2.7. Term Enrichment Analysis of Gene Ontologies
The next stage of the presented study was the functional characterisation of the investigated set of differentially expressed (for the transcription stage) and differentially translated genes (for translation, respectively).
KEGG
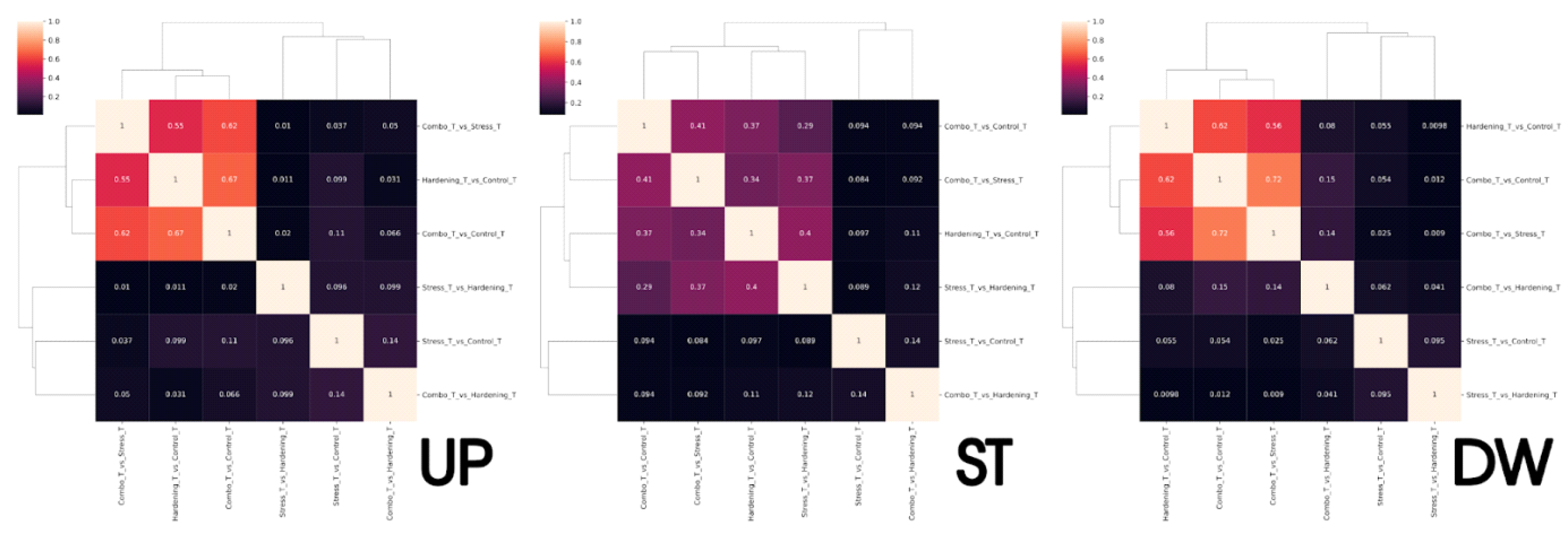
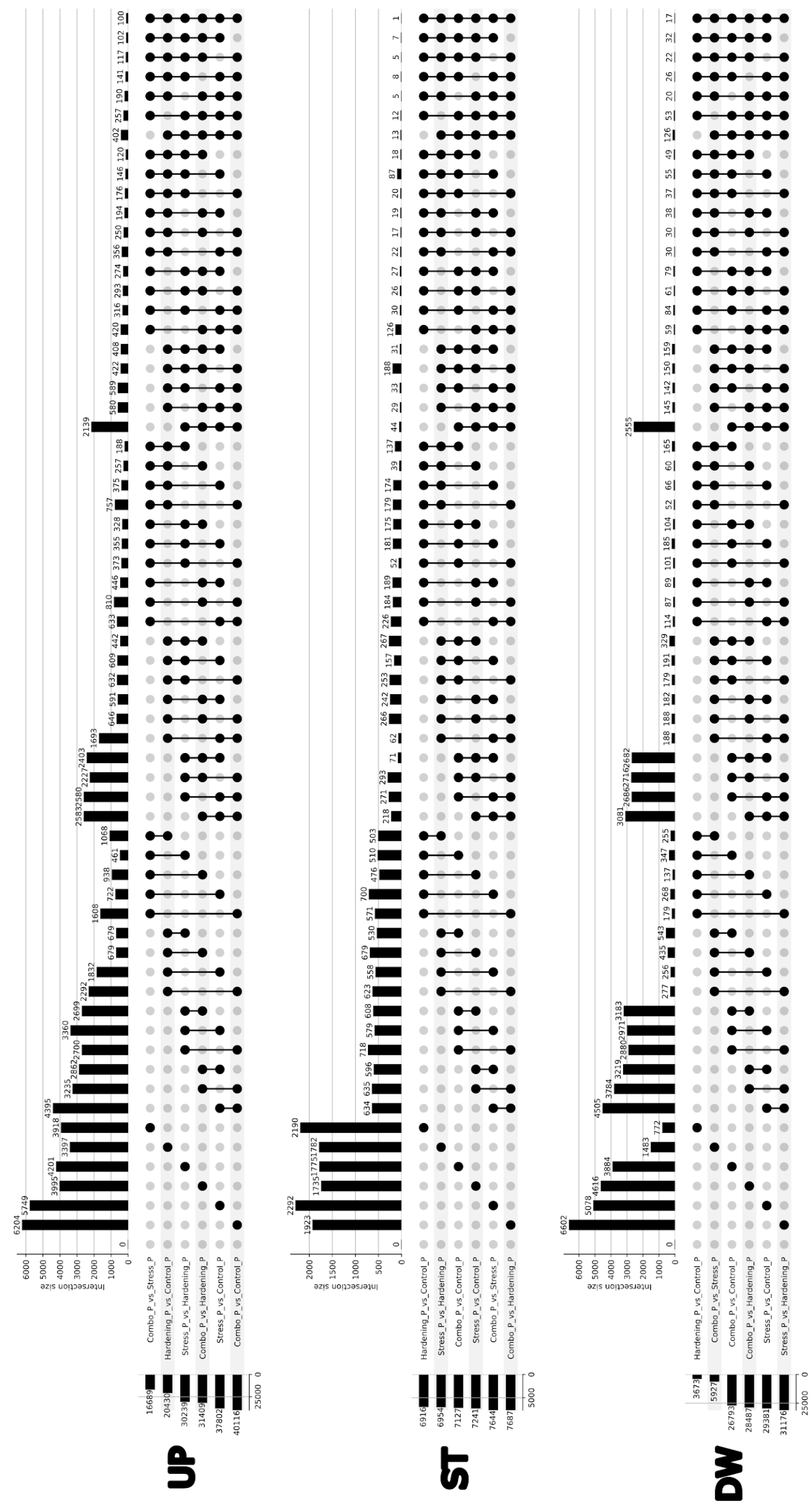
Up: pathways associated with ribosomal activity and regulation of protein synthesis (KEGG:03010, 03050, 03008) are predominant in the transcription step, particularly pronounced in Stress_T_vs_Hardening_T, Combo_T_vs_Control_T and Hardening_T_vs_Control_T comparisons. This reflects the activation of genetic programmes that prepare the cell for stress. In contrast, the translation step is dominated by metabolic pathways (KEGG:01100, 01110) related to energy metabolism, biosynthesis of secondary metabolites and carbohydrate metabolism. This is particularly expressed under Combo_vs_Stress, Combo_vs_Control and Hardening_vs_Control conditions. A comparison of expression levels within the experimental groups showed that under stress conditions transcription and translation are regulated independently: active transcription of ribosomal components is combined with metabolic compensation at the level of translation. In hardened plants, a stable combination of regulatory activity at the level of transcription and metabolic rearrangement in translation was observed. Under combined conditions (Combo), transcriptional responses are attenuated, whereas translation remains active, which may indicate the formation of stress memory and a switch of regulation to the posttranscriptional level. Thus, transcription reflects the preparatory phase of the response, while translation provides rapid metabolic adaptation. This emphasises the importance of integrated analysis of both expression levels for understanding the mechanisms of plant cold tolerance.
Stable: the KEGG:03010 ribosome-compared Stress_T_vs_Hardening_T, Combo_T_vs_Control_T, Combo_T_vs_Stress_T, and Hardening_T_vs_Control_T pathway is the most stable in transcription. This reflects the stable activity of the protein synthesis system. The KEGG:01200, 01230, 01100, and 01110 pathways associated with basic metabolism and degradation of substances are also repeated. At the level of translation, stability of metabolic processes prevails. KEGG:01230 is regularly active in several comparisons (Hardening_vs_Control, Combo_vs_Hardening, Stress_vs_Control), indicating maintenance of catabolic activity. Separate stable signals are observed at KEGG:03015, 00250, and 00330 (ribosome assembly, amino acid metabolism), whereas the ribosomal pathway (KEGG:03010) is conserved only in Combo_vs_Control and Combo_vs_Stress. Comparison of levels shows that transcription ensures stable expression of components of the protein machinery, while translation ensures stability of metabolic processes. The highest concordance is observed in the Combo group, which may reflect the physiological stability formed after hardening. Thus, stable genes represent a functional core that maintains basic cellular processes independent of external conditions.
Down: analysis of genes with reduced expression revealed that pathways related to general metabolism (KEGG:01100) and biosynthesis of secondary metabolites (KEGG:01110) are most pronouncedly repressed. These changes are particularly characteristic at the level of translation, where in most comparisons – including Stress_vs_Hardening, Combo_vs_Hardening and Stress_vs_Control – the number of annotated genes in these pathways exceeds 70. Simultaneously, a number of transcriptional comparisons such as Hardening_T_vs_Control_T, Combo_T_vs_Control_T and Combo_T_vs_Stress_T also show decreased activity of these same metabolic cascades as well as associated carbohydrate and amino acid metabolism pathways (00520, 00195, 00630, 00710, etc.). At the level of transcription, suppression of ribosomal and regulatory pathways – KEGG:03010, 03050, 03008 – was prominent, especially in the Stress_T_vs_Hardening_T comparison, which may reflect inhibition of synthesis of components of the translational machinery under conditions of transient or repeated stress. However, ribosomal pathways were suppressed less frequently and less significantly during the translation step, indicating that the main protein synthesising activity was retained even with a general decrease in metabolism. Thus, the reduced expression encompasses both key metabolic processes and individual elements of the translation system. This indicates the transition of cells into the energy-saving mode, in which transcription of most metabolic genes is reduced and translation becomes selective. The most pronounced and complex changes are noted in comparisons involving stressors after hardening, emphasising the significance of the sequence of exposure and the presence of adaptive metabolic rearrangement.
GO:MF
Functional annotation of genes by GO:MF category revealed systematic differences in molecular functions depending on the direction of expression change. The genes with increased expression are characterised by the predominance of functions providing translational response, regulation of protein metabolism and energy-dependent interactions. In particular, activities related to translation factors (GO:0008135), rRNA binding (GO:0019843), translation initiators (GO:0003743), and GTP- and ATP-binding proteins are enriched. These patterns are particularly represented when comparing stressed and hardened plants, reflecting enhanced translational activity and mobilisation of the protein synthesising apparatus as a key adaptive strategy. The group with unchanged expression exhibits predominantly general molecular functions lacking strict context specificity. Prominent among these are functions of binding and non-specific catalytic activity, including binding profiles and general-purpose enzymatic activities that show neither stress nor metabolic orientation. This spectrum of function appears to correspond to a basic molecular pool that is not subject to dynamic regulation in response to changes in conditions. Genes with reduced expression are predominantly associated with metabolic and signalling functions requiring coenzyme support and ion transport. Enriched activities include metal and ion binding (including calmodulin-dependent), functions related to biotin and iron-sulfur clusters, and redox enzyme activities. In particular, suppression of the activities of NAD(P)H-dependent reductases, flavin-binding enzymes and carboxylases may indicate systemic inhibition of certain energy and anabolic fluxes under stress conditions. Such functional shifts are particularly noticeable when comparing stressed plants with controls and indicate a prioritised shutdown of part of the metabolic and transduction cascades. Thus, the spectrum of molecular functions strictly correlates with the directionality of the transcriptional response: stress is associated with activation of protein synthesising and translational-regulatory functions, hardening is associated with rearrangement of enzymatic and coenzyme-dependent pathways, and stable functions remain the background of constant activity.
GO:CC
The functional distribution of transcripts and translationally active mRNAs across cellular components (GO:CC) revealed distinct differences in the spatial organisation of the genetic response to cold stress, hardening and their combination. At the transcriptional level, genes with reduced expression were predominantly localised in chloroplast and photosynthetic structures, including photosystems I and II, thylakoid membranes and the plastid envelope, reflecting systemic suppression of light-dependent processes under stress and, in part, in hardered and combined variants. At the same time, suppression of mitochondrial and ribosomal components was observed, especially in the Stress_T_vs_Hardening_T comparison, where structures related to protein transport (vesicles, EPR and Golgi complex compartments) are also involved. This indicates a broader impact of stress on the energy and protein synthetic infrastructure of cells. In contrast, genes with increased transcriptional activity showed enrichment in terms related to ribosomes, proteasomes, peptidase complexes and structures involved in protein degradation and translation regulation (e.g., preribosome, small-subunit processome). This is particularly pronounced in the Combo_T_vs_Control_T and Hardening_T_vs_Control_T comparisons. Variants involving stress (Stress_T_vs_Hardening_T) also activated chloroplast photosystems, indicating a possible recovery of photosynthetic function under some conditions. At the level of translation, trends were generally maintained but were more selective. Increased expression was predominantly associated with ribosomal subunits and proteasome complexes in the Stress_vs_Hardening, Combo_vs_Hardening, and Hardening_vs_Control comparisons, emphasising the activation of rapid protein regulation and degradation systems under conditions of recovery and adaptation. Variants showing increased expression of genes encoding chloroplast components (Combo_vs_Stress, Hardening_vs_Control) indicated the possibility of reactivation of photosynthetic processes under certain pretreatment. In turn, the reduced translational activity was concentrated in chloroplast-related structures including photosystems, thylakoids and plastid membranes, especially in the Stress_vs_Control and Combo_vs_Stress comparisons. This further emphasises the repressive role of cold on photosynthetic activity. Interestingly, genes with unchanged expression at both transcriptional and translational levels were consistently associated with ribosomes and transport vesicles (e.g., ER-to-Golgi vesicles) as well as spliceosomal complexes. This may indicate that a baseline level of transcriptional and translational activity is maintained to ensure cellular homeostasis even under stress. Thus, GO analysis by cell component revealed a multilevel rearrangement of intracellular architecture in response to stress and adaptive interventions.
GO: BP
Under conditions of stress and hardening, decreased expression of genes related to carbohydrate transport (GO:0008643) and pigment metabolism (GO:0042440, GO:0046148) was observed. This may indicate a redistribution of resources under stress conditions aimed at maintaining vital functions. Increased expression of genes involved in the localisation of proteins to organelles (GO:0033365, GO:0072599) indicates activation of processes that ensure proper functioning of cellular structures in response to stress effects. Decreased expression of genes related to amino acid catabolism (GO:0009063) and photosynthetic processes (GO:0009767) may reflect adaptation of metabolic pathways to altered environmental conditions. This allows the plant to efficiently utilise available resources and maintain energy balance. Genes with unchanged expression associated with key metabolic processes, such as amino acid synthesis and photosynthesis regulation, show resistance to stress. This emphasises the importance of maintaining basic cellular functions for plant survival. Thus, a comprehensive analysis of differential gene expression at the transcriptional and translational level under various stresses and hardening has revealed key biological processes involved in plant adaptation. The obtained data may serve as a basis for further studies of molecular mechanisms of plant resistance to unfavourable environmental factors.
WGCNA
To conclude the study, we analysed weighted gene co-expression networks (WGCNA). Following the scheme presented above, data corresponding to transcription and translation were analysed separately.
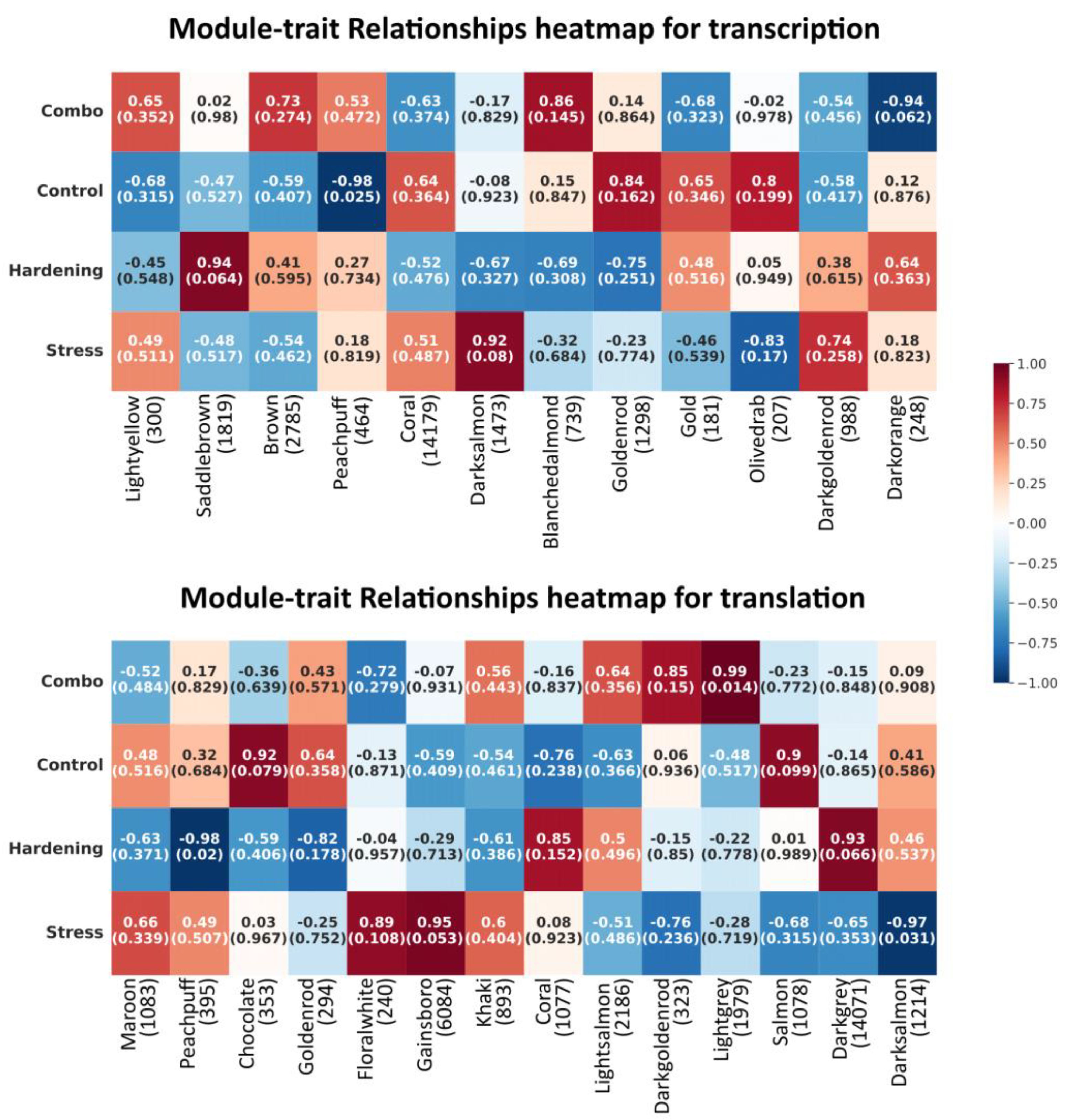
For transcriptional data, 12 modules (groups of genes whose expression shows correlation) were obtained; for data obtained for the translation step, 15 modules were obtained. For each of the steps (transcription and translation), the correlation between module and experimental conditions was examined (summarised data are presented as heat maps in
Appendix A). As can be seen for each condition, distinct co-expression modules were identified, differing both in the degree of correlation and in the power of these modules. It is worth noting, however, that in the vast majority of cases the p-value is quite large (values indicated in brackets in the heat maps). Therefore, even if the observed regularities do occur, they cannot be considered statistically reliable, and additional experiments may be required to confirm them. Nevertheless, a certain number of comparisons are successful on p-value. For these modules, we performed additional functional enrichment analyses, which showed the following.
For the transcriptional samples, the correlation between the control conditions and the peachpuff module can be considered statistically significant. The correlation coefficient in this case is -0.98 (p-value = 0.025), indicating a general decrease in gene expression of this module in control conditions (for other processing conditions, the data are not statistically significant but show a weak positive correlation within 0.2–0.5). This module includes 464 genes. Functionally, these genes are annotated with high-level terms such as cellular component organisation or biogenesis, plastid organisation, cellular component organisation, thylakoid membrane organisation, response to light stimulus, plastid membrane organisation, mitotic cell size control checkpoint signalling, etc. For enrichment with GO:BP terms, in case of KEGG, these are Ribosome, Ribosome biogenesis in eukaryotes, and TCA cycle Krebs cycle (
Appendix A).
For the translational samples, 3 modules with p-value < 0.05 were identified: peachpuff (395 genes), correlation coefficient -0.98 (p-value = 0.02); lightgrey (1979 genes), correlation coefficient -0.99 (p-value = 0.014); and darksalmon (1214 genes), correlation coefficient -0.97 (p-value = 0.031).
Modules were associated with the following experimental conditions: peachpuff – hardening, lightgrey – a combination of hardening and stress, and darksalmon – stress.
Functional enrichment analysis of the genes of the peachpuff module (for translation) shows the predominance of high-level terms such as translation, macromolecule biosynthetic process, gene expression, biosynthetic process, protein metabolic process, macromolecule metabolic process, negative regulation of gene expression via chromosomal CpG island methylation, protein targeting to ER, etc. For enrichment with GO:BP terms, in the case of KEGG and WP, they are Ribosome, Ribosome biogenesis in eukaryotes, and TCA cycle Krebs cycle. These processes are suppressed in hardered plants.
For the light grey module, closely positively correlated with combined effects, genes are annotated with the following terms: positive regulation of ubiquitin protein ligase activity, positive regulation of post-translational protein modification, positive regulation of protein modification by small protein conjugation or removal, positive regulation of transferase activity, regulation of post-translational protein modification, regulation of protein modification by small protein conjugation or removal, positive regulation of protein metabolic process, transcription preinitiation complex assembly, lipid X metabolic process. For enrichment of KEGG terms: Lipoic acid metabolism, ubiquitin-mediated proteolysis, amino sugar and nucleotide sugar metabolism.
For the darksalmon module, whose constituent genes are repressed in plants under stress, enrichment is observed for the following terms: transport, acetyl-CoA metabolic process, cellular process, regulation of transport, cellular localisation, regulation of monoatomic ion transmembrane transport, organophosphate biosynthetic process, organophosphate metabolic process, purine-containing compound metabolic process, membrane lipid metabolic process, acyl-CoA metabolic process, purine-containing compound biosynthetic process, nitrogen compound transport, protein transport, lipid metabolic process, regulation of transmembrane transport, positive regulation of translational elongation, positive regulation of translational termination, amide biosynthetic process, lipid biosynthetic process, carboxylic acid metabolic process, regulation of translational elongation, organic acid biosynthetic process. KEGG term enrichment: Biosynthesis of unsaturated fatty acids and Valine, leucine and isoleucine biosynthesis.
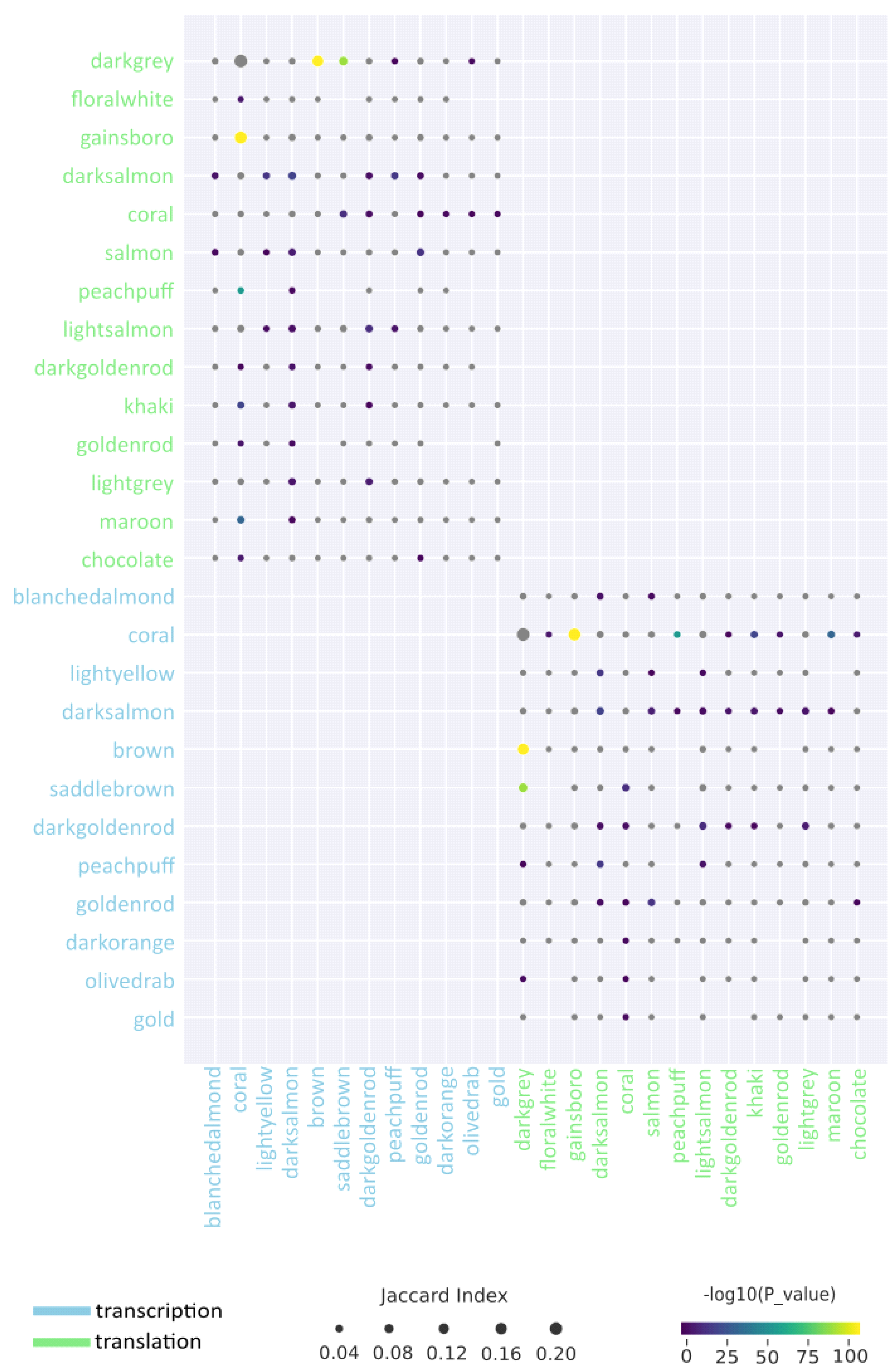
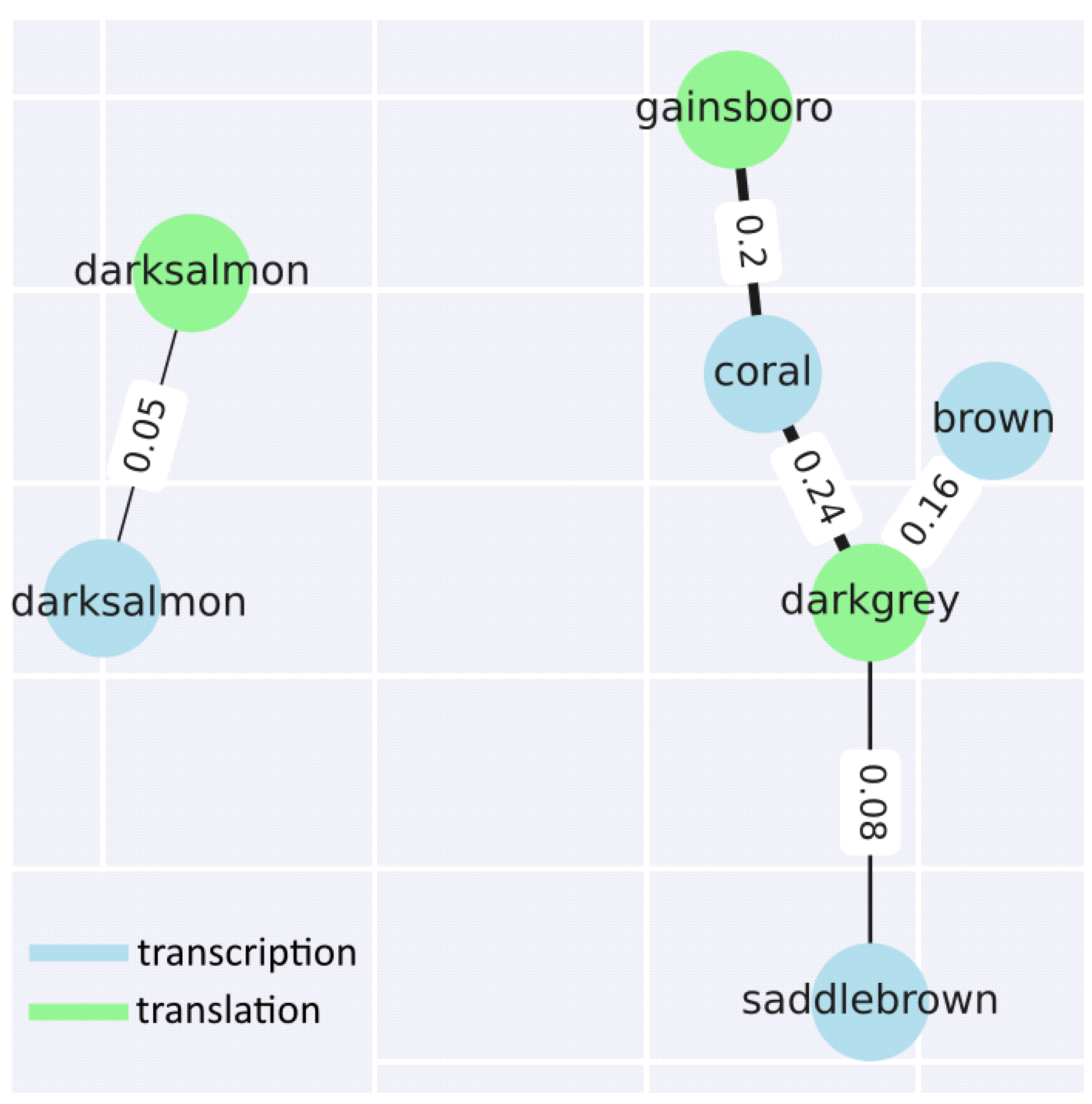
In addition to identifying and analysing the modules within each step, two groups of modules corresponding to each step were compared. As expected, based on the data described above, only 2 groups of modules show similarities (as expressed using the Jaccard coefficient) greater than 5%. If we look in more detail, the darksalmon modules for transcription and translation show insignificant similarity at the level of 5% (recall that the names do not indicate similarity, commonality, etc., and are chosen by the programme used for the analysis). The translational modules gainsboro and darkgrey and the transcriptional modules coral, brown and saddlebrown are more related. Excluding statistically insignificant comparisons, the following 3 comparisons were identified: gainsboro (translation) vs. coral (transcription); darkgrey (translation) vs. brown (transcription); and darkgrey (translation) vs. saddlebrown (transcription) (
Figure 6,
Appendix B).
Next, functional enrichment analysis was performed for common genes of similar modules. The comparison of gainsboro vs. coral demonstrates the predominance of such terms as MAPK cascade, acyl-CoA metabolic process, membrane fusion, negative regulation of abscisic acid-activated signalling pathway, heat acclimation, regulation of intracellular signal transduction, proteasome-mediated ubiquitin-dependent protein catabolic process, energy derivation by oxidation of organic compounds, positive regulation of signalling, proteasomal protein catabolic process, translational initiation, and carbohydrate derivative catabolic process.
The leading terms for darkgrey vs. brown are oxylipin biosynthetic process, glucan catabolic process, photosynthesis, light harvesting, nitrate metabolic process, polysaccharide catabolic process, galacturonan metabolic process, and response to light intensity. For the last comparison, steroid metabolic process, carboxylic acid catabolic process, cellular response to toxic substances, cellular oxidant detoxification, response to jasmonic acid, and response to fatty acids were identified.