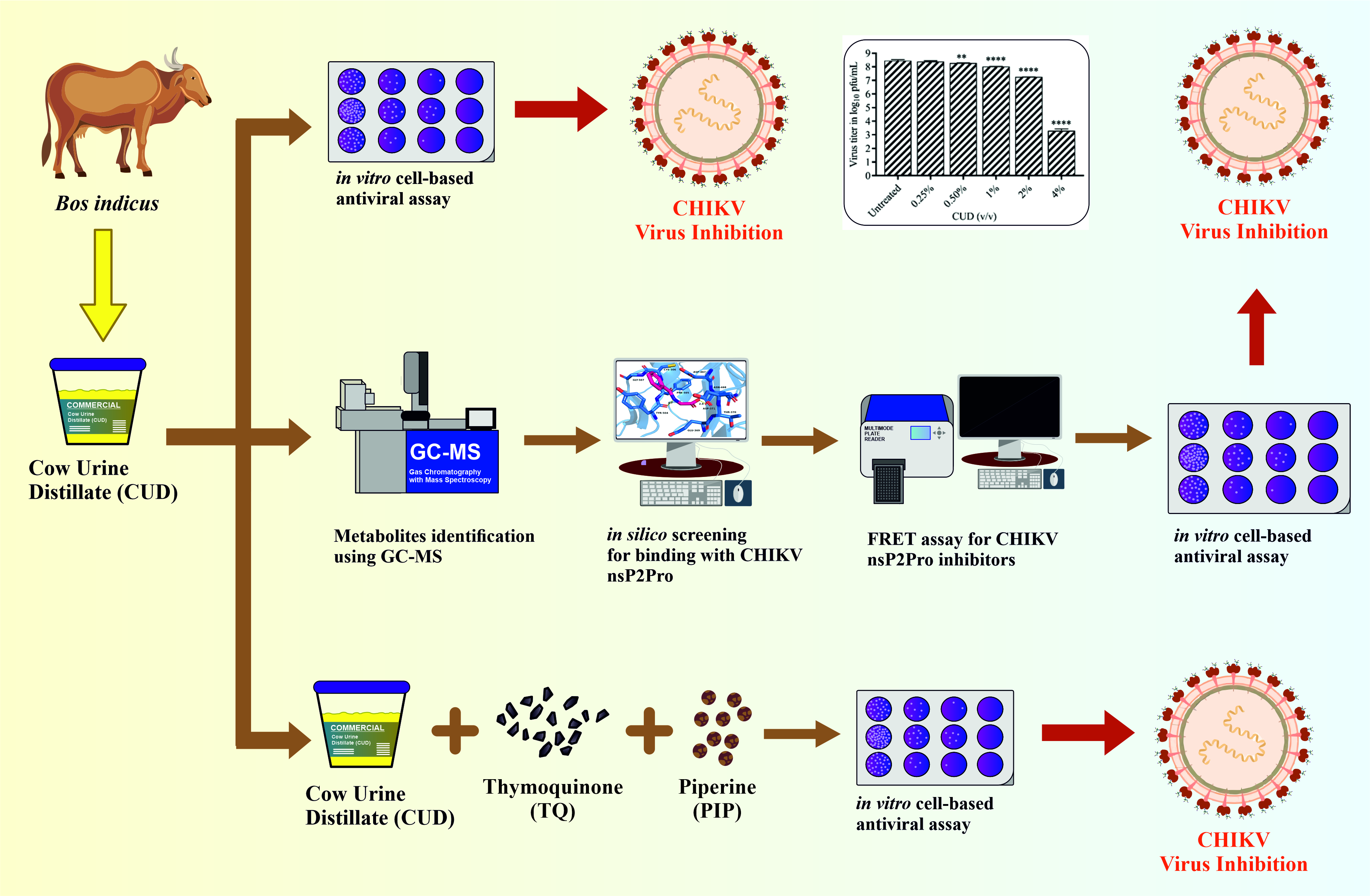
1. Introduction
Enveloped RNA viruses have an RNA genome enclosed by an envelope made of glycoproteins and the host-derived lipid membrane. These viruses are grouped into families based on differences in mechanisms of host cell penetration, genome replication, transcription, virion assembly, and structural organization. Chikungunya virus (CHIKV), transmitted by
Aedes aegypti and
Aedes albopictus species, belongs to the genus
Alphavirus in the family Togaviridae [
1,
2]. The alphavirus genome is a 42S single-stranded RNA of ~11.8 kb in size, with a 5′ capped terminus and a polyadenosine tail at the 3′ end. The genome comprises of two open reading frames (ORFs), an approximate 7 kb ORF located at the 5′ end that encodes the non-structural polyprotein, and an approximately 4 kb ORF at the 3′ end that encodes the structural polyprotein. [
3,
4,
5].
Alphaviruses are positive-sense enveloped RNA viruses. These viruses deliver their viral RNA into the host cell through membrane fusion, facilitated by membrane-anchored glycoproteins that recognise the host cell receptors. Alphaviruses utilise receptor-mediated endocytosis as their entry pathway [
6]. The two glycoproteins on the viral surface, E1 and E2, facilitate host cell invasion, with E1 aiding in membrane fusion and E2 assisting in receptor binding. One strategy to control alphaviruses is by blocking the virus’s attachment to the host receptor [
7]. Alphavirus capsid protein is a versatile protein essential for the encapsulation of the genome, virus budding, and assembly of virions[
8]. Post nucleocapsid disassembly, the RNA genome is directly translated into viral replication proteins, followed by structural proteins. Eventually, the host cell releases infectious viral particles. The viral genome translation and virus budding stages are critical for developing antiviral preparations that disrupt viral protein molecular interactions[
7,
9,
10,
11]. The viral replication enzymes nsP1, nsP2, nsP3 and nsP4 of alphaviruses, along with the host factors, drive viral genome replication in the host cytoplasm and are promising antiviral therapeutic targets[
5,
12,
13].
For thousands of years, India has practiced and used the five ingredients of Panchagavya in multiple ways in everyday life and in the traditional Ayurvedic system of medicine to heal the human body[
14].Cow urine distillate (CUD), popularly known as gaumutra ark (GA) of indigenous breeds of Indian cows, has been used in ancient India in various forms due to its medicinal and healing properties. Oral intake of CUD has been practiced in India for years because of its proposed usefulness in various ailments, i.e. Anorexia, indigestion, constipation, piles, worm infestations, oedema, bloating, colic pain, obesity, ascites, leukoderma, anemia, and various skin disorders etc. as per the different ancient Ayurvedic texts[
15]. CUD is sold commercially for consumption, with multiple suggested health benefits [
14]. Researchers have noted the diabetic wound healing properties of cow urine distillate during animal experiments[
16]. There are tremendous, reported health benefits of cow urine components as a bio-enhancer[
17]. Numerous studies have reported that CUD exhibits antimicrobial, antioxidant, and antifungal activities, highlighting its potential as a complementary therapeutic agent. [
18,
19]. Studies have also revealed the antiviral potential of cow urine on Canine Parvovirus (CPV)[
20].
Several compounds identified in cow urine distillate utilizing GC-MS (gas chromatography-mass spectrometry) have showcased anti-microbial activity[
15,
21]. Among these, compound precursors like Cinnamic acid have showcased antiviral potential against Zika virus (ZIKV), tested against RNA-dependent RNA polymerase (RdRp) [
22]. Furthermore, synergistic combination with CUD has been explored for therapeutic potential, for example, CUD along with
Zingiber officinale, enhanced the pharmacological efficacy of levetiracetam in an epileptic rat model[
23]. Thymoquinone (TQ), a monoterpene present in the seeds of
Nigella sativa L. (family Ranunculaceae) demonstrates a variety of beneficial biological and medicinal qualities and has been researched for its antiviral efficacy against the capsid protein of CHIKV [
24]. Likewise, another natural compound, Piperine (PIP), an alkaloid compound present in black pepper (
Piper nigrum L.), aids the bioavailability of drugs, nutrients, and vitamins. PIP showed various biological activities, such as anti-inflammatory [
25], anti-asthmatic [
26], and has showcased antiviral potential against Hepatitis-B virus (HBV) and CHIKV[
25,
27]
Urbanisation has contributed to the emergence of many zoonotic arboviral infections. These viruses persist in the environment and may undergo mutations, potentially re-emerging as more pathogenic strains[
28], which may result in significant morbidity and epidemics. Present-day treatment is targeted at alleviation of the symptoms of the disease, as no approved antiviral therapeutics are available against alphaviruses. Preventive measures against mosquito bites and vector control are the sole means to mitigate the risk of viral infection. There is a critical demand for an efficacious and non-toxic antiviral medication to mitigate CHIKV illness. The present study highlights the antiviral efficacy of CUD-derived components and provides a combinatorial therapy approach for developing a novel antiviral strategy against the CHIKV.
2. Materials and Methods
2.1. Cell Line, Virus and Compounds
Vero cells were obtained from the National Centre for Cell Science (NCCS), Pune and grown in Dulbecco’s Modified Eagle’s medium (DMEM; HiMedia), supplemented with 10% fetal bovine serum (FBS; Gibco), 1% penicillin-streptomycin solution (PenStrep; HiMedia) in a humidified incubator at 37 °C and 5% CO
2. Regular passaging using 1X trypsin-EDTA solution was performed at periodic intervals of 3 to 4 days to maintain the cell line. For all antiviral assays, CHIKV (Accession No. KY057363.1) was propagated and titrated in Vero cells according to the protocol described by Singh et al. (2018) [
29] . Hippuric acid (Sigma Aldrich), p-Aminohippuric acid (Sigma Aldrich), Benzoic acid (Sigma Aldrich), and Oelic acid (99%, SRL) were dissolved in tissue culture grade water. Thymoquinone (Sigma Aldrich) and Piperine (Sigma Aldrich) were dissolved in 100% dimethyl sulfoxide (DMSO). Cow urine distillate (CUD) was purchased from the Maharashtra Ayurved Center, Bavdhan Khurd, Pune-411021, India. CUD and compound dilutions were syringe filtered using a 0.2 μm filter.
2.2. In Vitro Cell-Based Antiviral Assay for Cow-Urine Distillate (CUD)
The concentrations of CUD and test compounds that maintain cell viability ≥ 70% were selected for subsequent antiviral evaluation. For evaluating CUD cytotoxicity, standard 3-(4,5- Dimethylthiazol- 2-yl)-2,5- Diphenyltetrazolium Bromide (MTT) assay was performed. Briefly, 96-well plates were seeded using Vero cells at an approximate density of 1x104 cells/well and incubated for 24 h at 37oC and 5% CO2. At 90% confluency, the media was removed and CUD dilutions of varying concentrations prepared in DMEM containing 2% FBS were added to each well. The cells were then incubated for 48 h with the compounds. Post incubation, MTT dissolved in PBS was added to each well and incubated for 4 h at 37oC. Media containing MTT was then removed, followed by the addition of 100% DMSO to the wells. The absorbance reading was taken at 570 nm with the help of Cytation 3 multi-mode plate reader (BioTek Instruments, Inc.). The data was analyzed, and percentage cell viability was calculated relative to untreated control (without compound). The graphs were plotted using GraphPad non-linear regression curve fit.
The titer determination of the progeny virus in the supernatant of the treated or untreated cells infected with CHIKV was used to analyze the antiviral activity of the compounds. Different concentrations of CUD (4, 2, 1, 0.5, and 0.25% v/v) were used in the antiviral assay. For this, 24-well culture plates were seeded with Vero cells at a cell density of approx. 1x105 cells/well and incubated at 37 °C for 24 h. Virus infection at multiplicity of infection (MOI) 1, including different concentrations of compounds, was done, and cells were incubated at 37 °C for 1.5 h. The media was then removed, and compound dilutions in maintenance media were added to each well and incubated for 24 h at 37 °C. At 24 h post-infection, the viral particles were collected from the supernatant and used for quantitative analysis of reduction in viral-titer on compound addition by plaque-reduction assay.
The conventional plaque-reduction assay was carried out to quantify the viral titer in the presence of various concentrations of CUD. Vero cells were seeded in 24-well plates and incubated until reaching 90% confluency. Ten-fold serial dilutions of the harvested virus samples were prepared, followed by infection of the Vero cells and incubation for 1.5 hours. The culture medium was aspirated and substituted with an overlay medium composed of a 1:1 mixture of 2X MEM (supplemented with 5% FBS) and 2% (w/v) carboxymethyl cellulose (CMC). Post 48 h incubation, the overlay media was removed, and the cells were fixed using 10% (v/v) formaldehyde and incubated for 3-4 h in the dark at room temperature. The cells were stained with 1% (w/v) crystal violet, to visualize plaques. The number of plaques was counted, and pfu/mL (plaque-forming units per mL) were calculated using the formula- pfu/mL = (No. of plaques)/ (Dilution factor x volume of diluted virus per well)
The reduction in viral titer was then determined, and graphs were plotted, which were further analyzed using the GraphPad non-linear regression curve fit method to determine the EC50 values. All experiments were performed in triplicates.
2.3. Gas Chromatography-Mass Spectrometry (GC-MS) Analysis of CUD
GC-MS analysis was performed on Agilent 7890 gas chromatograph coupled with an Agilent 5977C mass detector (Agilent technologies, CA, USA) for identification of major compounds/ metabolites in CUD. 2 mL of cow urine distillate was dried in vacuum concentrator (Eppendorf Concentrator plus™, Eppendorf; USA) at 37°C until completely dried. Finally, dried material in a micro-centrifuge tube was derivatized with N-methyl-N-(trimethylsilyl)-trifluoroacetamide (MSTFA) for GC-MS analyses. Derivatization was performed by adding 70 μl of MSTFA, followed by incubation at 37 °C for 30 minutes. A parallel derivatization reaction prepared in an empty tube without sample served as the control. The sample volume was 1 µl which was injected in GC-MS by an automatic sampler (G4513A Agilent technologies) with split ratio of 10:1. The injected sample was separated on fused silica capillary column DB-5 MS (5% phenyl methyl poly siloxane: 30 m × 0.25 mm i.d. × 0.25 µm, Agilent technologies). The temperature program used was 70°C for 5 min, then the temperature was increased to the final temperature of 300°C with a ramp rate of 10°C/min. The inlet temperature and interface temperature were set to 200°C and 120°C, respectively. The MS unit was set to its maximum sensitivity, and the total ion current was recorded for a mass range of m/z 25-780. The detector voltage was set at 1700 eV, and a 5 min solvent delay was given each time when the sample was run. The scan was done with a scan frequency of 4 S-1 (2.0 HZ). The entire sample was replicated three times.
2.4. Structure-Based Virtual Screening Targeting CHIKV nsP2Pro
The three-dimensional protein structure of CHIKV nsP2Pro was retrieved from the RCSB PDB data bank in .pdb format (PDB: 4ZTB). Virtual screening was performed using AutoDock vina module of PyRx 0.8 algorithm on a Windows i5 system. For virtual screening, macromolecules were prepared by removing crystallographic water, bound ligand, and ions from the protein PBD. The 3D structures of all the compounds were downloaded from PubChem in .sdf file format. These molecules were converted to pbdqt format using the Open Babel module of PyRx 0.8 after being energy minimized using the Universal Force Field (UFF). The active site residue used for preparing the grid and grid parameters are listed in a table (Supplement 2).
2.5. Screening Compounds Inhibiting the Proteolytic Activity of CHIKV nsP2Pro via FRET Assay
The CHIKV nsP2
Pro is a significant drug target as it is essential for processing viral non-structural polypeptide precursors, which release enzymes necessary for viral replication [
30]. It cleaves the polyprotein complex (nsP1234) into functional proteins associated with the formation of the replication complex[
29]. nsP2
Pro activity involves deprotonation of the thiol group of cysteine, which is a residue in the catalytic dyad of the active site (Cys478 and His548) [
30]. FRET-based assay leverages the physical principle where energy transfer occurs between two fluorophores when they are in proximity. For nsP2
Pro activity measurement, this involves using an internally quenched fluorogenic peptide substrate where a donor fluorophore (DABCYL) and quencher (EDANS) are positioned on opposite sides of the protease cleavage site (DRAGG ↑ YISF) [
30]. The protease cleavage site lies between the C-terminal of nsP3 and the N-terminal of nsP4. When intact, the quencher suppresses fluorescence from the donor, but upon cleavage by nsP2
Pro, the donor and quencher separate, resulting in detectable fluorescence.
An optimized concentration of 5µM of active protein was used for the analysis of proteolysis activity. The purified protein was resuspended in reaction buffer containing 20 mM bis-tris propane at pH 7.5. Compounds dilutions were prepared and added in a Corning® 96 Well Half-Area Microplate (flat bottom, black polystyrene, Corning USA). The compounds were analysed for intrinsic fluorescence, and compounds exhibiting higher fluorescence than the protein control were not utilized for the assay. The modified substrate (DABCYL)- DRAGG↑YISF-(EDANS) was diluted to 100µM. To determine the optimal concentration for effective inhibition, various concentrations of inhibitors were tested against 5 µM of nsP2Pro, and a concentration of 500 µM was found to substantially suppress proteolytic activity, making it a suitable concentration for inhibition. Following the addition of substrate and inhibitor, the samples were incubated for 10 minutes. Fluorescence was then measured at an excitation and emission wavelength of 360 nm and 460 nm respectively using the BioTek Synergy H1 multimode microplate reader. All sample readings were taken in duplicates to ensure accuracy and data acquisition was performed using Gen5 software. The fluorescence reading obtained from the substrate control was subtracted from the measurement of fluorescence corresponding to proteolytic activity. The compounds exhibiting significant inhibition were documented and plotted for comparative analysis for inhibition in fluorescence against protein control. All result graphs were plotted using GraphPad Prism 8.
2.6. In Vitro Antiviral Assay of Compounds Selected After In Silico Studies:
For evaluating the cytotoxicity of Benzoic acid, Hippuric acid, p-Aminohippuric acid, and Oleic acid, a standard MTT assay was performed, as described above. The antiviral potency of the compounds was evaluated through titer determination of progeny virus in the supernatant of cells, both treated and untreated, infected with CHIKV. Briefly, 24-well plates were seeded with Vero cells at a cell density of approx. 1x105 cells/well and incubated for 24 h at 37 °C in a CO2 incubator. The monolayer of cells was treated with compound dilutions for 2 h, the compounds were removed, followed by CHIKV infection at MOI 0.1 for 1.5 h. The media was then removed, and compound dilutions in maintenance media were added to each well and incubated for 24 h at 37 °C. At 24 hours of post-infection in cells, the viral particles were collected from the supernatant and used for quantitative analysis of reduction in viral titer on compound addition by plaque-reduction assay. Plaque assay was performed, and the viral titer was calculated as mentioned above. The percentage inhibition was determined. All the data was plotted using GraphPad Prism 8 software. Each experiment was performed in triplicate.
2.7. Combinatorial Antiviral Assay: Assessment of Synergistic Effect
To assess the synergistic effect of natural compounds PIP and TQ in combination with CUD, against CHIKV, the studies were performed as described above. A range of concentrations of TQ (1.25, 2.5, 5, 10 and 20 µM), PIP (1.25, 2.5, 5, 10 and 20 µM) and CUD (0.25, 0.5, 1, and 2% v/v) was used as working concentrations in-combinations for this experiment. Briefly, 24-well plates were seeded with Vero cells and incubated for 90% confluency. Infection with the virus at a MOI of 1 was performed alongside the compounds for 1.5 h. Subsequently, the virus inoculum was removed, and compound dilutions were added again with incubation for an additional 24 h. Samples were then collected for plaque assay, and viral titer determination was done employing the standard plaque assay to analyze the reduction in viral titer using GraphPad Prism. All experiments were performed in triplicates.
3. Results
3.1. Assessment of Antiviral Activity of CUD
The plaque-reduction assay was performed to test the antiviral activity of CUD post MTT assay (
Figure 1a). The percentage reduction in CHIKV viral titer treated with CUD was evaluated and promising results were obtained (
Figure 1b). More than 90% and 99.9% reduction in CHIKV titer was observed with 2% v/v and 4% v/v CUD, respectively. (
Figure 1b). The effective concentration at which 50% reduction in the virus titer was observed i.e., EC50 was 0.423% v/v of CUD.
3.2. Identification of Metabolites in CUD
Data acquisition, automated peak detection, mass spectral deconvolution, and library searches were performed using Agilent ChemStation™ software and W Search Pro (
www.wsearch.com.au). Metabolite identification was achieved by comparing the mass spectra of target metabolites with entries in the NIST-17 mass spectral library (National Institute of Standards and Technology) as well as an in-house mass spectral database. Metabolites with a matching similarity of 80 % or more in the library search were only considered. The final calculation did not consider all artifact peaks like plasticizers, column bleed, and derivatizing agent peaks. Unique quantification masses were taken and specified to obtain accurate peak areas for each metabolite. Each mass spectrum was carefully analyzed for co-elution detection.
The CUD was characterized by GC-MS for identification of metabolites/substances present in it and was summarized in (
Table 1 and
Figure 2). This was done to identify the potential active metabolites/substances present in CUD. The presence of some of these molecules in CUD or a combination of these metabolites/substances makes CUD an effective antiviral in the composition.
3.3. In Silico Binding Study of Identified Compounds from CUD
The virtual screening of CHIKV nsP2
Pro was performed using AutoDock Vina. The 3D structure of CHIKV nsP2
Pro was downloaded from the RCSB PDB data bank (PDB:4ZTB) and used for computer-aided structure-based binding studies. Ligand preparation involved energy minimization and conversion to. pdbqt format, while grid parameters were optimized for accurate docking. Binding energy of docking, polar and non-polar interactions formed by compound with CHIKV nsP2
Pro is given in table (
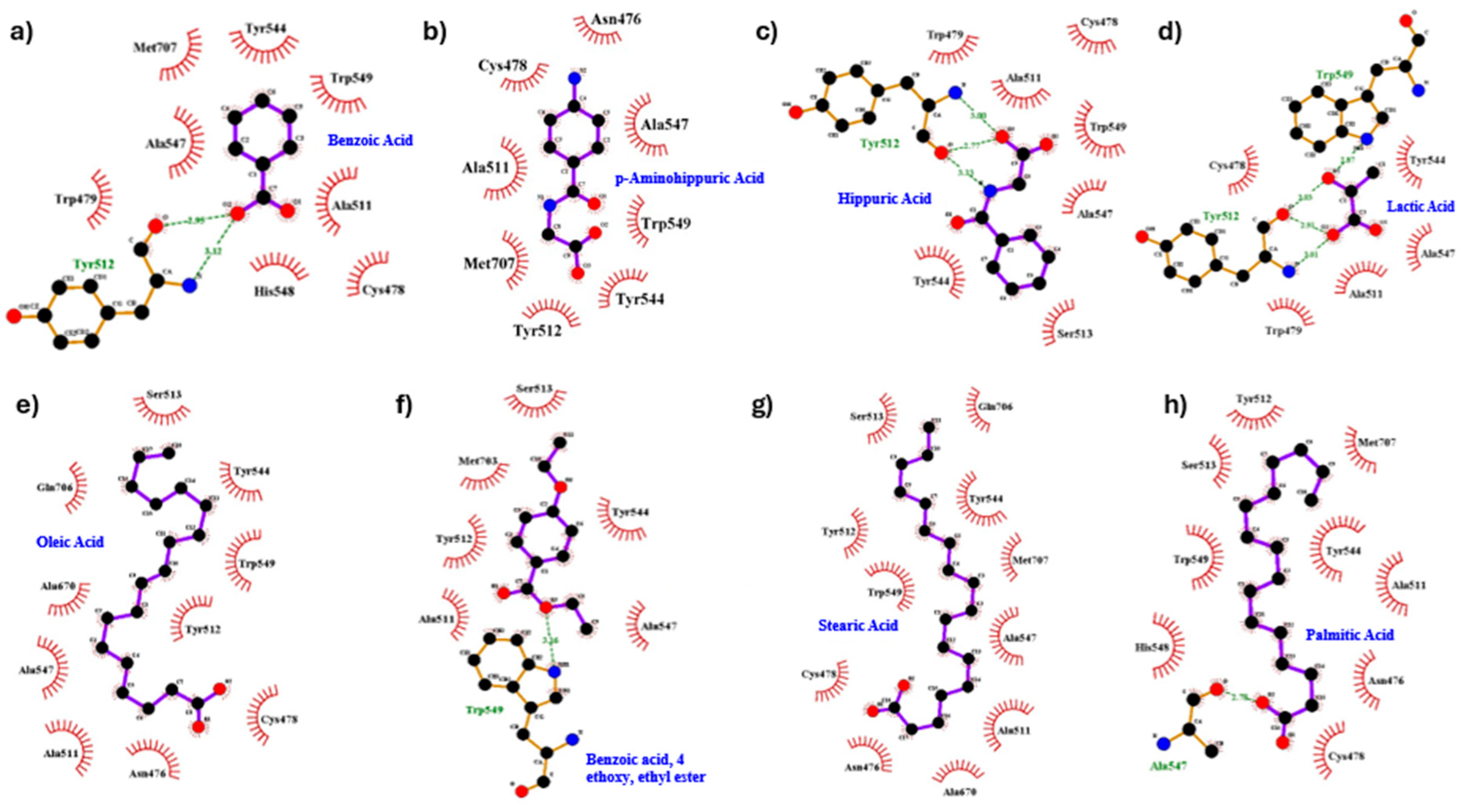
Table 2). Binding affinities ranged from -4.5 to -5.6 kcal/mol. Hippuric acid showed the strongest binding (-5.6 kcal/mol), forming three hydrogen bonds with Tyr512, while others exhibited fewer or no hydrogen bonds (
Figure 3). Hydrophobic interactions involved residues such as Cys478, Ala511, Trp549, and Met707, providing significant stabilization for the ligands within the active site.
3.4. Inhibition of the Proteolytic Activity of nsP2Pro:
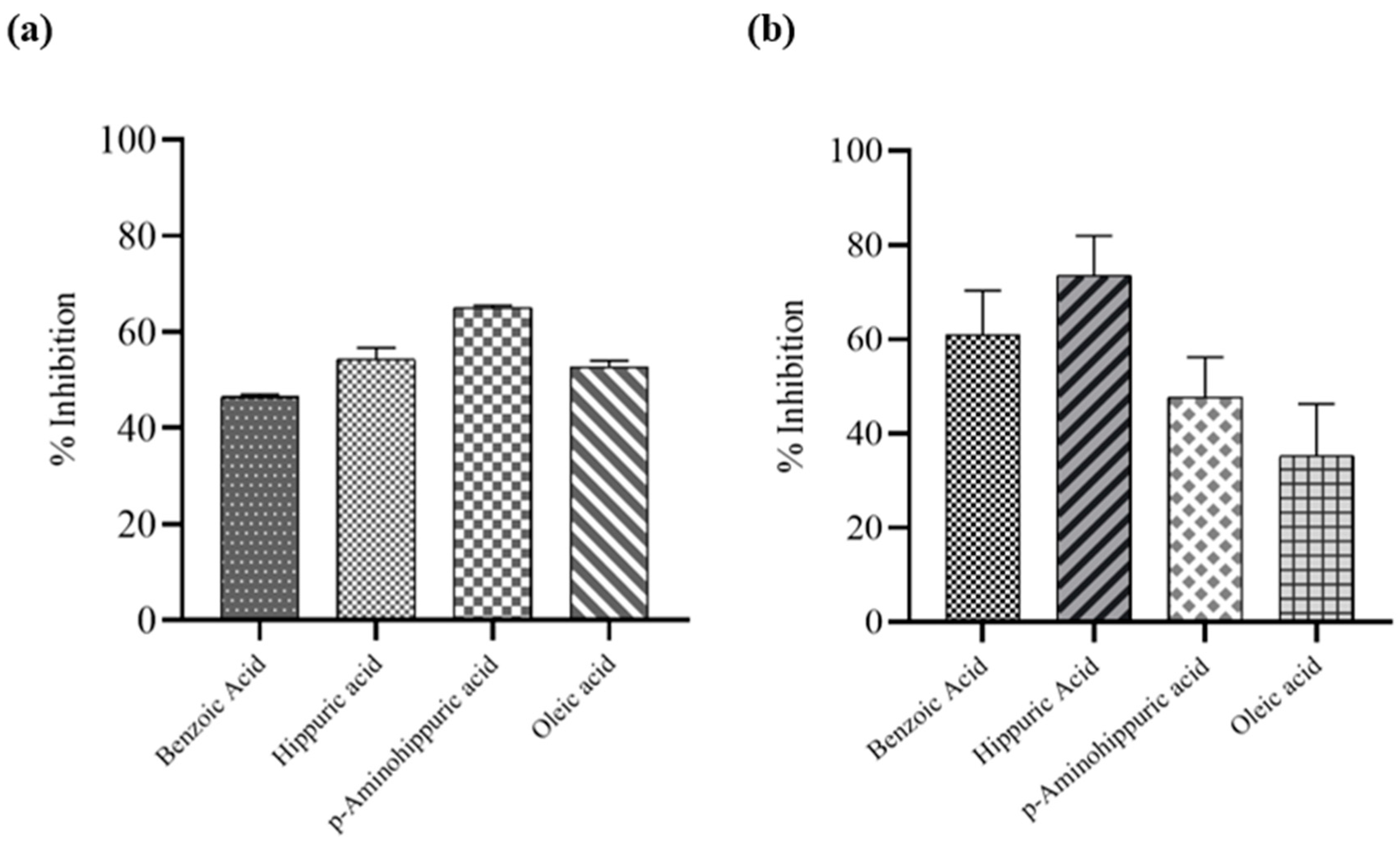
For determining the inhibition potential of proteolytic activity, various concentrations of the protein were used, and fluorescence signals were measured. The fluorogenic peptide substrate was stimulated at a 360 nm wavelength and emission was measured at 460 nm using BioTek Synergy H1 multimode-plate reader. In the case of inhibition of nsP2
Pro proteolytic activity against cleavage of nsP3/4, the fluorophore EDANS and the quencher DABCYL would remain in proximity due to the intact peptide substrate. As a result, the fluorescence of EDANS would be quenched by DABCYL, and minimal fluorescence would be emitted by the sample. The cleavage of the peptide substrate results in the separation of the FRET pair and a reduction in energy transfer, leading to an increase in the fluorescence signal. An optimized concentration of 500 µM for all test compounds resulted in a marked reduction in fluorescence, indicative of protease inhibition. Quantitative analysis across multiple substrate concentrations revealed that the test compounds exhibited inhibition ranging from 45% to 65%. The percentage inhibition was observed to be 46.45 ± 0.6 %, 54.27 ± 2.45 %, 65.01 ± 0.52 %, and 52.63 ± 1.43 % for Benzoic acid, Hippuric acid, p-Aminohippuric acid, and Oleic acid, respectively. (
Figure 4a)
3.5. Assessment of Antiviral Activity of Identified Metabolites:
Antiviral activity of Benzoic acid, Hippuric acid, p-Aminohippuric acid, and Oleic acid was assessed against CHIKV. Cytotoxicity was assessed in vitro in Vero cells employing the standard MTT assay protocol. (Supplement 5). The concentration of compounds at which cell viability remained above 80% was selected for antiviral assessment. Plaque-reduction assay was performed to test the antiviral activities of Benzoic acid, Hippuric acid, and p-aminohippuric acid at 250 µM concentration, and Oleic acid at 31.25 µM concentration. The percentage reduction in CHIKV viral titer treated with individual compounds was evaluated, and promising results were obtained (
Figure 4b). The percentage inhibition was observed to be 60.88 ± 9.43 %, 73.51 ± 8.45 %, 47.64 ± 8.57 %, and 35.24 ± 11.0 % for benzoic acid, for hippuric acid, p-aminohippuric acid, and oleic acid, respectively.
3.6. Synergistic Effect of TQ and PIP in Combination with CUD
Prior to conducting the subsequent experiments, an MTT assay was performed to assess the cytotoxicity of TQ, PIP, and their combination with CUD (Supplement 1a, 1b, 1c). Additionally, preliminary antiviral assays were conducted for TQ and PIP to evaluate their inhibitory potential (Supplement 1d, 1e). The antiviral effect of different concentrations of CUD (0.25 to 2% v/v), TQ (1.25 to 20 µM), and PIP (1.25 to 20 µM) in combination was evaluated by plaque-reduction assay. The combination of 2%v/v CUD, 20 µM TQ and 20 µM PIP and lower concentrations were used for the combinatorial studies. The compounds TQ, PIP, and CUD, when added in combination, showed better viral inhibition compared to their individual inhibitory effect in an additive and synergistic manner (
Figure 5a-e). There was a significant reduction in the CHIKV titer when the cells were treated with a combination of 1.25 µM TQ + 1.25 µM PIP + 0.25% v/v GA (
Figure 5a), 2.5 µM TQ + 2.5 µM PIP + 0.25% v/v GA (
Figure 5b), 5 µM TQ + 5 µM PIP + 0.5% v/v GA (
Figure 5c), 10 µM TQ + 10 µM PIP + 1% v/v GA (
Figure 5d), and 20 µM TQ + 20 µM PIP + 2% v/v GA (
Figure 5e). The compound concentrations were more effective and synergistic against CHIKV in 20 µM TQ + 20 µM PIP + 2% v/v GA combination (
Figure 5e) with ~ 4-log or 99.99% reduction in CHIKV titer.
4. Discussion
Alphaviruses are known to cause debilitating diseases in both humans and animals. These are transmitted by arthropods, typically mosquitoes that feed on blood [
31]. In the last decade, research on alphaviruses has considerably intensified, partially due to the re-emergence and expansion of the CHIKV in Asia, Europe, and the Americas [
32]. No effective antiviral drugs or therapies are available to combat Chikungunya disease. Therefore, there is an urgent need to develop an effective antiviral preparation. Developing a novel antiviral prophylactic treatment against the re-emerging viruses is quite challenging and requires extensive expertise for antiviral experiments [
33]. India, since time immemorial, has practiced and used natural drugs, molecules, and substances as traditional Ayurvedic medicines in various ways in everyday life and in healing the human body. The natural drug molecule antiviral combination reduces the economic burden to a greater extent, especially in developing countries like India. The large quantities of easily available natural drug molecule antiviral combination make the process viable. The five components of Panchagavya have been utilized in India since ancient times in various aspects of daily life and within the Ayurvedic system of medicine. Cow urine distillate (CUD), commonly referred to as gaumutra ark (GA) derived from indigenous Indian cattle breeds, is one of the components historically employed via oral administration with several reported health benefits. CUD in Ayurveda is believed to be one of the most effective animal-origin substances, with the ability to improve general health, and is established as an antimicrobial substance scientifically. The current study scientifically explores and unravels its antiviral effect against emerging CHIKV.
To commence the study, the antiviral activity of the CUD was investigated both as monotherapies and in combinatorial treatments, against Chikungunya virus (CHIKV), utilizing
in vitro cell-based assay post MTT assay to determine cytotoxicity (
Figure 1a). CUD exhibited a significant reduction in CHIKV virus titer with a prominent EC50 value, i.e., 0.423 % v/v (
Figure 1b). It showed potent antiviral effect with more than 99.9% reduction in CHIKV titer at 4% v/v concentration (
Figure 1b). This confirmed the antiviral property of CUD against CHIKV.
Several compounds identified in cow urine distillate using Gas Chromatography coupled with mass spectrometry (GC-MS) have showcased anti-microbial activity in previous studies. (reference 22). GC-MS was performed in the current study to characterize and identify the metabolites/substances (
Figure 4) of CUD responsible for the antiviral activity. The metabolites identified have been listed in
Table 1. The compounds were further explored for their antiviral potential against CHIKV nsP2
Pro. Among CHIKV proteins, nsP2
Pro is a key therapeutic target due to its essential role in viral replication and transcription. It is a protease that exerts its proteolytic function via a conserved Cys478-His548 catalytic dyad. As the nsP2
Pro plays a critical role in viral replication, its inhibition presents a possible opportunity for therapeutic intervention. Computational approaches have emerged as indispensable tools in the initial phases of drug discovery and development [
34]. The virtual screening of CHIKV nsP2
Pro demonstrated that the identified compounds bind effectively to CHIKV nsP2
Pro, with binding affinities ranging from -4.5 to -5.6 kcal/mol (
Table 2). Hydrogen bonds and hydrophobic interactions with critical residues in nsP2 were observed, indicating stable complex formation (
Figure 3).
Four candidate compounds, Benzoic acid, Hippuric acid, p-Aminohippuric acid, and Oleic acid, exhibited pronounced inhibitory potential against nsP2
Pro, as substantiated by their inhibitory activity in a FRET-based biophysical assay. The observed inhibition likely arises from the compounds’ capacity to disrupt the enzymatic cleavage of the substrate, demonstrating their potential for inhibition (
Figure 4a). Subsequently, these four compounds were tested for antiviral activity against CHIKV via
in vitro plaque reduction assay to further substantiate their antiviral activity. The substantial decrease in viral plaques demonstrates their capacity to inhibit viral replication and dissemination within host cells (
Figure 4b).
Natural products are a valuable resource for identifying new antivirals and formulating effective preventive and therapeutic approaches to combat viral infections. Combination therapies utilising natural agents alongside standard therapeutics may synergistically decrease the possibility of developing drug-resistant viruses. The research group has previously demonstrated the role of natural compounds and their derivatives in the inhibition of CHIKV, including chitinase (chi)-like lectin from Tamarind (TCLL) against viral entry [
7], rugosaflavonoid derivative against nsP3 [
35], herbacetin (HC) and caffeic acid phenethyl ester (CAPE) against nsP1 [
36]. The natural drug molecules TQ and PIP are known to possess antiviral activity against the enveloped RNA viruses[
11,
25,
37] TQ, a natural compound, targets the hydrophobic pocket of CHIKV capsid protein (CP) and inhibited CHIKV replication by affecting the CP-cdE2 interactions in the viral life cycle[
11]. While PIP, an alkaloid compound, targets the RdRp protein of alphaviruses and efficiently inhibits the replication of alphaviruses at an early stage of the viral life cycle[
25,
37]. The combinatorial antiviral effect of CUD, TQ and PIP was estimated against CHIKV. TQ, PIP, and CUD showed better viral inhibition when added in combination compared to their individual inhibitory effect (
Figure 5, Supplement 1). The significant reduction in the CHIKV titer was due to the additive and synergistic properties of natural compounds and CUD. The compound concentrations were found to be more effective and synergistic against CHIKV in 20 µM TQ + 20 µM PIP + 2% v/v GA combination with ~ 4 log or 99.99% reduction in CHIKV titer (
Figure 5). The results validated that the combinatorial antiviral effect exerted by the compounds was significant when used in combination against CHIKV as compared to the viral reduction observed with monotherapy.
In conclusion, crude CUD and its components- Benzoic acid, Hippuric acid, p-Aminohippuric acid and Oleic acid show substantial antiviral activity against CHIKV. From the screening for composition of CUD for identifying potential inhibitors, four compounds out of the selected eight compounds showed significant inhibition of alphavirus, at micromolar non-toxic concentrations as confirmed by in vitro cell-based antiviral assay. Moreover, CUD in combination with TQ and PIP showed a significant reduction in CHIKV virus titre when tested in Vero cells. The emergence of resistance to antiviral drugs is a concern as it poses a significant challenge to the sustained use of antiviral drugs in clinical usage. A combinatorial approach by leveraging the synergistic effect of CUD in combination with TQ and PIP, with different viral targets, might mitigate the risk of antiviral resistance and circumvent the development of escape mutants observed during the administration of monotherapy. The present study provides a novel method for improving antiviral activity and bioavailability of natural drug molecules using CUD in different formulations. The developed composition may be explored for broad-spectrum antiviral therapy in the future, as there is a necessity for an antiviral formulation that can provide broad-spectrum antiviral treatment against other developing enveloped RNA viruses, including Dengue, Zika, and West Nile virus.
Supplementary Material
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Supplementary File 1
Author Contributions
Conceptualization: Ravi Kumar, Tanuj Handa, Ankita Saha, Vishakha Singh, Vedita Anand Singh, Shashank Sagar Saini, Arvind Kumar, Sumer Singh, Debabrata Sircar, Pravindra Kumar and Shailly Tomar; Formal analysis: Ravi Kumar, Tanuj Handa, Ankita Saha and Shailly Tomar; Methodology: Ravi Kumar, Tanuj Handa, Ankita Saha, Vishakha Singh, Vedita Anand Singh, Shashank Sagar Saini, Arvind Kumar, Sumer Singh, Debabrata Sircar, Pravindra Kumar and Shailly Tomar; Validation: Ravi Kumar, Tanuj Handa, Ankita Saha, Vishakha Singh, Vedita Anand Singh, Shashank Sagar Saini, Arvind Kumar, Sumer Singh, Debabrata Sircar, Pravindra Kumar and Shailly Tomar; Investigation: Shailly Tomar; Writing - original draft: Ankita Saha, Ravi Kumar and Tanuj Handa; Writing - review & editing: Ravi Kumar, Tanuj Handa, Ankita Saha, Vishakha Singh, Vedita Anand Singh, Shashank Sagar Saini, Arvind Kumar, Sumer Singh, Debabrata Sircar, Pravindra Kumar and Shailly Tomar
Funding
This research was financially supported by the Ministry of AYUSH (Project ref. no.: S-14011/8/2022-SCHEME)
Institutional Review Board Statement
The study was conducted under the approval by the Institutional Biosafety Committee, Indian Institute of Technology Roorkee (Approval number: IITR/IBSC/02/12/2024).”
Acknowledgement
ST and PK thank the Department of Biotechnology, Govt. of India for supporting the Translational and Structural Bioinformatics- BIC at Department of Biosciences and Bioengineering, IIT Roorkee (BT/PR40141/BTIS/137/16/2021). RK is thankful to the Ministry of Health and Human Resources (MHRD) for the Post-Doctoral Fellowship. VAS and TH gratefully acknowledge MHRD for providing research fellowship. AS gratefully acknowledges Prime Minister’s Research Fellows (PMRF) scheme, Ministry of Education, India for research fellowship. Authors also thank Ashok Soota Molecular Medicine Facility at the Indian Institute of Technology Roorkee (IIT Roorkee).
Conflict of Interest
The authors declare that there is no conflict of interests.
Declaration of Generative AI
No AI tools were used in this article.
References
- Vega-Rúa, A., Zouache, K., Girod, R., Failloux, A.-B., Lourenço-de-Oliveira, R.: High Level of Vector Competence of Aedes aegypti and Aedes albopictus from Ten American Countries as a Crucial Factor in the Spread of Chikungunya Virus. J Virol. 88, 6294–6306 (2014). [CrossRef]
- Weaver, S.C., Lecuit, M.: Chikungunya Virus and the Global Spread of a Mosquito-Borne Disease. New England Journal of Medicine. 372, 1231–1239 (2015). [CrossRef]
- Bergren, N.A., Auguste, A.J., Forrester, N.L., Negi, S.S., Braun, W.A., Weaver, S.C.: Western Equine Encephalitis Virus: Evolutionary Analysis of a Declining Alphavirus Based on Complete Genome Sequences. J Virol. 88, 9260–9267 (2014). [CrossRef]
- Strauss, E.G., Rice, C.M., Strauss, J.H.: Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology. 133, 92–110 (1984). [CrossRef]
- Tomar, S., Hardy, R.W., Smith, J.L., Kuhn, R.J.: Catalytic Core of Alphavirus Nonstructural Protein nsP4 Possesses Terminal Adenylyltransferase Activity. J Virol. 80, 9962–9969 (2006). [CrossRef]
- Helenius, A., Kartenbeck, J., Simons, K., Fries, E.: On the entry of semliki forest virus into BHK-21 cells. J Cell Biol. 84, 404–420 (1980). [CrossRef]
- Kaur, R., Neetu, Mudgal, R., Jose, J., Kumar, P., Tomar, S.: Glycan-dependent chikungunya viral infection divulged by antiviral activity of NAG specific chi-like lectin. Virology. 526, 91–98 (2019). [CrossRef]
- Strass, J.H., Strauss, E.G., Kuhn, R.J.: Budding of alphaviruses. Trends Microbiol. 3, 346–350 (1995). [CrossRef]
- Aggarwal, M., Kaur, R., Saha, A., Mudgal, R., Yadav, R., Dash, P.K., Parida, M., Kumar, P., Tomar, S.: Evaluation of antiviral activity of piperazine against Chikungunya virus targeting hydrophobic pocket of alphavirus capsid protein. Antiviral Res. 146, 102–111 (2017). [CrossRef]
- Sharma, R., Fatma, B., Saha, A., Bajpai, S., Sistla, S., Dash, P.K., Parida, M., Kumar, P., Tomar, S.: Inhibition of chikungunya virus by picolinate that targets viral capsid protein. Virology. 498, 265–276 (2016). [CrossRef]
- Kumar, R., Nehul, S., Singh, A., Tomar, S.: Identification and evaluation of antiviral potential of thymoquinone, a natural compound targeting Chikungunya virus capsid protein. Virology. 561, 36–46 (2021). [CrossRef]
- Mudgal, R., Mahajan, S., Tomar, S.: Inhibition of Chikungunya virus by an adenosine analog targeting the SAM-dependent nsP1 methyltransferase. FEBS Lett. 594, 678–694 (2020). [CrossRef]
- Puranik, N. V., Rani, R., Singh, V.A., Tomar, S., Puntambekar, H.M., Srivastava, P.: Evaluation of the Antiviral Potential of Halogenated Dihydrorugosaflavonoids and Molecular Modeling with nsP3 Protein of Chikungunya Virus (CHIKV). ACS Omega. 4, 20335–20345 (2019). [CrossRef]
- Bajaj, K.K., Chavhan, V., Raut, N.A., Gurav, S.: Panchgavya: A precious gift to humankind. J Ayurveda Integr Med. 13, 100525 (2022). [CrossRef]
- Pant, L., Thapa, S., Dahal, B., Khadka, R., Biradar, M.S.: In Silico and In Vitro Studies of Antibacterial Activity of Cow Urine Distillate (CUD). Evidence-Based Complementary and Alternative Medicine. 2024, 1–10 (2024). [CrossRef]
- Hirapara, H., Ghori, V., Anovadiya, A., Tripathi, C.: Evaluation of wound healing activity of cow urine ark in diabetic Wistar albino rats. J Intercult Ethnopharmacol. 5, 434 (2016). [CrossRef]
- Khanuja, S.P.S., Kumar, S., Shasany, A.K., Arya, J.S., Darokar, M.P., Singh, M., Dawle, S.H.: U.S. Patent No. 6,410,059, (2002).
- Randhawa, G.K., Sharma, R.: Chemotherapeutic potential of cow urine: A review. J Intercult Ethnopharmacol. 4, 180–6 (2015). [CrossRef]
- Hoh, J.M., Dhanashree, B.: Antifungal effect of cow’s urine distillate on Candida species. J Ayurveda Integr Med. 8, 233–237 (2017). [CrossRef]
- Ravi, H., Dhar, P., Awasthi, A., Verma, S., Bharadwaj, M., Chahota, R.: In vitro Evaluation of Antiviral Efficacy of Himachali Pahari Cattle Urine against Canine Parvovirus. Indian J Anim Res. (2025). [CrossRef]
- Nautiyal, V., Dubey, R.C.: FT-IR and GC-MS analyses of potential bioactive compounds of cow urine and its antibacterial activity. Saudi J Biol Sci. 28, 2432–2437 (2021). [CrossRef]
- Chen, Y., Li, Z., Pan, P., Lao, Z., Xu, J., Li, Z., Zhan, S., Liu, X., Wu, Y., Wang, W., Li, G.: Cinnamic acid inhibits Zika virus by inhibiting RdRp activity. Antiviral Res. 192, 105117 (2021). [CrossRef]
- Solanki, N., Patel, H., Patel, M., Patel, Y., Shukla, P., Kakadiya, J., Maheshwari, R., Chauhan, P.: Alleviating Potential of Zingiber officinale and Cow Urine Distillate Co-administered with Levetiracetam in Epileptic Rats: A Pharmacokinetic and Pharmacodynamics Approach. Journal of Natural Remedies. 677–685 (2023). [CrossRef]
- Kumar, R., Nehul, S., Singh, A., Tomar, S.: Identification and evaluation of antiviral potential of thymoquinone, a natural compound targeting Chikungunya virus capsid protein. Virology. 561, 36–46 (2021). [CrossRef]
- Pareek, A., Kumar, R., Mudgal, R., Neetu, N., Sharma, M., Kumar, P., Tomar, S.: Alphavirus antivirals targeting RNA-dependent RNA polymerase domain of nsP4 divulged using surface plasmon resonance. FEBS J. 289, 4901–4924 (2022). [CrossRef]
- Kumar, S., Singhal, V., Roshan, R., Sharma, A., Rembhotkar, G.W., Ghosh, B.: Piperine inhibits TNF-α induced adhesion of neutrophils to endothelial monolayer through suppression of NF-κB and IκB kinase activation. Eur J Pharmacol. 575, 177–186 (2007). [CrossRef]
- Jiang, Z.-Y., Liu, W.-F., Zhang, X.-M., Luo, J., Ma, Y.-B., Chen, J.-J.: Anti-HBV active constituents from Piper longum. Bioorg Med Chem Lett. 23, 2123–2127 (2013). [CrossRef]
- Neiderud, C.-J.: How urbanization affects the epidemiology of emerging infectious diseases. Infect Ecol Epidemiol. 5, 27060 (2015). [CrossRef]
- Singh, H., Mudgal, R., Narwal, M., Kaur, R., Singh, V.A., Malik, A., Chaudhary, M., Tomar, S.: Chikungunya virus inhibition by peptidomimetic inhibitors targeting virus-specific cysteine protease. Biochimie. 149, 51–61 (2018). [CrossRef]
- Saha, A., Acharya, B.N., Priya, R., Tripathi, N.K., Shrivastava, A., Rao, M.K., Kesari, P., Narwal, M., Tomar, S., Bhagyawant, S.S., Parida, M., Dash, P.K.: Development of nsP2 protease based cell free high throughput screening assay for evaluation of inhibitors against emerging Chikungunya virus. Sci Rep. 8, 10831 (2018). [CrossRef]
- Lim, E., Lee, W., Madzokere, E., Herrero, L.: Mosquitoes as Suitable Vectors for Alphaviruses. Viruses. 10, 84 (2018). [CrossRef]
- Tomar, S., Mahajan, S., Kumar, R.: Advances in structure-assisted antiviral discovery for animal viral diseases. In: Genomics and Biotechnological Advances in Veterinary, Poultry, and Fisheries. pp. 435–468. Elsevier (2020).
- Singh, V.A., Kumar, C.S., Khare, B., Kuhn, R.J., Banerjee, M., Tomar, S.: Surface decorated reporter-tagged chikungunya virus-like particles for clinical diagnostics and identification of virus entry inhibitors. Virology. 578, 92–102 (2023). [CrossRef]
- Handa, T., Saha, A., Narayanan, A., Ronzier, E., Kumar, P., Singla, J., Tomar, S.: Structural Virology: The Key Determinants in Development of Antiviral Therapeutics. Viruses. 17, 417 (2025). [CrossRef]
- Puranik, N. V., Rani, R., Singh, V.A., Tomar, S., Puntambekar, H.M., Srivastava, P.: Evaluation of the Antiviral Potential of Halogenated Dihydrorugosaflavonoids and Molecular Modeling with nsP3 Protein of Chikungunya Virus (CHIKV). ACS Omega. 4, 20335–20345 (2019). [CrossRef]
- Bhutkar, M., Kumar, A., Rani, R., Singh, V., Saha, A., Pathak, A., Kothiala, A., Mahajan, S., Waghmode, B., Verma, S., Kumar, R., Mudgal, R., Sircar, D., Kumar, P., Tomar, S.: Structure-based identification of herbacetin and caffeic acid phenethyl ester as inhibitors of S-adenosylmethionine-dependent viral methyltransferase. FEBS Lett. 599, 1531–1555 (2025). [CrossRef]
- Kumar, S., Singhal, V., Roshan, R., Sharma, A., Rembhotkar, G.W., Ghosh, B.: Piperine inhibits TNF-α induced adhesion of neutrophils to endothelial monolayer through suppression of NF-κB and IκB kinase activation. Eur J Pharmacol. 575, 177–186 (2007). [CrossRef]
Figure 1.
(a) Evaluation of the cell cytotoxicity of cow urine distillate (CUD)in Vero cells: Cytotoxicity was measured using the MTT assay. The absorbance for cell viability was recorded at 570 nm. The percentage cell viability of treated Vero cells at different concentrations was compared with that of the untreated cell control. The graph was plotted via the non-linear regression method with 95% confidence intervals. (b)Assessment of the reduction in CHIKV titer on treatment with CUD) by plaque-reduction assay; CHIKV (MOI 1) was cultivated in Vero cells with the treatment of different concentrations of CUD for 24 h. The virus titer was determined in the form of PFU/mL, and the graph was plotted using GraphPad Prism Version 8. The graphs showed the reduction in CHIKV titer plotted between virus titer in log10 pfu/mL versus CUD concentration. Values are mean and error bars represent standard deviation from triplicate experiments. The statistical analysis was done by one-way - ANOVA and Dunnett’s method. All the data were statistically significant as the p values are less than 0.05 (**; p ≤ 0.01, ***; p ≤ 0.001, ****; p ≤ 0.0001).
Figure 1.
(a) Evaluation of the cell cytotoxicity of cow urine distillate (CUD)in Vero cells: Cytotoxicity was measured using the MTT assay. The absorbance for cell viability was recorded at 570 nm. The percentage cell viability of treated Vero cells at different concentrations was compared with that of the untreated cell control. The graph was plotted via the non-linear regression method with 95% confidence intervals. (b)Assessment of the reduction in CHIKV titer on treatment with CUD) by plaque-reduction assay; CHIKV (MOI 1) was cultivated in Vero cells with the treatment of different concentrations of CUD for 24 h. The virus titer was determined in the form of PFU/mL, and the graph was plotted using GraphPad Prism Version 8. The graphs showed the reduction in CHIKV titer plotted between virus titer in log10 pfu/mL versus CUD concentration. Values are mean and error bars represent standard deviation from triplicate experiments. The statistical analysis was done by one-way - ANOVA and Dunnett’s method. All the data were statistically significant as the p values are less than 0.05 (**; p ≤ 0.01, ***; p ≤ 0.001, ****; p ≤ 0.0001).
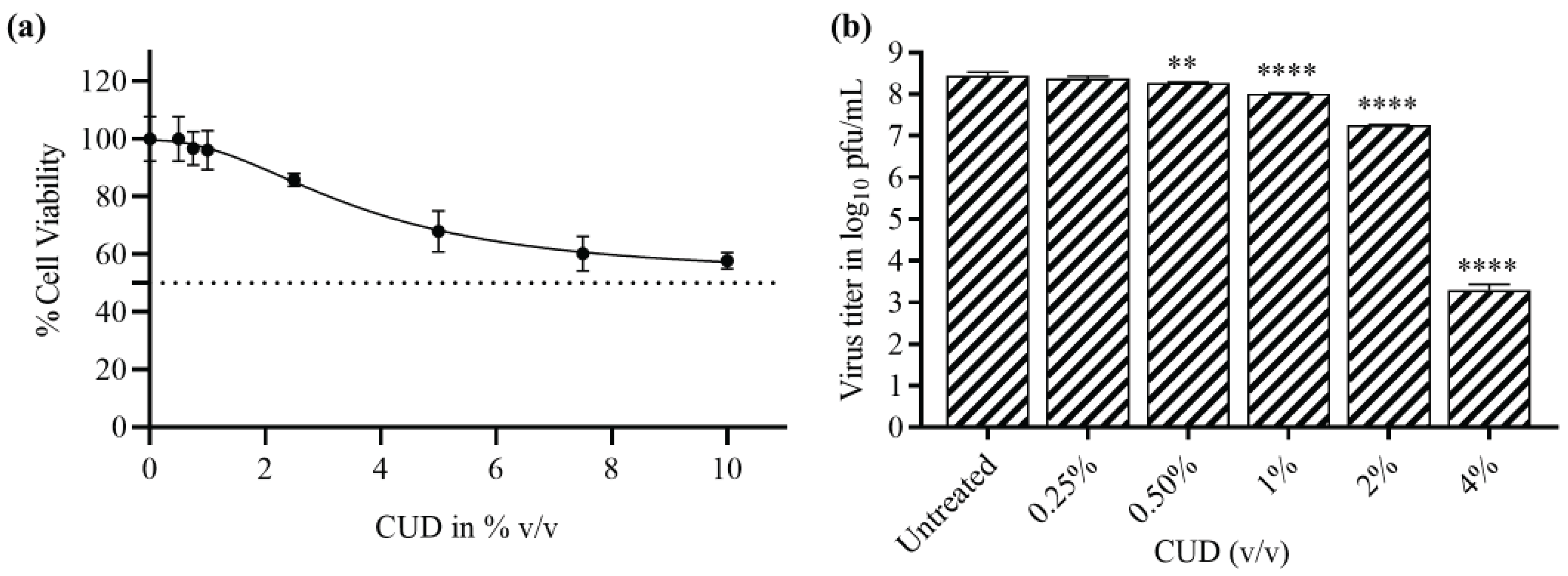
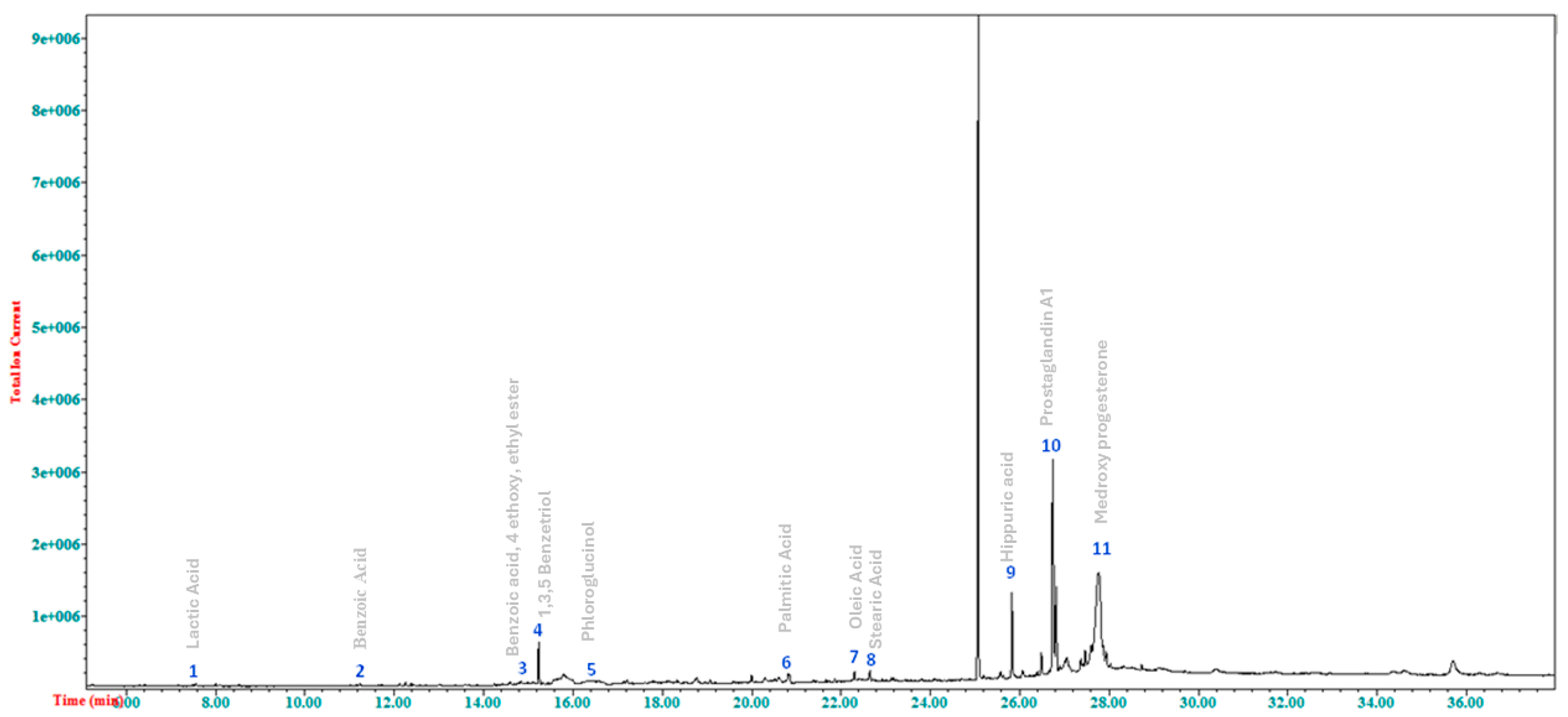
Figure 2.
Estimation Total Ion Chromatogram (TIC) of the Gas chromatography–mass spectrometry of Cow urine distillate (CUD). Peaks showed the major substances/metabolites in CUD.
Figure 2.
Estimation Total Ion Chromatogram (TIC) of the Gas chromatography–mass spectrometry of Cow urine distillate (CUD). Peaks showed the major substances/metabolites in CUD.
Figure 3.
Interaction analysis of docked complexes of nsP2Pro with different ligands, with their docking score and 2D interaction analysis of docked complexes generated using Ligplot. 2D interactions are show for compounds a. Benzoic acid, b. p-Aminohippuric acid c. Hippuric acid d. Lactic acid e. Oleic acid f. Benzoic acid, 4 ethoxy, ethyl ester g. Stearic acid h. Palmitic acid.
Figure 3.
Interaction analysis of docked complexes of nsP2Pro with different ligands, with their docking score and 2D interaction analysis of docked complexes generated using Ligplot. 2D interactions are show for compounds a. Benzoic acid, b. p-Aminohippuric acid c. Hippuric acid d. Lactic acid e. Oleic acid f. Benzoic acid, 4 ethoxy, ethyl ester g. Stearic acid h. Palmitic acid.
Figure 4.
Evaluating the inhibition of the proteolytic activity of CHIKV nsP2Pro. The inhibitory potential of selected candidate compounds was measured using (a). FRET-based proteolytic activity inhibition and (b) Cell-based Plaque reduction assay. Among the tested molecules, Benzoic acid, Hippuric acid, p-Aminohippuric acid, and Oleic acid demonstrated notable inhibition of nsP2pro activity. Data points represent the mean, and error bars indicate the standard deviation (SD) between replicates.
Figure 4.
Evaluating the inhibition of the proteolytic activity of CHIKV nsP2Pro. The inhibitory potential of selected candidate compounds was measured using (a). FRET-based proteolytic activity inhibition and (b) Cell-based Plaque reduction assay. Among the tested molecules, Benzoic acid, Hippuric acid, p-Aminohippuric acid, and Oleic acid demonstrated notable inhibition of nsP2pro activity. Data points represent the mean, and error bars indicate the standard deviation (SD) between replicates.
Figure 5.
Assessment of the reduction in viral titer determined by plaque-reduction assay using Vero cells infected with CHIKV and treated with CUD, TQ, PIP in combination. CHIKV-infected Vero cells were treated with different combinations of CUD, TQ and PIP compounds for 24 h. Untreated virus control was kept as a reference. The Virus titer was calculated as log10 pfu/mL by plaques-reduction assay. and the graph was plotted using Graph Pad Prism Version 8. The graphs showed the reduction in CHIKV titer. (a) Viral titer (log10 pfu/mL) after treatment of CHIKV infected cells with 1.25 μM TQ, 1.25 μM PIP, 0.25% GA (v/v) and a combination of 1.25 μM TQ + 1.25 μM PIP + 0.25% GA (v/v); (b) Viral titer (log10 pfu/mL) after treatment of CHIKV infected cells with 2.5 μM TQ, 2.5 μM PIP, 0.25% GA (v/v) and a combination of 2.5 μM TQ + 2.5 μM PIP + 0.25% GA (v/v); (c) Viral titer (log10 pfu/mL) after treatment of CHIKV infected cells with 5 μM TQ, 5 μM PIP, 0.5% GA (v/v) and a combination of 5 μM TQ + 5 μM PIP + 0.5% GA (v/v); (d) Viral titer (log10 pfu/mL) after treatment of CHIKV infected cells with 10 μM TQ, 10 μM PIP, 1% GA (v/v) and a combination of 10 μM TQ + 10 μM PIP + 1% GA (v/v); (e) Viral titer (log10 pfu/mL) after treatment of CHIKV infected cells with 20 μM TQ, 20 μM PIP, 2% GA (v/v) and a combination of 20 μM TQ + 20 μM PIP + 2% GA (v/v). Values are mean and error bars represent standard deviation from triplicate experiments. The statistical analysis was done by one-way - ANOVA and Dunnett’s method. All the data were statistically significant as the p values are less than 0.05 (**; p ≤ 0.01, ***; p ≤ 0.001, ****; p ≤ 0.0001).
Figure 5.
Assessment of the reduction in viral titer determined by plaque-reduction assay using Vero cells infected with CHIKV and treated with CUD, TQ, PIP in combination. CHIKV-infected Vero cells were treated with different combinations of CUD, TQ and PIP compounds for 24 h. Untreated virus control was kept as a reference. The Virus titer was calculated as log10 pfu/mL by plaques-reduction assay. and the graph was plotted using Graph Pad Prism Version 8. The graphs showed the reduction in CHIKV titer. (a) Viral titer (log10 pfu/mL) after treatment of CHIKV infected cells with 1.25 μM TQ, 1.25 μM PIP, 0.25% GA (v/v) and a combination of 1.25 μM TQ + 1.25 μM PIP + 0.25% GA (v/v); (b) Viral titer (log10 pfu/mL) after treatment of CHIKV infected cells with 2.5 μM TQ, 2.5 μM PIP, 0.25% GA (v/v) and a combination of 2.5 μM TQ + 2.5 μM PIP + 0.25% GA (v/v); (c) Viral titer (log10 pfu/mL) after treatment of CHIKV infected cells with 5 μM TQ, 5 μM PIP, 0.5% GA (v/v) and a combination of 5 μM TQ + 5 μM PIP + 0.5% GA (v/v); (d) Viral titer (log10 pfu/mL) after treatment of CHIKV infected cells with 10 μM TQ, 10 μM PIP, 1% GA (v/v) and a combination of 10 μM TQ + 10 μM PIP + 1% GA (v/v); (e) Viral titer (log10 pfu/mL) after treatment of CHIKV infected cells with 20 μM TQ, 20 μM PIP, 2% GA (v/v) and a combination of 20 μM TQ + 20 μM PIP + 2% GA (v/v). Values are mean and error bars represent standard deviation from triplicate experiments. The statistical analysis was done by one-way - ANOVA and Dunnett’s method. All the data were statistically significant as the p values are less than 0.05 (**; p ≤ 0.01, ***; p ≤ 0.001, ****; p ≤ 0.0001).
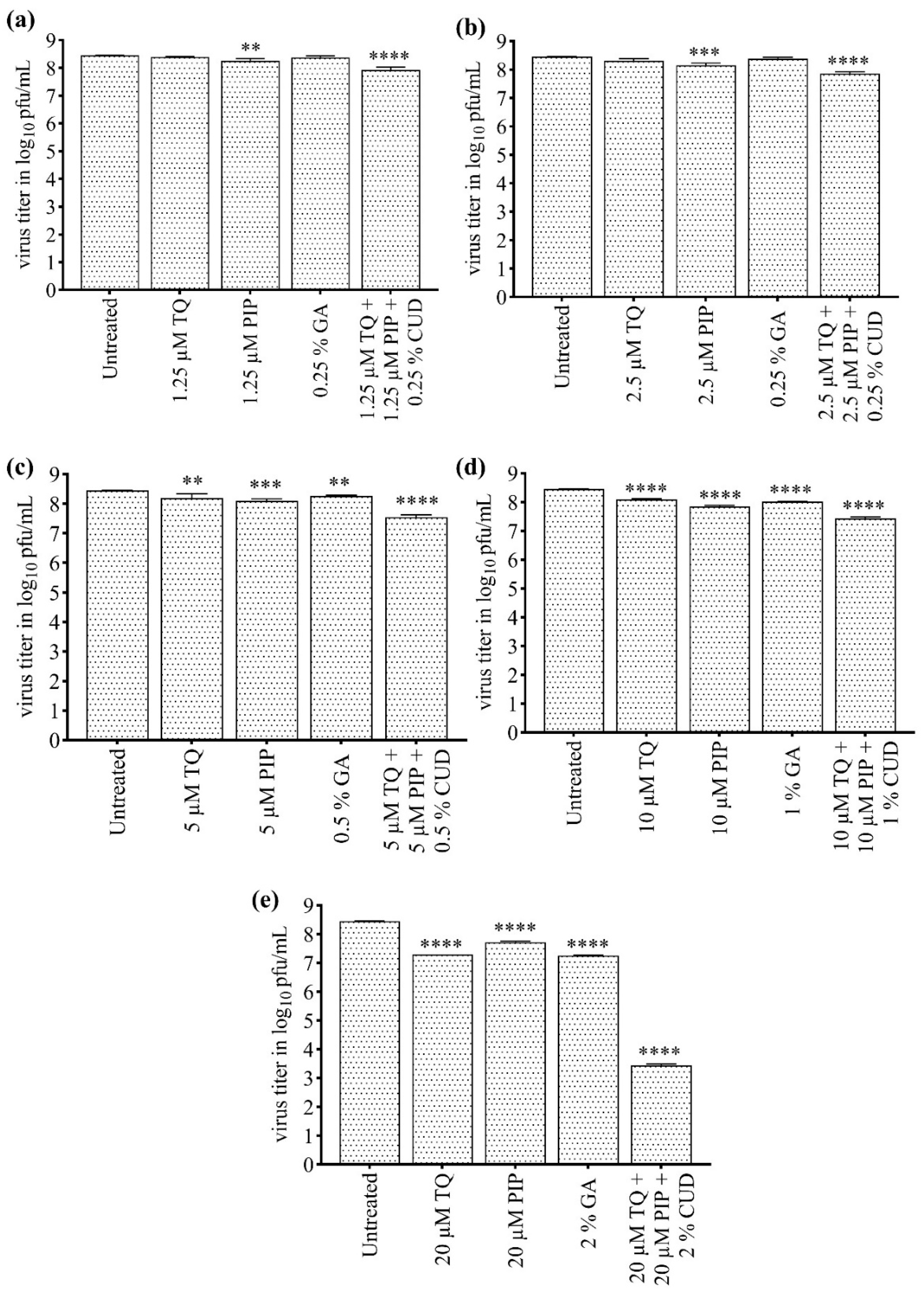
Table 1.
Characterized major substances/metabolites in CUD by GC-MS.
Table 1.
Characterized major substances/metabolites in CUD by GC-MS.
| S. No |
Metabolite |
Derivative |
NIST ID |
Retention Time |
Qualification Ions |
| 1. |
Lactic Acid |
2 TMS |
78865 |
7.562 |
234,219 |
| 2. |
Benzoic Acid |
1 TMS |
64182 |
11.236 |
194,179 |
| 3. |
Benzoic acid, 4
ethoxy, ethyl ester |
2 TMS |
107721 |
15.229 |
194,175 |
| 4. |
1,3,5Benzetriol |
3 TMS |
79582 |
15.573 |
342,327 |
| 5. |
Phloroglucinol |
2 TMS |
118676 |
16.58 |
270,255 |
| 6 |
Palmitic Acid |
1 TMS |
333711 |
20.824 |
328,313 |
| 7 |
Oleic Acid |
1 TMS |
30824 |
22.477 |
355,339 |
| 8 |
Stearic Acid |
1 TMS |
333710 |
22.635 |
356,341 |
| 9 |
Hippuric acid |
2 TMS |
333154 |
25.91 |
323,208 |
| 10 |
Prostaglandin A1 |
2 TMS |
395599 |
27.77 |
480,409 |
| 11. |
Medroxy progesterone |
3 TMS |
55071 |
27.81 |
560, 487 |
Table 2.
Details of the binding energy and interactions for the docked complexes with CHIKV nsP2Pro.
Table 2.
Details of the binding energy and interactions for the docked complexes with CHIKV nsP2Pro.
| Ligand |
Binding Affinity |
Hydrogen bond |
Hydrophobic Bond |
| Benzoic Acid |
-4.5
|
1(Tyr512) |
Cys478, Trp479, Ala511, Tyr544, Ala547, His548, Trp549, Met707
|
Benzoic acid, 4
ethoxy, ethyl ester |
-5.1
|
1(Trp549) |
Ala511, Tyr512, Ser513, Tyr544, Ala547, Met703
|
| Hippuric Acid |
-5.6
|
3(Tyr512) |
Cys478, Trp479, Ala511, Ser513, Tyr544, Ala547, Trp549 |
| Lactic Acid |
-3.6
|
3(Tyr512), 1(Trp549) |
Cys478, Trp479, Ala511, Tyr544, Ala547 |
| Oleic Acid |
-4.5
|
0 |
Asn476, Cys478, Ala511, Tyr512, Ser513, Ala547, Tyr544, Trp549, Ala670, Gln706, Met707 |
| Palmitic Acid |
-4.5
|
1(Ala547) |
Asn476, Cys478, Ala511, Tyr512, Ser513, Tyr544, His548, Trp549, Met707 |
| Stearic Acid |
-4.6
|
0 |
Asn476, Cys478, Ala511, Tyr512, Ser513, Tyr544,Ala547, Trp549, Met707 |
| p-Aminohippuric acid |
-4.5 |
0 |
Cys478 , Ala511, Met707, Tyr512, Asn476, Ala547, Trp549, Tyr544 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).