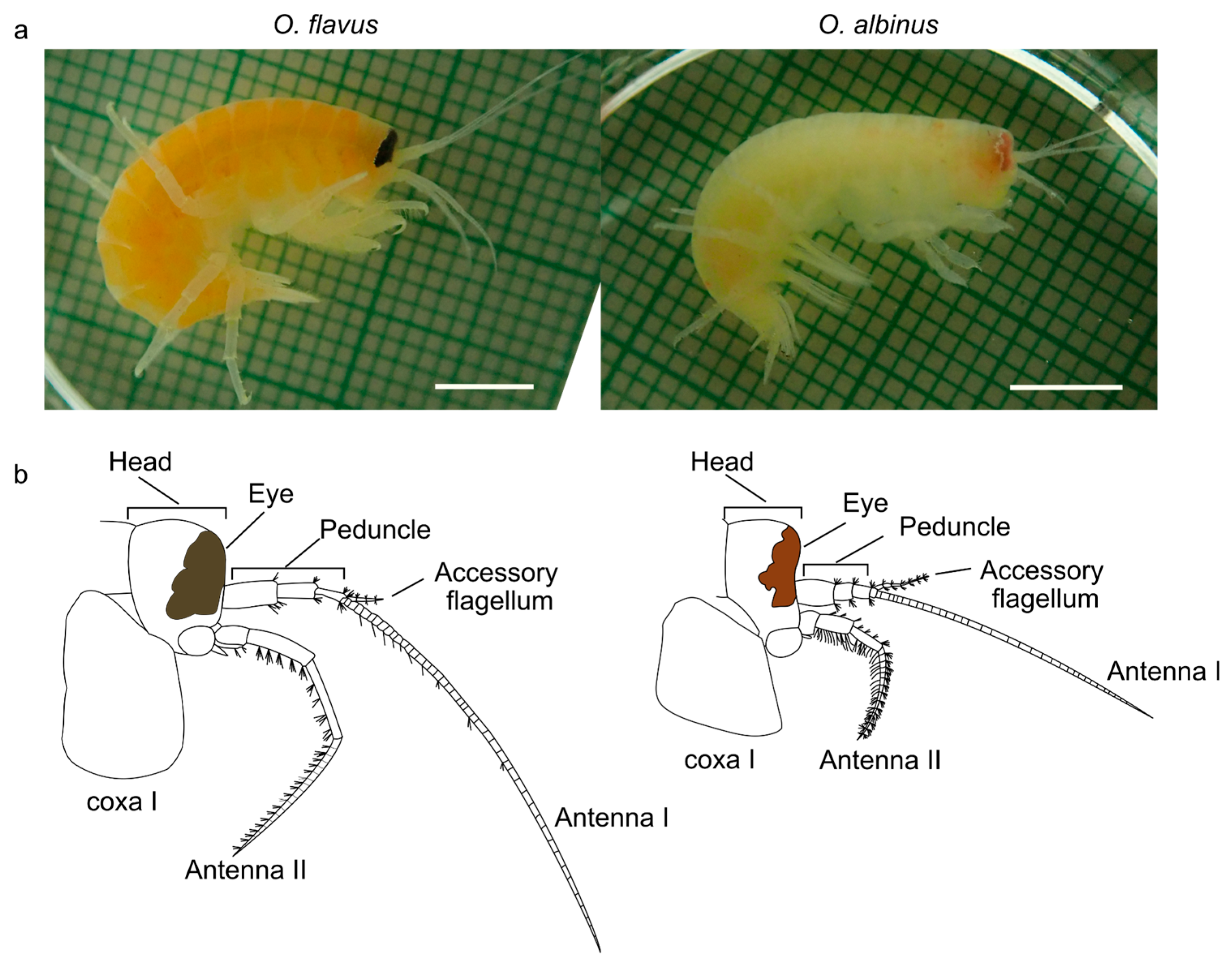
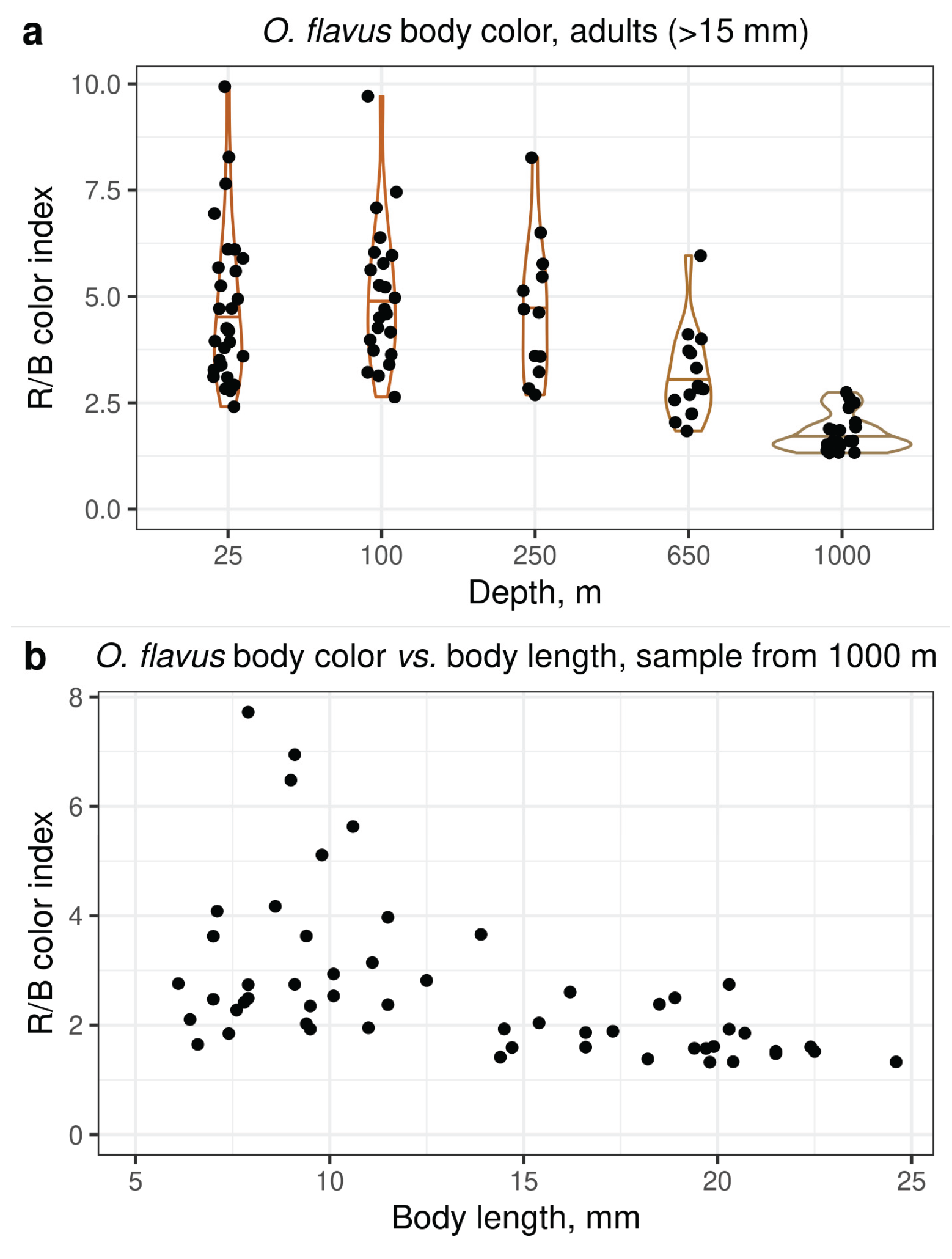
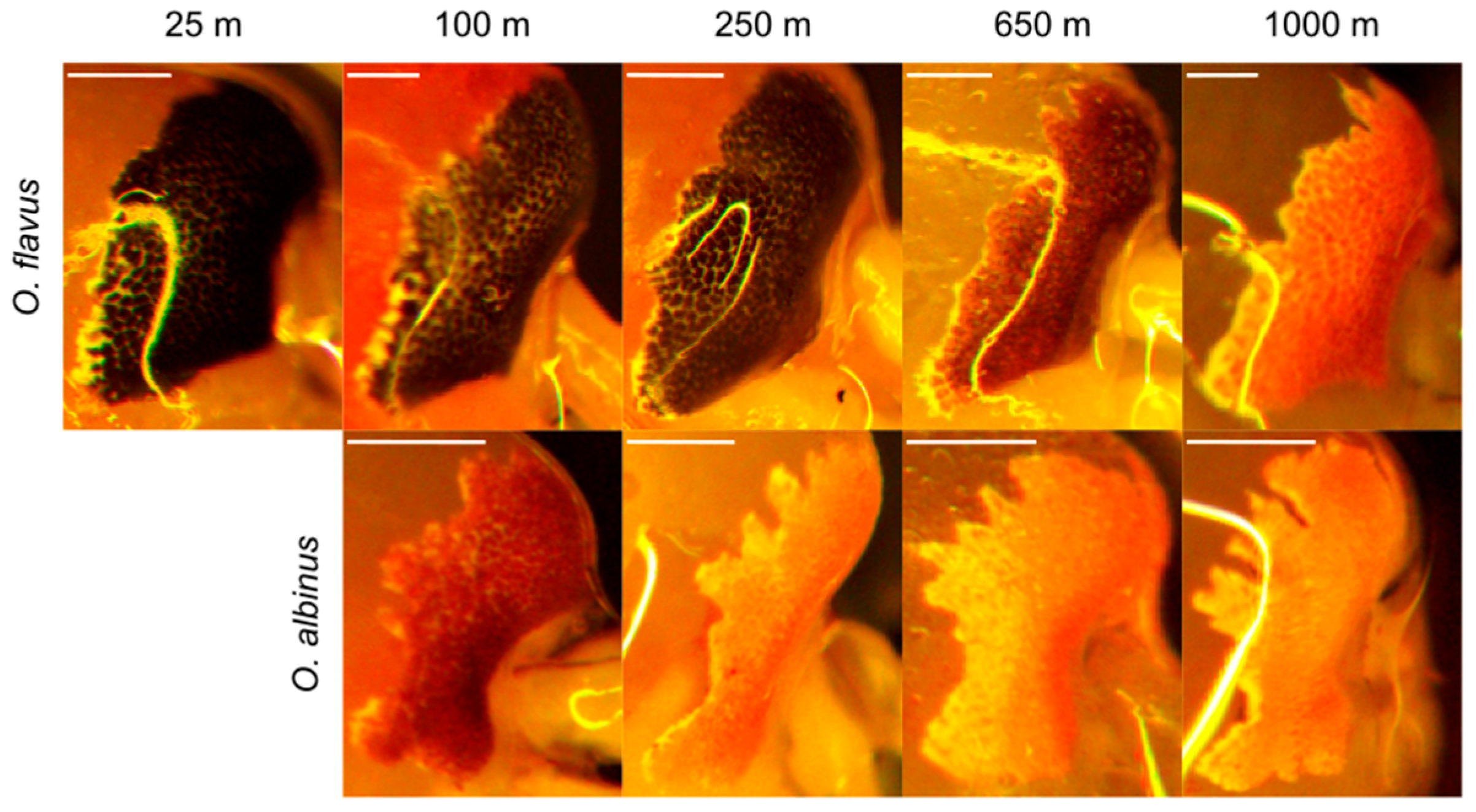
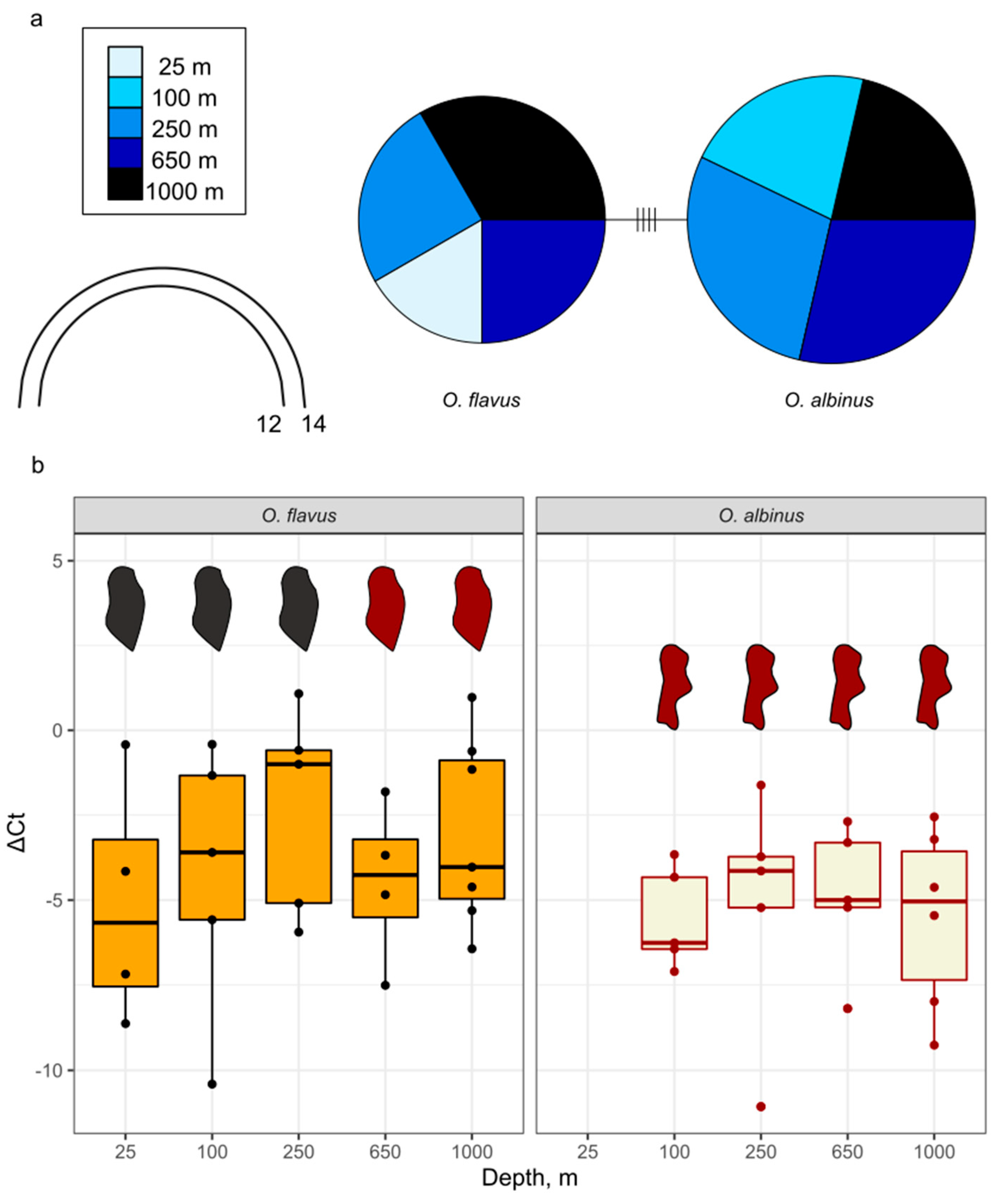
3.2. Analysis of Coloration and Eye Shape Reveals Depth-Related Phenotypes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Thurber, A.R.; Sweetman, A.K.; Narayanaswamy, B.E.; Jones, D.O.B.; Ingels, J.; Hansman, R.L. Ecosystem Function and Services Provided by the Deep Sea. Biogeosciences 2014, 11, 3941–3963. [Google Scholar] [CrossRef]
- Taylor, M.L.; Roterman, C.N. Invertebrate Population Genetics across Earth’s Largest Habitat: The Deep-Sea Floor. Mol. Ecol. 2017, 26, 4872–4896. [Google Scholar] [CrossRef]
- Webb, T.J.; Vanden Berghe, E.; O’Dor, R. Biodiversity’s Big Wet Secret: The Global Distribution of Marine Biological Records Reveals Chronic Under-Exploration of the Deep Pelagic Ocean. PLOS ONE 2010, 5, e10223. [Google Scholar] [CrossRef]
- Danovaro, R.; Snelgrove, P.V.R.; Tyler, P. Challenging the Paradigms of Deep-Sea Ecology. Trends Ecol. Evol. 2014, 29, 465–475. [Google Scholar] [CrossRef]
- Feng, J.-C.; Liang, J.; Cai, Y.; Zhang, S.; Xue, J.; Yang, Z. Deep-Sea Organisms Research Oriented by Deep-Sea Technologies Development. Sci. Bull. 2022, 67, 1802–1816. [Google Scholar] [CrossRef]
- Schön, I.; Martens, K. Adaptive, Pre-Adaptive and Non-Adaptive Components of Radiations in Ancient Lakes: A Review. Org. Divers. Evol. 2004, 4, 137–156. [Google Scholar] [CrossRef]
- Piccolroaz, S.; Toffolon, M. The Fate of Lake Baikal: How Climate Change May Alter Deep Ventilation in the Largest Lake on Earth. Clim. Change 2018, 150, 181–194. [Google Scholar] [CrossRef]
- Chapelle, G.; Peck, L.S. Polar Gigantism Dictated by Oxygen Availability. Nature 1999, 399, 114–115. [Google Scholar] [CrossRef]
- Shimaraev, M.N.; Domysheva, V.M. Trends in Hydrological and Hydrochemical Processes in Lake Baikal under Conditions of Modern Climate Change. Clim. Change Glob. Warm. Inland Waters Impacts Mitig. Ecosyst. Soc. 2013, 43–66. [Google Scholar] [CrossRef]
- Hampton, S.E.; Gray, D.K.; Izmest’eva, L.R.; Moore, M.V.; Ozersky, T. The Rise and Fall of Plankton: Long-Term Changes in the Vertical Distribution of Algae and Grazers in Lake Baikal, Siberia. PLoS One 2014, 9, e88920. [Google Scholar] [CrossRef]
- Arfianti, T.; Wilson, S.; Costello, M.J. Progress in the Discovery of Amphipod Crustaceans. PeerJ 2018, 6, e5187. [Google Scholar] [CrossRef]
- Horton, T.; De Broyer, C.; Bellan-Santini, D.; Coleman, C.O.; Copilaș-Ciocianu, D.; Corbari, L.; Daneliya, M.E.; Dauvin, J.-C.; Decock, W.; Fanini, L.; et al. The World Amphipoda Database: History and Progress. Rec. Aust. Mus. 2023, 75, 329–342. [Google Scholar] [CrossRef]
- Bowen, B.W.; Forsman, Z.H.; Whitney, J.L.; Faucci, A.; Hoban, M.; Canfield, S.J.; Johnston, E.C.; Coleman, R.R.; Copus, J.M.; Vicente, J.; et al. Species Radiations in the Sea: What the Flock? J. Hered. 2020, 111, 70–83. [Google Scholar] [CrossRef]
- Chenuil, A.; Saucède, T.; Hemery, L.G.; Eléaume, M.; Féral, J.-P.; Améziane, N.; David, B.; Lecointre, G.; Havermans, C. Understanding Processes at the Origin of Species Flocks with a Focus on the Marine Antarctic Fauna. Biol. Rev. 2018, 93, 481–504. [Google Scholar] [CrossRef]
- Drozdova, P.B.; Madyarova, E.V.; Gurkov, A.N.; Saranchina, A.E.; Romanova, E.V.; Petunina, J.V.; Peretolchina, T.E.; Sherbakov, D.Y.; Timofeyev, M.A. Lake Baikal Amphipods and Their Genomes, Great and Small. Vavilov J. Genet. Breed. 2024, 28, 317–325. [Google Scholar] [CrossRef]
- Copilaș-Ciocianu, D.; Sidorov, D. Taxonomic, Ecological and Morphological Diversity of Ponto-Caspian Gammaroidean Amphipods: A Review. Org. Divers. Evol. 2022. [Google Scholar] [CrossRef]
- Cristescu, M.E.; Adamowicz, S.J.; Vaillant, J.J.; Haffner, D.G. Ancient Lakes Revisited: From the Ecology to the Genetics of Speciation. Mol. Ecol. 2010, 19, 4837–4851. [Google Scholar] [CrossRef]
- Jamieson, A.J.; Weston, J.N.J. Amphipoda from Depths Exceeding 6,000 Meters Revisited 60 Years On. J. Crustac. Biol. 2023, 43, ruad020. [Google Scholar] [CrossRef]
- Kamaltynov, R.M. Amphipods (Amphipoda: Gammaroidea). In Index of animal species inhabiting Lake Baikal and its catchment area; Novosibirsk Branch of “Nauka” Publishing House: Novosibirsk, 2001; ISBN 978-0-02-018978-2. [Google Scholar]
- Kozhova, O.M.; Izmest’eva, L.R. Lake Baikal: Evolution and Biodiversity; Backhuys Publishers: Leiden, 1998; p. 225. [Google Scholar]
- Palumbi, S.R. Genetic Divergence, Reproductive Isolation, and Marine Speciation. Annu. Rev. Ecol. Syst. 1994, 547–572. [Google Scholar] [CrossRef]
- Dupont, D.W.E.; Patel, T.; Kochzius, M.; Schön, I. Evidence for a Single Population Expansion Event across 24,000 Km: The Case of the Deep-Sea Scavenging Amphipod Abyssorchomene distinctus. Hydrobiologia 2024, 851, 2309–2327. [Google Scholar] [CrossRef]
- Jażdżewska, A.M.; Horton, T.; Hendrycks, E.; Mamos, T.; Driskell, A.C.; Brix, S.; Arbizu, P.M. Pandora’s Box in the Deep Sea–Intraspecific Diversity Patterns and Distribution of Two Congeneric Scavenging Amphipods. Front. Mar. Sci. 2021. [Google Scholar] [CrossRef]
- Weston, J.N.J.; Jamieson, A.J. The Multi-Ocean Distribution of the Hadal Amphipod, Hirondellea dubia Dahl, 1959 (Crustacea, Amphipoda). Front. Mar. Sci. 2022. [Google Scholar] [CrossRef]
- Havermans, C.; Sonet, G.; d’Udekem d’Acoz, C.; Nagy, Z.T.; Martin, P.; Brix, S.; Riehl, T.; Agrawal, S.; Held, C. Genetic and Morphological Divergences in the Cosmopolitan Deep-Sea Amphipod Eurythenes gryllus Reveal a Diverse Abyss and a Bipolar Species. PloS One 2013, 8, 1–15. [Google Scholar] [CrossRef]
- Havermans, C. Have We so Far Only Seen the Tip of the Iceberg? Exploring Species Diversity and Distribution of the Giant Amphipod Eurythenes. Biodiversity 2016, 17, 12–25. [Google Scholar] [CrossRef]
- Weston, J.N.J.; Jensen, E.L.; Hasoon, M.S.R.; Kitson, J.J.N.; Stewart, H.A.; Jamieson, A.J. Barriers to Gene Flow in the Deepest Ocean Ecosystems: Evidence from Global Population Genomics of a Cosmopolitan Amphipod. Sci. Adv. 2022, 8, eabo6672. [Google Scholar] [CrossRef]
- Daneliya, M.E.; Kamaltynov, R.M.; Väinölä, R. Phylogeography and Systematics of Acanthogammarus s. Str., Giant Amphipod Crustaceans from Lake Baikal. Zool. Scr. 2011, 40, 623–637. [Google Scholar] [CrossRef]
- Drozdova, P.; Saranchina, A.; Madyarova, E.; Gurkov, A.; Timofeyev, M. Experimental Crossing Confirms Reproductive Isolation between Cryptic Species within Eulimnogammarus verrucosus (Crustacea: Amphipoda) from Lake Baikal. Int. J. Mol. Sci. 2022, 23, 10858. [Google Scholar] [CrossRef]
- Gomanenko, G.V.; Kamaltynov, R.M.; Kuzmenkova, Zh.V.; Berenos, K.; Sherbakov, D.Yu. Population Structure of the Baikalian Amphipod Gmelinoides Fasciatus (Stebbing). Russ. J. Genet. 2005, 41, 907–912. [Google Scholar] [CrossRef]
- Gurkov, A.; Rivarola-Duarte, L.; Bedulina, D.; Fernández Casas, I.; Michael, H.; Drozdova, P.; Nazarova, A.; Govorukhina, E.; Timofeyev, M.; Stadler, P.F.; et al. Indication of Ongoing Amphipod Speciation in Lake Baikal by Genetic Structures within Endemic Species. BMC Evol. Biol. 2019, 19, 138. [Google Scholar] [CrossRef]
- Petunina, J.V.; Vavrishchuk, N.V.; Romanova, E.V. Variabel’nost’ Morfologicheskikh i Geneticheskikh Priznakov Macrohectopus Branickii [Variability of Morphological and Genetic Traits of Macrohectopus Branickii]. In Proceedings of the Development of physicochemical biology, bioengineering and bioinformatics at the present stage; Publishing house of ISU: Irkutsk, October 25, 2023; pp. 111–113. [Google Scholar]
- Zaidykov, I.Y.; Naumova, E.Y.; Sukhanova, L.V. MtDNA Polymorphism of Macrohectopus Branickii Dybowsky, 1974 (Amphipoda) – An Endemic Pelagic Key Species of Lake Baikal. In Proceedings of the Complex Investigation of the World Ocean (CIWO-2023); Chaplina, T., Ed.; Springer Nature Switzerland: Cham, 2023; pp. 223–229. [Google Scholar]
- Bazikalova, A.Y. Amfipody Ozera Baikala [Amphipods of Lake Baikal]. Proc. Baikal Limnol. Stn. 1945, 11, 1–440. [Google Scholar]
- Takhteev, V.V. Ocherki o Bokoplavakh Ozera Baikal (sistematika, sravnitel’naya ekologiya, evolyutsiya) [Essays on the amphipods of Lake Baikal (systematics, comparative ecology, evolution)]; Irkutsk University Publishing House: Irkutsk, 2000; ISBN 5-7430-0123-5. [Google Scholar]
- Karaman, G.S. New Genus of Family Gammaridae from Baikal Lake, Abludogammarus, n. Gen. with References to Genus Ommatogammarus Stebb. (Contribution to the Knowledge of the Amphipoda 108). Glas. Od. Prir. Nauka Crnog. Akad. Nauka Umjet. Titogr. 1980, 3, 149–169. [Google Scholar]
- Macdonald III, K.S.; Yampolsky, L.; Duffy, J.E. Molecular and Morphological Evolution of the Amphipod Radiation of Lake Baikal. Mol. Phylogenet. Evol. 2005, 35, 323–343. [Google Scholar] [CrossRef]
- Shirokova, Y.A.; Saranchina, A.E.; Shatilina, Zh.M.; Kashchuk, N.D.; Timofeyev, M.A. Comparison of Olfactory Sensilla Structure in Littoral and Deep-Water Amphipods from the Baikal Region. Inland Water Biol. 2023, 16, 873–883. [Google Scholar] [CrossRef]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; Mcveigh, R.; O’Neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A Comprehensive Update on Curation, Resources and Tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef]
- Horton, T.; Lowry, J.; Broyer, C.D.; Bellan-Santini, D.; Copilaș-Ciocianu, D.; Corbari, L.; Costello, M.J.; Daneliya, M.; Dauvin, J.-C.; Gasca, R.; et al. World Amphipoda Database 2025. Available online: https://www.marinespecies.org/amphipoda/ (accessed on 10th May, 2025).
- Ratnasingham, S.; Wei, C.; Chan, D.; Agda, J.; Agda, J.; Ballesteros-Mejia, L.; Boutou, H.A.; El Bastami, Z.M.; Ma, E.; Manjunath, R.; et al. BOLD v4: A Centralized Bioinformatics Platform for DNA-Based Biodiversity Data. In DNA Barcoding: Methods and Protocols; DeSalle, R., Ed.; Springer US: New York, NY, 2024; ISBN 978-1-0716-3581-0. [Google Scholar]
- Dybowsky, B.N. Beiträge Zur Näheren Kenntniss Der in Dem Baikal-See Vorkommenden Niederen Krebse Aus Der Gruppe Der Gammariden [Contributions to a more detailed knowledge of the lower crustaceans of the gammarid group found in Lake Baikal]; Buchdr. von W. Besobrasoff: St. Petersburg, 1874. [Google Scholar]
- Madyarova, E.; Shirokova, Y.; Gurkov, A.; Drozdova, P.; Baduev, B.; Lubyaga, Y.; Shatilina, Z.; Vishnevskaya, M.; Timofeyev, M. Metabolic Tolerance to Atmospheric Pressure of Two Freshwater Endemic Amphipods Mostly Inhabiting the Deep-Water Zone of the Ancient Lake Baikal. Insects 2022, 13. [Google Scholar] [CrossRef]
- Kondrateva, E.; Gurkov, A.; Rzhechitskiy, Y.; Saranchina, A.; Diagileva, A.; Drozdova, P.; Vereshchagina, K.; Shatilina, Z.; Sokolova, I.; Timofeyev, M. UV Sensitivities of Two Littoral and Two Deep-Freshwater Amphipods (Amphipoda, Crustacea) Reflect Their Preferred Depths in the Ancient Lake Baikal. Biology 2024, 13. [Google Scholar] [CrossRef]
- Takhteev, V.; Didorenko, S. Fauna i ekologiya bokoplavov ozera Baikal [Fauna and ecology of amphipods of Lake Baikal]; Publ. House Inst Geog VB Sochavy SB RAS: Irkutsk, Russia, 2015; ISBN 978-5-94797-244-3. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Drozdova, P.; Saranchina, A.; Morgunova, M.; Kizenko, A.; Lubyaga, Y.; Baduev, B.; Timofeyev, M. The Level of Putative Carotenoid-Binding Proteins Determines the Body Color in Two Species of Endemic Lake Baikal Amphipods. PeerJ 2020, 8, e9387. [Google Scholar] [CrossRef]
- Saranchina, A.; Mutin, A.; Govorukhina, E.; Rzhechitskiy, Y.; Gurkov, A.; Timofeyev, M.; Drozdova, P. Genetic Diversity in a Baikal Species Complex Eulimnogammarus verrucosus (Amphipoda: Gammaroidea) in the Angara River, the Only Outflow of Lake Baikal. Zool. Scr. 2024, 53, 867–879. [Google Scholar] [CrossRef]
- Drozdova, P.; Rivarola-Duarte, L.; Bedulina, D.; Axenov-Gribanov, D.; Schreiber, S.; Gurkov, A.; Shatilina, Z.; Vereshchagina, K.; Lubyaga, Y.; Madyarova, E.; et al. Comparison between Transcriptomic Responses to Short-Term Stress Exposures of a Common Holarctic and Endemic Lake Baikal Amphipods. BMC Genomics 2019, 20, 712. [Google Scholar] [CrossRef]
- Naumenko, S.A.; Logacheva, M.D.; Popova, N.V.; Klepikova, A.V.; Penin, A.A.; Bazykin, G.A.; Etingova, A.E.; Mugue, N.S.; Kondrashov, A.S.; Yampolsky, L.Y. Transcriptome-Based Phylogeny of Endemic Lake Baikal Amphipod Species Flock: Fast Speciation Accompanied by Frequent Episodes of Positive Selection. Mol. Ecol. 2017, 26, 536–553. [Google Scholar] [CrossRef]
- Drozdova, P.; Kizenko, A.; Saranchina, A.; Gurkov, A.; Firulyova, M.; Govorukhina, E.; Timofeyev, M. The Diversity of Opsins in Lake Baikal Amphipods (Amphipoda: Gammaridae). BMC Ecol. Evol. 2021, 21, 81. [Google Scholar] [CrossRef]
- Slater, G.S.C.; Birney, E. Automated Generation of Heuristics for Biological Sequence Comparison. BMC Bioinformatics 2005, 6, 31. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA Primers for Amplification of Mitochondrial Cytochrome c Oxidase Subunit I from Diverse Metazoan Invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Minton, J.A.L.; Flanagan, S.E.; Ellard, S. Mutation Surveyor: Software for DNA Sequence Analysis. In PCR Mutation Detection Protocols; Theophilus, B.D.M., Rapley, R., Eds.; Humana Press: Totowa, NJ, 2011; ISBN 978-1-60761-946-8. [Google Scholar]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A Unified Bioinformatics Toolkit. Bioinforma. Oxf. Engl. 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- R Core Team R: A Language and Environment for Statistical Computing 2024.
- Paradis, E. Pegas: An R Package for Population Genetics with an Integrated–Modular Approach. Bioinformatics 2010, 26, 419–420. [Google Scholar] [CrossRef]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for Primary Species Delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef]
- Puillandre, N.; Brouillet, S.; Achaz, G. ASAP: Assemble Species by Automatic Partitioning. Mol. Ecol. Resour. 2021, 21, 609–620. [Google Scholar] [CrossRef]
- Miralles, A.; Ducasse, J.; Brouillet, S.; Flouri, T.; Fujisawa, T.; Kapli, P.; Knowles, L.L.; Kumari, S.; Stamatakis, A.; Sukumaran, J.; et al. SPART: A Versatile and Standardized Data Exchange Format for Species Partition Information. Mol. Ecol. Resour. 2022, 22, 430–438. [Google Scholar] [CrossRef]
- GEBCO Compilation Group. GEBCO 2024 Grid. [CrossRef]
- Pante, E.; Simon-Bouhet, B.; Irisson, J. Marmap: Import, Plot and Analyze Bathymetric and Topographic Data. R Package Version 1.0. Available online at https://CRAN.R-project.org/package=marmap. (accessed on 14 March 2025).
- Massicotte, P.; South, A. Rnaturalearth: World Map Data from Natural Earth. R Package Version 03. Available online at https://CRAN.R-project.org/package=rnaturalearth. (accessed on 14 March 2025).
- Hothorn, T.; Hornik, K.; Wiel, M.A. van de; Zeileis, A. Implementing a Class of Permutation Tests: The Coin Package. J. Stat. Softw. 2008, 28. [Google Scholar] [CrossRef]
- Holm, S. A Simple Sequentially Rejective Multiple Test Procedure. Scand. J. Stat. 1979, 6, 65–70. [Google Scholar]
- Paradis, E.; Schliep, K. Ape 5.0: An Environment for Modern Phylogenetics and Evolutionary Analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer Verlag, 2016.
- Wickham, H. Forcats: Tools for Working with Categorical Variables (Factors) Available online:. Available online: https://cran.r-project.org/web/packages/forcats/index.html (accessed on 14 March 2025).
- Slowikowski, K.; Schep, A.; Hughes, S.; Lukauskas, S.; Irisson, J.-O.; Kamvar, Z.N.; Ryan, T.; Christophe, D.; Hiroaki, Y.; Gramme, P. ggrepel: Automatically Position Non-Overlapping Text Labels with 'ggplot2' https://CRAN.R-project.org/package=ggrepel. (accessed on 14 March 2025).
- Sherstyankin, P.P.; Alekseev, S.P.; Abramov, A.M.; Stavrov, K.G.; De Batist, M.; Hus, R.; Canals, M.; Casamor, J.L. Computer-Based Bathymetric Map of Lake Baikal. Dokl. Earth Sci. 2006, 408, 564–569. [Google Scholar] [CrossRef]
- Wade, N.M.; Gabaudan, J.; Glencross, B.D. A Review of Carotenoid Utilisation and Function in Crustacean Aquaculture. Rev. Aquac. 2017, 9, 141–156. [Google Scholar] [CrossRef]
- Maoka, T. Carotenoids as Natural Functional Pigments. J. Nat. Med. 2020, 74, 1–16. [Google Scholar] [CrossRef]
- Stebbing, T.R.R. Amphipoda; Friedlaender (R.), Berlin, 1906.
- Mats, V.D.; Shcherbakov, D.Y.; Efimova, I.M. Late Cretaceous-Cenozoic History of the Lake Baikal Depression and Formation of Its Unique Biodiversity. Stratigr. Geol. Correl. 2011, 19, 404–423. [Google Scholar] [CrossRef]
- Martin, P.; Martens, K.; Goddeeris, B. Oligochaeta from the Abyssal Zone of Lake Baikal (Siberia, Russia). Hydrobiologia 1999, 406, 165–174. [Google Scholar] [CrossRef]
- Takhteev, V.V. Trends in the Evolution of Baikal Amphipods and Evolutionary Parallels with Some Marine Malacostracan Faunas. In Advances in Ecological Research; Ancient Lakes: Biodiversity, Ecology and Evolution; Academic Press, 2000. [Google Scholar]
- Fazalova, V.; Nevado, B.; Peretolchina, T.; Petunina, J.; Sherbakov, D. When Environmental Changes Do Not Cause Geographic Separation of Fauna: Differential Responses of Baikalian Invertebrates. BMC Evol. Biol. 2010, 10, 320. [Google Scholar] [CrossRef]
- Kaygorodova, I.A.; Sherbakov, D.Y.; Martin, P. Molecular Phylogeny of Baikalian Lumbriculidae (Oligochaeta): Evidence for Recent Explosive Speciation. Comp. Cytogenet. 2007, 1, 71–84. [Google Scholar]
- Khlystov, O.M.; Kononov, E.E. Mesorelief of the Submarine Academician Ridge in Lake Baikal (Based on Newly Gathered Instrument Readings). Geogr. Nat. Resour. 2024, 45, 390–396. [Google Scholar] [CrossRef]
- Saranchina, A.; Drozdova, P.; Mutin, A.; Timofeyev, M. Diet Affects Body Color and Energy Metabolism in the Baikal Endemic Amphipod Eulimnogammarus Cyaneus Maintained in Laboratory Conditions. Biol. Commun. 2021, 66. [Google Scholar] [CrossRef]
- Thoen, H.H.; Johnsen, G.; Berge, J. Pigmentation and Spectral Absorbance in the Deep-Sea Arctic Amphipods Eurythenes Gryllus and Anonyx Sp. Polar Biol. Polar Biol. 2011, 34, 83–93. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




